Introduction
Salmonella is one of the four key global causes of diarrhoeal diseases [1]. According to an estimate by Majowicz and co-workers, 93.8 million cases of gastroenteritis due to Salmonella spp. may occur globally each year, with 155 000 deaths [Reference Majowicz2]. Globally, Salmonella Typhimurium was the most commonly isolated serovar from people for many years, but S. Enteritidis has outnumbered S. Typhimurium in many regions, including Europe, in recent years [Reference Lan, Reeves and Octavia3]. Since the beginning of the 1990s, Salmonella clonal complexes that are resistant to a range of antimicrobials have emerged and are now a serious public health concern [1].
In Great Britain, S. Typhimurium was the second most frequently isolated serovar from human salmonellosis cases for several years, according to the PHE Gastrointestinal Infections Data [4] and the Zoonoses report published by Health Protection Scotland [5], and several different phage types of S. Typhimurium have dominated over the years. Salmonella Typhimurium became the most common serovar causing human salmonellosis during the late 1970s, replacing S. Agona.
Between 1981 and 1991, the incidence of non-typhoidal salmonellosis in the UK rose by >170%, driven primarily by an epidemic of S. Enteritidis phage type (PT) 4, which peaked in 1993 [Reference Lane6, Reference O'Brien7]. In 1988, S. Enteritidis overtook S. Typhimurium as the most common serovar in humans and has been the most common serovar causing human illness ever since. However, since 1997, when vaccination of large flocks of commercial laying hens was introduced, the number of human illnesses cases caused by S. Enteritidis has declined considerably [Reference O'Brien7].
The number of reported human salmonellosis cases across the EU in 2015 was 94 625, which is around 28.0% less compared with 2008 but 14.4% higher than in 2013, when the lowest number was observed [8]. However, the inclusion of data from new member states and changes in reporting systems in some member states mean that it may be difficult to compare data across Europe from one year to another. Overall, the number of human cases has reduced considerably over the past ten years, with S. Enteritidis remaining the most commonly isolated serovar, followed by S. Typhimurium and monophasic strains of S. Typhimurium in 2014 and the preceding years [8].
Salmonella Typhimurium includes a diverse group of genotypes, comprising both host-adapted strains and strains with a broad host range. Host-adapted strains, such as S. Typhimurium DT2 and DT99 in pigeons or S. Typhimurium DT8 in ducks, are rarely found in other species, whereas strains with a broad host range, such as S. Typhimurium DT193 or DT104, can be found in a variety of mammalian and avian hosts [Reference Rabsch9]. The large genetic diversity of S. Typhimurium was reviewed by Lan and co-workers [Reference Lan, Reeves and Octavia3] who grouped S. Typhimurium strains into 12 clonal complexes, based on their MLST types.
Traditionally, S.Typhimurium isolates have been grouped into phage types according to the phage typing scheme of Anderson and co-workers [Reference Anderson10]. While this system has been a useful tool for source attribution and outbreak investigations for many years [Reference Baggesen11, Reference Hald, Lo Fo Wong and Aarestrup12], neither serovars nor phage types necessarily represent a single clonal origin of the isolates involved [Reference Lan, Reeves and Octavia3]. Salmonella Typhimurium DT104 for example, and in particular the multi-drug resistant variant, is not genetically homogenous [Reference Mather13] and isolates belong to four different MLST sequence types, although they may have a single origin within one sequence type [Reference Lan, Reeves and Octavia3]. This observation, together with the fact that phage types can be variable, generally reduces the suitability of phage-typing for more in-depth or longer term epidemiological investigations. The rapid development of molecular methods, including Whole Genome Sequencing, is likely to lead to the replacement of conventional serotyping and phage typing in the future [Reference Bale14].
Salmonella surveillance in livestock in Great Britain has a long tradition and has been established for several decades. Because of the early establishment of veterinary surveillance centres across Great Britain and the linked laboratories at the beginning of the 20th century, data on Salmonella and other pathogens have been captured for many years and are readily available for further analysis and comparative studies. This situation is fairly unusual in Europe and offers the opportunity to analyse the dynamics of different Salmonella serovars and/or strains over several decades and across all relevant livestock species.
Several different S.Typhimurium clones have dominated in Great Britain over time, and often, they were found in both human and livestock populations at the same time.
Although the spread of various clones of S. Typhimurium in certain countries has been covered in several peer-reviewed publications, comprehensive surveillance data from livestock in different countries are not readily available.
In this retrospective study, we describe and analyse Salmonella data from four livestock species (cattle, pigs, chickens and turkeys) in Great Britain between 1983 and 2014, focussing on S. Typhimurium. These species are considered an important source of human salmonellosis in Great Britain. The surveillance system in Great Britain, in which every Salmonella isolate from food animals is sent to the National Reference Laboratory under the Zoonoses Order 1989 [15], facilitates investigation of common trends of increasing or decreasing prevalence, emerging phage types and changes in antimicrobial resistance patterns over time in different livestock species. Environmental and feed samples were not included in this study.
Materials and methods
Salmonella isolates from four livestock species (cattle, chickens, turkeys and pigs), received between the years 1983 and 2014, were included in this study. Data on antimicrobial resistance were available for isolates received from 1986 onwards. However, before 2002, our database does not distinguish between isolates which were not tested for antimicrobial resistance and isolates which were sensitive to all antimicrobials. Therefore, only resistance data from 2002 onwards were analysed to ensure comparability of results.
Statutory reporting of Salmonella isolates
In Great Britain, all Salmonella isolates from animals, their environment or animal feed must be reported to the Competent Authority, as required by the Zoonoses Order 1989. These reports are made to the Animal and Plant Health Agency (APHA), and Salmonella isolates are submitted to the APHA Salmonella Reference Laboratory for further analysis [16]. This includes serotyping, phage typing (if applicable) and tests for resistance to a panel of 16 antimicrobials.
Number and origin of isolates
A total of 96 044 Salmonella isolates were obtained during the study period, of which 45 336 originated from cattle, 31 492 from chickens, 11 507 from turkeys and 7709 from pigs. Due to much lower numbers of Salmonella isolates obtained from other livestock species, such as sheep, goats and other poultry, it was decided to focus on the above mentioned four livestock species only. Salmonella isolates were obtained through four different streams, which are: (i) statutory surveillance, (ii) voluntary surveillance, (iii) investigations of clinical disease and (iv) investigations under the Zoonoses Order. Isolates from cattle and pigs were mainly submitted to investigate clinical disease, whereas isolates from chickens and turkeys originated mainly from voluntary surveillance before 2007 and from statutory surveillance since the introduction of the National Control Programmes from 2007 onwards. In the case of clinical disease investigations, isolates were obtained during post-mortem examinations from internal organs, while surveillance isolates were mostly faecal samples.
Serotyping and phage typing methods
Serotyping was carried out according to the White-Kauffmann- Le Minor Scheme [Reference Grimont and Weill17] using micro-agglutination and slide/tube agglutination tests, plus additional biochemical testing for some serovars. Salmonella Typhimurium isolates and monophasic strains of S. Typhimurium were phage typed according to current versions of the Health Protection Agency (now Public Health England) phage-typing schemes [Reference Anderson10, Reference Bale14].
Antimicrobial susceptibility testing
Salmonella isolates were tested for their in vitro susceptibility to 16 antimicrobials. Prior to 1996, all submitted isolates were tested; since 1996, only the first isolate of a given serovar or phage type from a specific animal holding within a 4-week period has been tested to reduce potential surveillance bias associated with multiple isolates submitted from some holdings.
Before 2002, the database does not distinguish between isolates which were not tested for antimicrobial resistance and isolates which were sensitive to all antimicrobials. Therefore, only resistance data from 2002 onwards were analysed to ensure comparability of results.
Isolates were tested against a panel of 16 antimicrobials using the British Society for Antimicrobial Chemotherapy (BSAC) disc diffusion technique (www.bsac.org.uk) on isosensitest agar (Oxoid), using antimicrobial discs of amikacin (30 µg), amoxicillin/clavulanic acid (30 µg), ampicillin (10 µg), apramycin (15 µg), cefoperazone (30 µg) (replaced with 30 µg cefotaxime disc in 2004), cefuroxime (30 µg) (replaced with 30 µg ceftazidime disc in 2001), chloramphenicol (30 µg) (10 µg until 2008), tetracycline (10 µg), colistin (25 µg) (replaced with 1μg ciprofloxacin disc in 2004), furazolidone (15 µg), gentamicin (20 µg) (10 µg from 1999), nalidixic acid (30 µg), neomycin (10 µg), streptomycin (25 µg) (10 µg from 2008), trimethoprim/sulphamethoxazole (25 µg), sulphonamide compounds (500 µg until 1998, 300 µg from 1999; 80 µg sulphamerazine, 110 µg sulphadiazine, 110 µg sulphathiazole).
A 13 mm veterinary breakpoint was used up until 2007, where isolates were regarded as resistant if the size of growth inhibition zone was ⩽13 mm [Reference Jones18].
BSAC-recommended breakpoints were adopted for ciprofloxacin, amikacin, ceftazidime and cefotaxime from 2007 and for trimethoprim/ sulphamethoxazole, chloramphenicol, amoxicillin/calvulanate and gentamicin from 2008. This change resulted for example in an increased breakpoint of ⩽19 mm for ciprofloxacin. For the remaining compounds, the historical APHA veterinary breakpoint of resistant <13 mm was used throughout the period of study. The breakpoints used each year are published in the UK-Veterinary Antimicrobial Resistance and Sales Surveillance reports [19].
Results
Salmonella isolates from cattle
Cattle were the species with the highest number of isolates reported throughout the study period. Of the 96 044 isolates obtained from the four species, 47.2% (45 336 isolates) originated from cattle.
In cattle, the number of all Salmonella isolates showed a pronounced peak between 1993 and 1998, with the highest numbers reported in 1995 (3207 isolations), followed by a decline since the late 1990s (Fig. 1a). The decline has been maintained since, and in 2014, the number of reported Salmonella isolates fell below 500 for the first time during the study period (465 isolates in 2014).
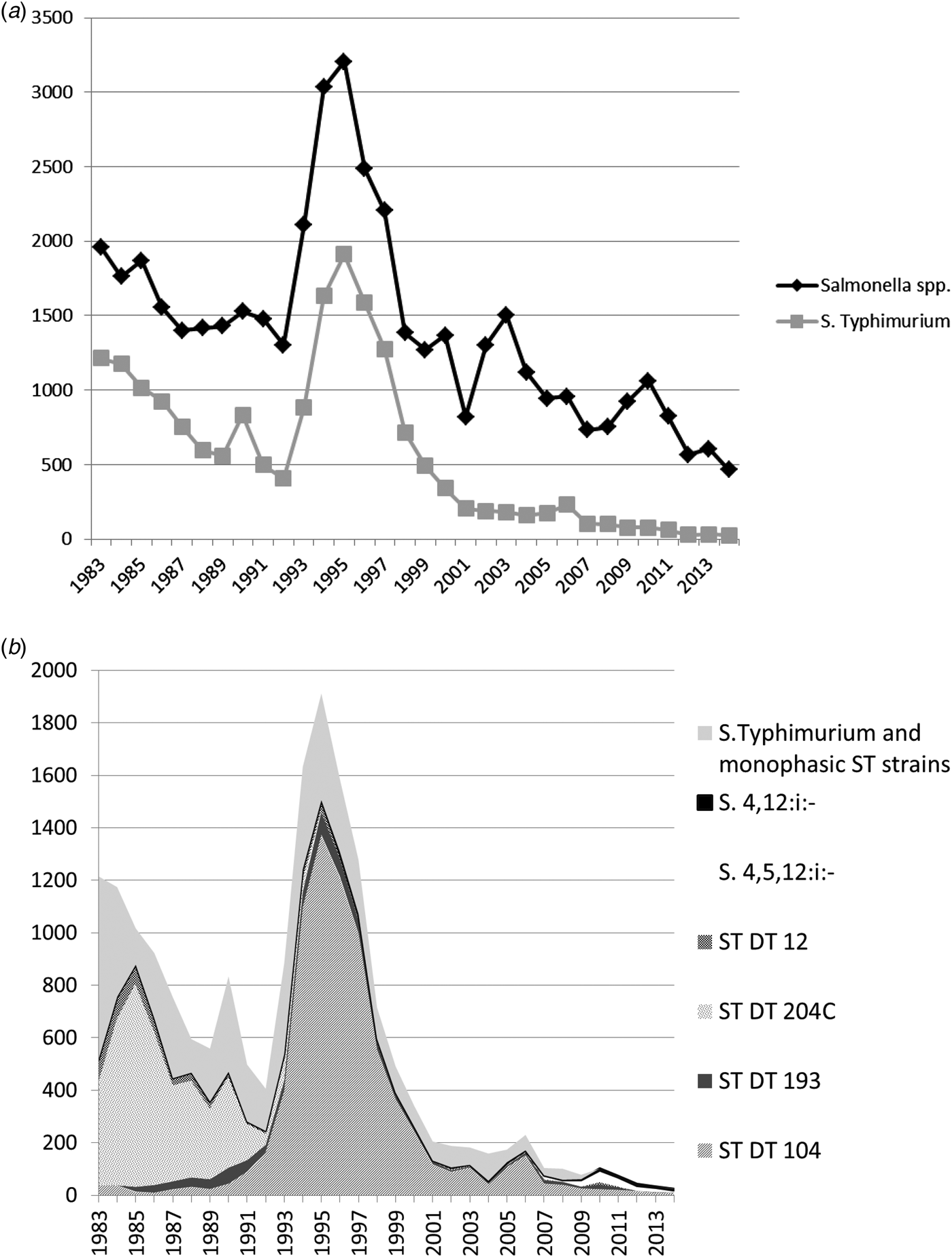
Fig. 1. (a) All Salmonella spp. and Salmonella Typhimurium isolates from cattle from Great Britain for the period 1983–2014. Monophasic strains of Salmonella Typhimurium are not included in the count of S. Typhimurium. (b) Overall numbers of Salmonella Typhimurium isolates, the most common phagetypes of S. Typhimurium and Monophasic strains of Salmonella Typhimurium isolated from cattle from Great Britain for the period 1983–2014 (ST, S. Typhimurium).
Looking at the whole time period, S. Typhimurium was the predominant serovar isolated from cattle and represented 40.7% (18 455/45 336) of isolates from this species. However, the relative contribution of S. Typhimurium has declined over the years, and while it accounted for 66.6% (1174/1763) of all Salmonella isolates in 1984 and for 63.8% (1585/2486) in 1996, its relative percentage fell consistently since 1996, and only accounted for 5.4% (25/465) of all Salmonella isolates from cattle in 2014.
Salmonella Typhimurium DT104 was the most commonly isolated phage type from cattle, accounting for 16.6% (7530/45 336) of all S. Typhimurium isolates, although it was mostly seen in the 1990s with declining numbers since (Fig. 1b).
Monophasic strains of S. Typhimurium have only been seen in small numbers in cattle and accounted for only 0.5% of isolates over the study period (234/45 336), with a relative maximum of 4.9% (52/1061) of all Salmonella isolates in 2010.
Salmonella isolates from pigs
A total of 7709 Salmonella isolates were obtained from pigs throughout the study period, representing 7.8% of all isolates.
Salmonella isolates from pigs showed a peak in the 1990s similar to cattle, although it was a more prolonged peak compared to the sharp peak seen in cattle (Fig. 2a). The number of isolates reported each year slowly started to increase from 1989 onwards, with the highest numbers observed in 1994 (445 isolates) and 1996 (457 isolates). Since 1996, numbers have generally declined, down to 159 isolates in 2014, although a smaller peak was observed in the year 2000.
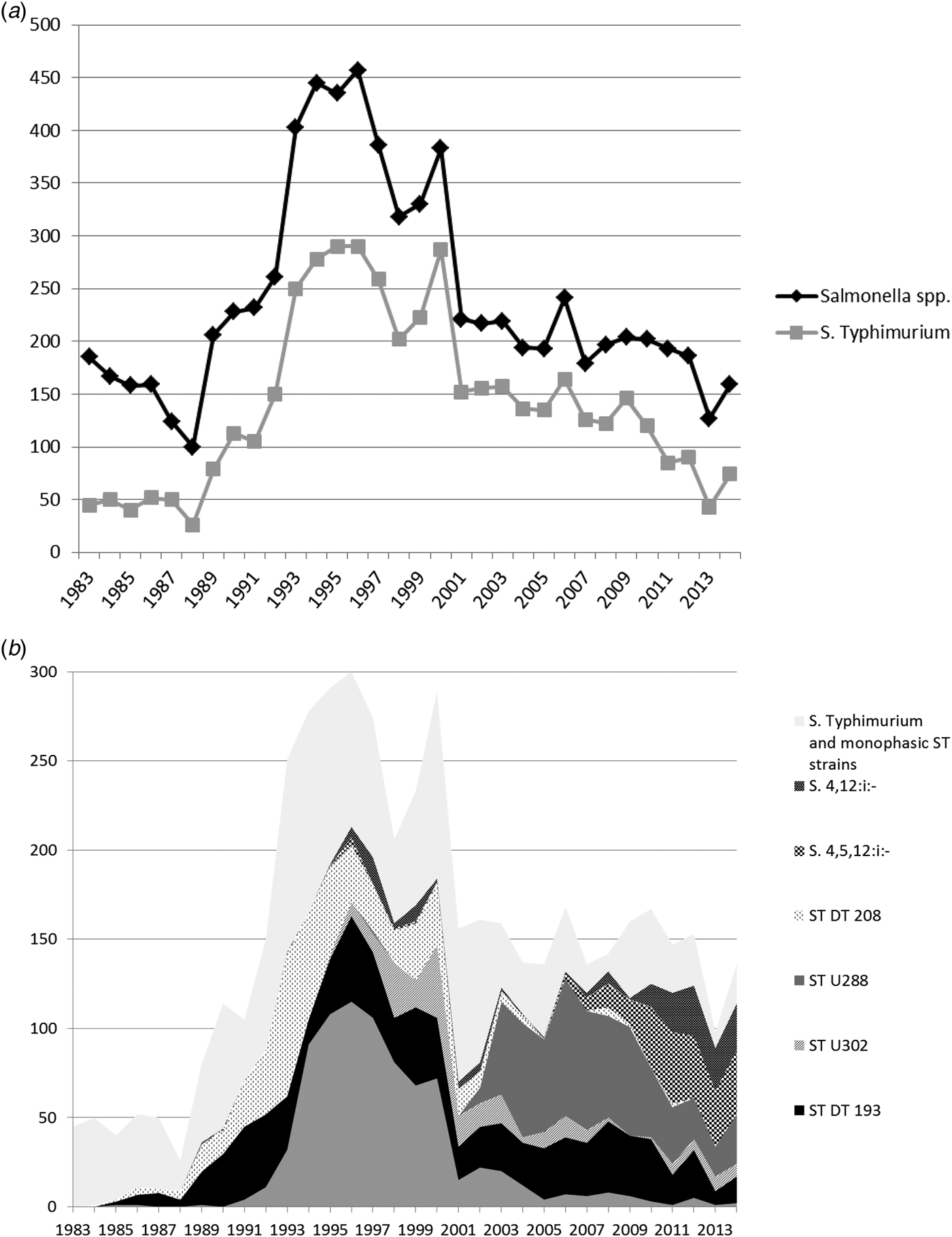
Fig. 2. (a) All Salmonella spp. and Salmonella Typhimurium isolates from pigs from Great Britain for the period 1983–2014. Monophasic strains of Salmonella Typhimurium are not included in the count of S. Typhimurium. (b) Overall numbers of Salmonella Typhimurium isolates, the most common phagetypes of S. Typhimurium and Monophasic strains of Salmonella Typhimurium isolated from pigs from Great Britain for the period 1983–014 (ST, S. Typhimurium).
At the beginning of the study period in the early 1980s, S. Derby was the predominant serovar found in pigs before the increase in S. Typhimurium isolates began. Over the whole time period, S. Typhimurium was the predominant serovar isolated from pigs and represented 58.3% (4495/7709) of isolates from this species. However, the relative importance of S. Typhimurium has declined again in recent years, and in 2014, it represented 46.5% (74/159) of all Salmonella isolates (Fig. 2a).
Predominant S. Typhimurium phage types seen in pigs during the study period were DT104 (803; 17.9% of all S. Typhimurium isolates), DT193 (774; 17.2%), undefined phage type U288 (580; 12.9%) and DT208 (479; 10.7%) (Fig. 2b).
Reports of monophasic strains of S. Typhimurium began to increase from 2009 onwards, with both S. 4,5,12:i:- and S. 4,12:i:- being reported frequently. In 2014, S. 4,5,12:i:- and S. 4,12:i:- accounted for 22.6% (36/159) and 16.4% (26/159) of all Salmonella isolates from pigs respectively, while in 2009, they had only accounted for 6.4% (13/204) and 0.5% (1/204), respectively. (Fig. 2b).
Salmonella isolates from chickens
A total of 31 492 Salmonella isolates were obtained from chickens during the study period, therefore accounting for 32.8% of isolates in the study.
In contrast to cattle and pigs, S. Typhimurium only accounted for a small proportion of all chicken isolates, i.e. 6.7% (2114/31 492) (Fig. 3a).
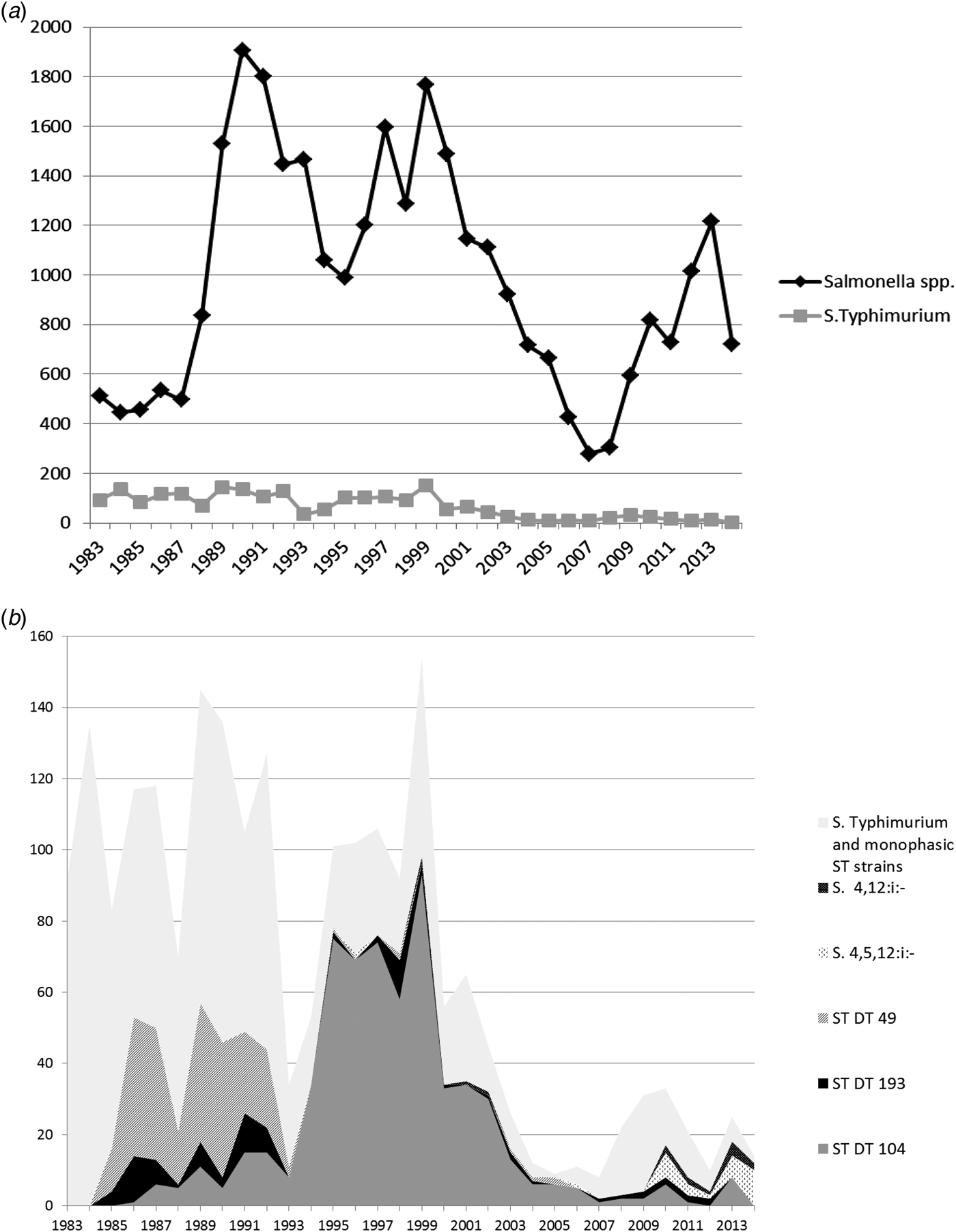
Fig. 3. (a) All Salmonella spp. and Salmonella Typhimurium isolates from chickens from Great Britain for the period 1983–2014. Monophasic strains of Salmonella Typhimurium are not included in the count of S. Typhimurium. (b) Overall numbers of Salmonella Typhimurium isolates, the most common phagetypes of S. Typhimurium and Monophasic strains of Salmonella Typhimurium isolated from chickens from Great Britain for the period 1983–2014 (ST, S. Typhimurium).
The relative importance of S. Typhimurium peaked in 1984 when 30.3% (135/446) of all chicken Salmonella isolates were S. Typhimurium, and a decline was seen afterward, reaching a nadir in 2012, when only 0.8% (8/1014) of Salmonella isolates were S. Typhimurium.
Throughout the study period, predominant phage types from chickens were DT104 (616; 29.1% of all S. Typhimurium isolates), DT49 (236; 11.2%) and DT193 (86; 4.1%).
Between 2008 and 2011, an increase in the number of reports of S. Typhimurium was attributable to a variety of phage types, including a number of wild-bird associated strains, such as DT40, DT41 and DT56 (data not shown).
Monophasic strains of S. Typhimurium were first detected in chickens in 2010, and in recent years, monophasic strains of S. Typhimurium either reached the same numbers as S. Typhimurium or even outnumbered them in some years (Fig. 3b). In 2014, the year with the highest number of monophasic strains of S. Typhimurium (12 isolates), only two isolates of S. Typhimurium were recorded.
Salmonella isolates from turkeys
11 507 Salmonella isolates were obtained from turkeys during the study period, accounting for 12.0% of isolates in the study. 926 of those isolates were S. Typhimurium, which accounted for only 8.1% of all Salmonella isolates from turkeys.
Salmonella Typhimurium was mainly reported between 1995 and 2006 and showed a plateau with a small increase in 1995 and no pronounced peak (Fig. 4a). Salmonella Typhimurium reports were already at a very low level in 2008, 2 years before statutory surveillance started and the National Control Programmes according to EU legislation were introduced.
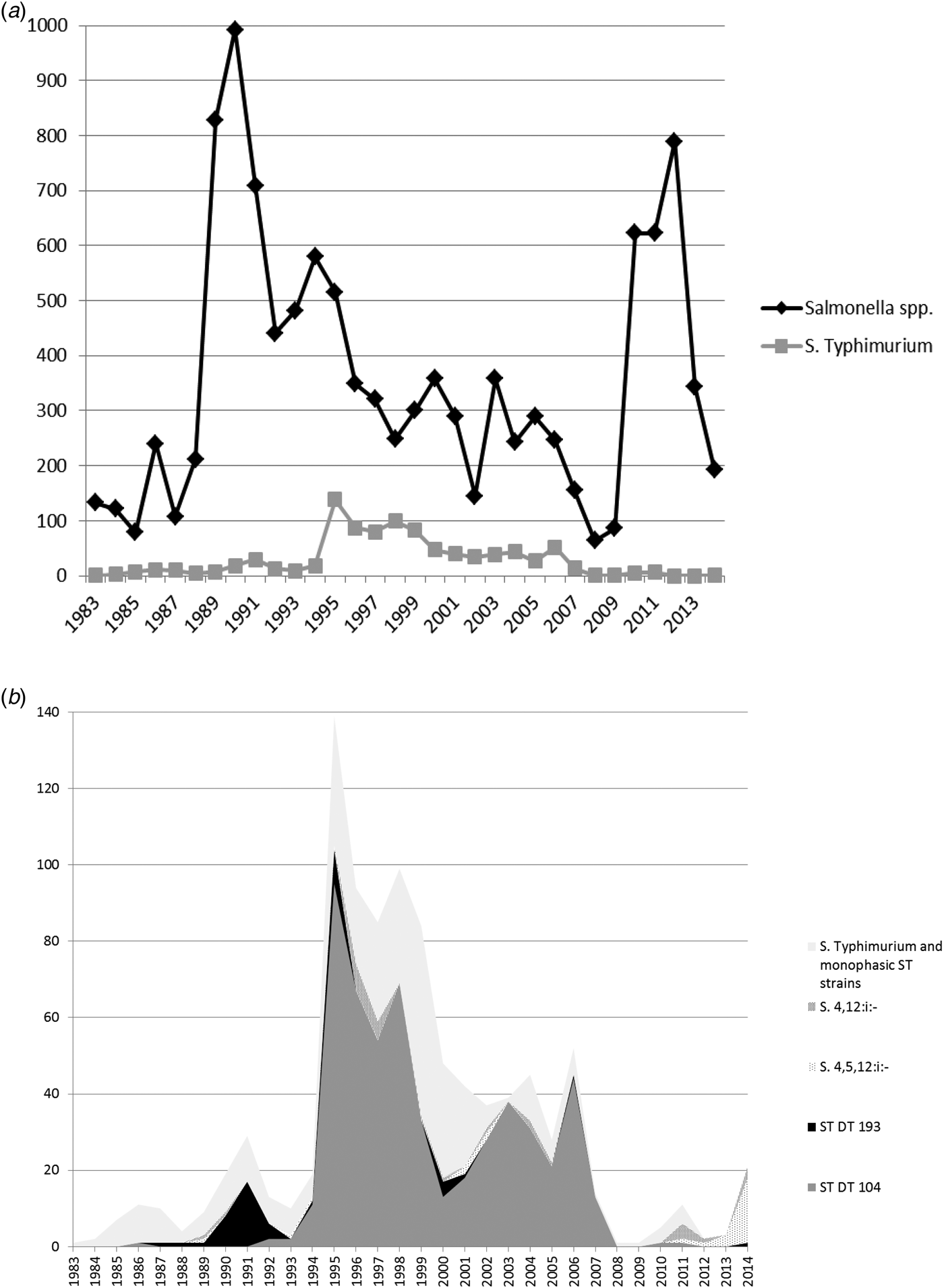
Fig. 4. (a) All Salmonella spp. and Salmonella Typhimurium isolates from turkeys from Great Britain for the period 1983–2014. Monophasic strains of Salmonella Typhimurium are not included in the count of S. Typhimurium. (b) Overall numbers of Salmonella Typhimurium isolates, the most common phagetypes of S. Typhimurium and Monophasic strains of Salmonella Typhimurium isolated from turkeys from Great Britain for the period 1983–2014 (ST, S. Typhimurium).
Monophasic strains of S. Typhimurium (mST) were occasionally reported between 1989 and 2005, but these are likely to be untypable rather than monophasic, and a substantial increase in PCR-confirmed mST was only observed from 2011 onwards. After the disappearance of S. Typhimurium DT104, there are currently few S. Typhimurium isolates reported from turkeys, and monophasic strains now outnumber biphasic strains of S. Typhimurium (Fig. 4b). In 2014, monophasic strains of S. Typhimurium accounted for 10.36% (20/193) of all turkey Salmonella isolates, while the equivalent figure for S. Typhimurium was 0.52% (1/193).
Antimicrobial resistance of Salmonella Typhimurium isolates
Tables 1–4 show the percentage of S. Typhimurium isolates that were fully susceptible to the panel of antimicrobials and the percentage of isolates resistant to each individual antimicrobial in the panel. Figures 5a–d show the same data, with added trend lines where applicable.

Fig. 5. (a) Percentage of Salmonella Typhimurium isolates from cattle showing resistance to individual antimicrobials. Trendlines are added for resistance to T, C, AM and SU. (b) Percentage of Salmonella Typhimurium isolates from pigs showing resistance to individual antimicrobials. (c) Percentage of Salmonella Typhimurium isolates from chickens showing resistance to individual antimicrobials. (d) Percentage of Salmonella Typhimurium isolates from turkeys showing resistance to individual antimicrobials.
Table 1. Percentage of Salmonella Typhimurium isolates from cattle showing resistance to individual antimicrobials
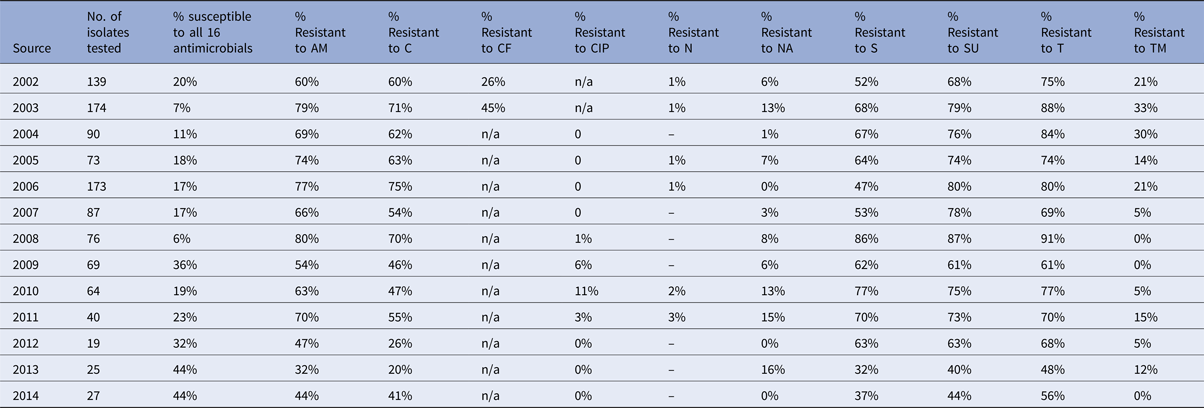
Table 2. Percentage of Salmonella Typhimurium isolates from pigs showing resistance to individual antimicrobials

Table 3. Percentage of Salmonella Typhimurium isolates from chickens showing resistance to individual antimicrobials
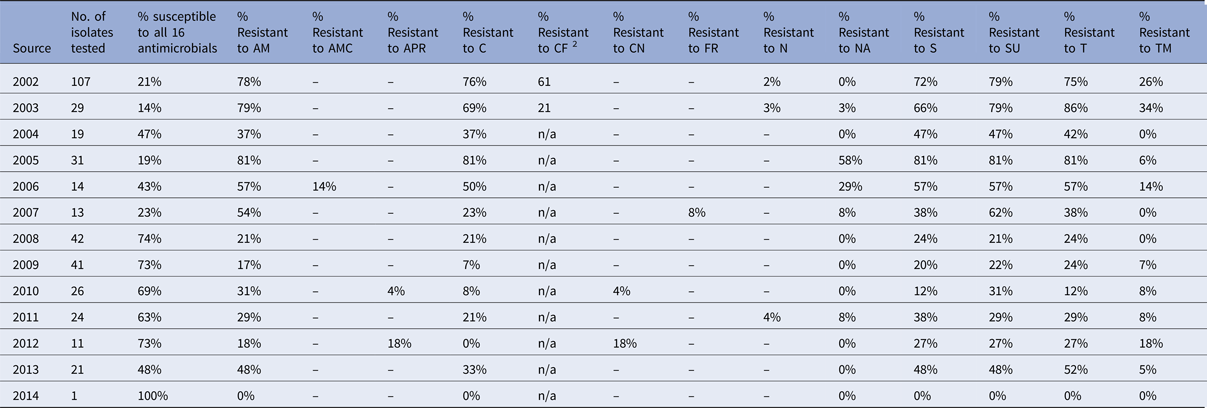
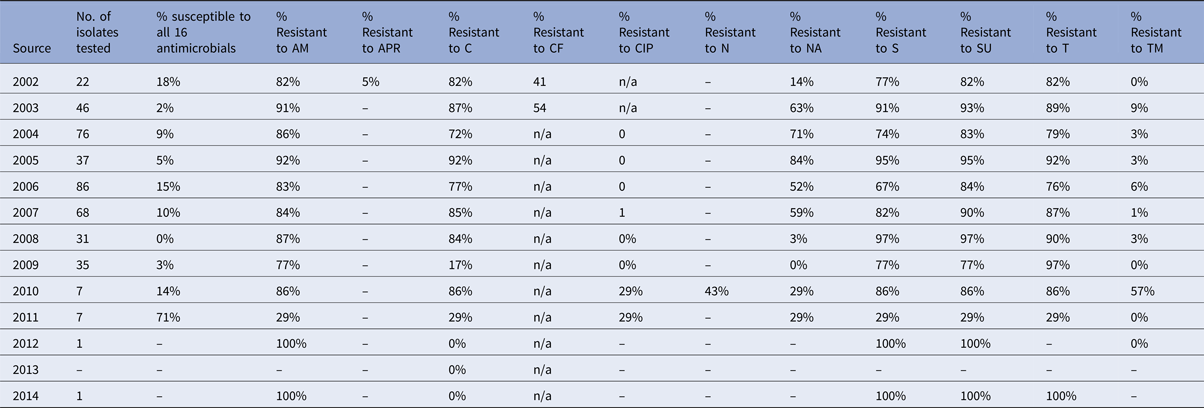
Table 4. Percentage of Salmonella Typhimurium isolates from turkeys showing resistance to individual antimicrobials
NA, Nalidixic acid; T, Tetracycline; N, Neomycin; AM, Ampicillin; FR, Furazolidone; CAZ, Ceftazidime; TM, Sulphamethoxazole/trimethoprim; C, Chloramphenicol; AK, Amikacin; AMC, Amoxicillin/clavulanic acid; CN, Gentamicin; S, Streptomycin; SU, Sulphonamide compounds.
In cattle, 44% of S. Typhimurium isolates from 2014 were fully susceptible to all antimicrobials tested, with an upwards trend observed since 2002, when 20% of isolates were fully susceptible (Table 1). During this time period, a reduction in resistance can be observed for chloramphenicol, ampicillin, sulphonamide compounds, streptomycin and tetracycline. While the proportion of isolates resistant to these antimicrobials was 60%, 60%, 68%, 52% and 74% respectively in 2002, it was only 41%, 44%, 44%, 37% and 56% in 2014 (Fig. 5a and Table 1). However, the number of isolates tested per year was substantially lower in 2014 (27 S. Typhimurium isolates) compared to 2002 (139 isolates). Unfortunately, it is not possible to compare the percentage of fully susceptible isolates from the 1990s, when the peak of S. Typhimurium isolates was reported, with the period after 2002, due to the changes in testing schedule.
In pigs, the percentage of fully sensitive S. Typhimurium isolates was considerably lower compared to cattle, with 10% of isolates being fully sensitive in 2014, but was as low as 1% in 2007 and 2013 (Table 2). Resistance to several antimicrobials was common throughout the period 2002 to 2014, and neither a clear upward nor a downward trend was observed (Fig. 5b). Highest levels of resistance were reported to ampicillin (up to 96%), streptomycin (up to 96%), sulphonamide compounds (up to 98%) and tetracycline (up to 93%). Resistance to trimethoprim was observed at a higher level in pigs compared to all other species, with up to 87% of isolates being resistant in one year (Table 2), but resistance levels to trimethoprim varied over the years. Salmonella Typhimurium isolates from chickens generally show low levels of antimicrobial resistance, with an increasing number of isolates being fully sensitive to the panel of antimicrobials (Table 3). In 2012, 73% of isolates were fully sensitive, but in 2013 there was a reduction down to 48%. The reason for this is that in 2013, 53.3% of S. Typhimurium isolates from chickens were DT104, which is usually resistant to ampicillin, chloramphenicol, streptomycin, sulphonamide compounds and tetracycline.
Salmonella Typhimurium isolates from turkeys show high levels of resistance to a number of antimicrobials, and in some years, no isolates tested were fully susceptible to the test panel. Resistance to ampicillin reached a maximum of 92% in a particular year, chloramphenicol 92%, nalidixic acid 84%, streptomycin 91% and tetracycline up to 92% (Table 4). For most of these antimicrobials, no downwards trend in resistance can be observed (Fig. 5d).
Discussion
Very little structured surveillance data are available on the occurrence of zoonotic Salmonella serovars in farm animals from around the world. The ‘European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks’, published by the European Food Safety Authority (EFSA) and the European Centre for Disease Prevention and Control (ECDC) each year [8] is the most comprehensive source of Salmonella data from livestock in EU member states and Iceland, Norway and Switzerland. However, the majority of data from species other than chickens and turkeys is not directly comparable between member states, due to different surveillance and reporting systems used and differences in sampling and testing methods.
Publication of longitudinal surveillance data in livestock is also scarce; some countries or local authorities publish their surveillance data annually; however, in many cases, these are not readily available in the English language. In other cases, these may only involve outbreak investigations of human salmonellosis rather than all Salmonella isolations from livestock. The data published in this work are therefore unique as they draw a longitudinal picture of the Salmonella situation in four livestock species for the whole of Great Britain.
However, as Salmonella surveillance in pigs and cattle is not regulated and depends on voluntary submissions, direct comparisons between cattle and pigs are difficult to make.
During the study period, S. Typhimurium DT104 was the most commonly isolated phage type from the four livestock species in Great Britain, representing 36.5% (9491/25 990) of all S. Typhimurium isolates. However, a clear upward and downward trends were seen over the years, with distinct differences between species, and it is not clear what the reason behind the rise and decline of DT104 was and why numbers peaked at different times in different species.
In cattle, S. Typhimurium DT104 was mostly responsible for the general peak in Salmonella reports, which began in the early 1990s, although S. Dublin, the second most frequently isolated serovar from cattle, also showed a peak around the same time. Over the whole study period, DT104 was the most commonly isolated phage type from cattle (40.8% of all S. Typhimurium isolates), followed by DT204C (21.9%) and DT193 (4.1%). DT204C was hardly seen in any of the other species, suggesting for example either a certain host-susceptibility of this particular phage type or a difference in exposure between the host species.
DT104 isolates from cattle peaked in 1995, with 1374 isolates, but numbers quickly fell to 244 in 2000 and 108 in 2005 (Fig. 1b). DT104 peaked in all species covered in this study, but in cattle, the peak was more pronounced compared to other species (see Figs 2b, Figs 4b). DT104 also caused a contemporaneous epidemic in humans in several countries worldwide [Reference Helms, Ethelberg and Molbak20, Reference Threlfall21] with human cases in the United Kingdom peaking in 1996/97 [Reference Helms, Ethelberg and Molbak20]. In pigs, DT104 showed a peak between the years 1994 and 2000, at the same time as the epidemic in cattle, and subsequently declined, with less than ten isolates per year since 2005.
Salmonella Typhimurium DT104 in chickens showed a pronounced peak between 1995 and 1999, at the same time as the prolonged peak in pigs, but a few years after the peak observed in cattle. Numbers declined rapidly after 1999, with the exception of two small peaks in 2010 and 2013 (see Fig. 3b). Similar to cattle and pigs, the main phage type causing the S. Typhimurium peak in turkeys in 1995 was also DT104 (Fig. 4b). However, S. Typhimurium DT104 was seen frequently in turkeys for a much longer period compared with the other species; in 2006, very little DT104 was seen in cattle, pigs and chickens, while this particular phage type still accounted for the vast majority of S. Typhimurium isolates from turkeys. Overall, DT104 accounted for a higher proportion of all S. Typhimurium isolates from turkeys (58.5% of all S. Typhimurium isolates) compared to the other species (40.8% in cattle, 17.9% in pigs and 29.1% in chickens). The reason for this is not clear; possible explanations could be prolonged recirculation within some turkey companies or greater susceptibility of turkeys in general.
Salmonella Typhimurium DT193 in cattle shows an interesting pattern, with a pronounced peak in 1978 and 1979 prior to the study period (data not shown), caused by reports that originated mainly from calves (APHA, data not shown). A similar peak was not observed in any of the other species included in this study (data not shown). Numbers declined from 1980 onwards, and despite some smaller peaks over the years, reports have never exceeded 87 per year from cattle during the study period. Salmonella Typhimurium DT193 in pigs, on the other hand, did not show a peak in the late 1970s as described for cattle, but numbers started to increase from 1989 onwards, reaching a plateau around 1991. Between 1991 and 2010, between 20 and 40 isolates were reported each year, until numbers started to decline. DT193 was rarely found in chickens and only accounted for more than 10 isolates per year on one occasion (11 isolates in 1998).
In 1985, a peak of S. Typhimurium definitive type (DT) 204C was observed in cattle, with 41.4% of all Salmonella isolates belonging to ST DT204C, while the average over the study period was only 8.9% (Fig. 1b). DT204C numbers declined subsequently, and it was not isolated again after 1997. It is not clear why this particular phage type disappeared from the cattle population and why it was not seen in other species.
Monophasic strains of S. Typhimurium were first detected in cattle in 2006, with the exception of a few sporadic cases, which were reported from 1989, but these were not confirmed genetically at the time and are therefore likely to be non-typable strains rather than true monophasic isolates. Although a number of cases were linked to high mortality in cattle, overall, the number of monophasic strains of S. Typhimurium found in cattle was never as great as in pigs. After a peak in 2010 involving 45 isolates of S. 4,5,12:i:- and seven isolates of S. 4,12:i:-, numbers declined to eight and five isolates respectively in 2014 (see Fig. 1b). In 2010, monophasic strains of S. Typhimurium also reached the highest relative proportion of all Salmonella isolates in cattle (4.9%), compared with 2.8% in 2014.
Compared to cattle, monophasic strains have been of significant importance in the pig population ever since they were first seen in 1995, and in 2013, 43.3% of all Salmonella isolates from pigs were monophasic strains.
Monophasic strains were first detected in chickens in 2010, with a few possible exceptions reported previously, which are likely to be untypable strains rather than true monophasic strains. Similar to the situation described for DT104, they started to emerge a few years later than in any of the other species investigated, including cattle, pigs and turkeys.
In pigs, phage type DT208 was frequently seen in the 1990s and peaked in 1993, but has disappeared over the past few years and was never seen in significant numbers in any other species. The same is true for undefined phage type U302, which is closely related to PT104, and which was only seen in pigs between 1998 and 2003.
Reports of undefined phage type U288 from pigs peaked between 2003 and 2010 and this is still the most commonly reported phage type in pigs (Fig. 2b), which is very rarely seen in other livestock species.
In chickens, the number of different serovars reported was considerably greater than for cattle and pigs. In 2014, for example, 22 different serovars were reported from cattle, 12 from pigs, 38 from chickens and 15 from turkeys. In 2014, the most common serovars from chickens were S. Mbandaka, S. Senftenberg, S. 13,23:i:-, S. Kedougou and S. Montevideo. These five serovars together accounted for 73.4% of all Salmonella isolates detected in 2014. Some of these serovars are known to be common contaminants of poultry feed, which might be an explanation why they are not seen in cattle and pigs, but other reasons, such as host susceptibility may play a role as well.
A number of different serovars have also been found in turkeys over the years; in the 1980s, S. Agona, S. Hadar and S. Heidelberg were frequently found in turkeys. In the 1990s, S. Newport was the dominant serovar, but S. Indiana and S. Senftenberg were also frequently reported. The remarkable increase in isolates from 1988 onwards, with a peak in 1990, was caused by cumulative high numbers of different serovars, such as S. Newport, S. Heidelberg, S. Indiana, S. Agona and S. Senftenberg. From 1996 onwards, numbers of S. Derby isolates slowly started to increase, and the peak from 2010 onwards was almost exclusively caused by S. Derby. Feed-associated serovars are not seen to the same extent in turkeys as they are seen in chicken flocks, and it is not clear why this might be the case. However, a number of resident Salmonella serovars have become established in the turkey industry or in particular integrated companies, and some of them are difficult to eliminate from the farm and/or hatchery environment, which might explain their high numbers compared to other livestock species.
The introduction of EU legislation, which targets S. Typhimurium and S. Enteritidis in chickens and turkeys, did not result in a pronounced reduction of S. Typhimurium reports from chickens and turkeys as one may have expected, mainly because the number of isolates was already at a low level when the control programmes started, due to high awareness and voluntary control programmes already in place in Great Britain.
When interpreting data over such a long period, it has to be kept in mind that minor changes to the surveillance system were made over the years, and therefore, comparison of absolute numbers of isolates over the whole time period covered in this publication has to be carried out with care. In some years, more isolates may have been obtained from a particular species; this does not necessarily reflect an increase in prevalence, but could, for example, be the result of enhanced surveillance activities due to other reasons, such as outbreaks of unrelated diseases. On the contrary, the outbreak of Foot and Mouth Disease in Great Britain in 2001 reduced surveillance activities in the country for several months and probably even in the following year, and therefore resulted in fewer surveillance isolates compared with other years.
Surveillance for Salmonella in cattle and pigs in GB relies mostly on voluntary submission of samples, samples collected from post-mortem investigations and samples from clinically diseased animals. Therefore, events such as the DT104 epidemic in cattle and other livestock species probably led to heightened awareness and resulted in higher overall numbers of submissions from cattle, which in turn resulted in higher numbers of other Salmonella serovars isolated, such as S. Dublin.
Contrary to that, statutory surveillance for Salmonella in all commercial-scale flocks of chickens and turkeys became mandatory from 2007 onwards under EU legislation. This has led to an increase in the number of Salmonella isolates reported in most EU member states, although in some only for a short period of time [22]. On the other hand, the introduction of a mandatory surveillance programme for chickens and turkeys was accompanied by increased awareness and improved Salmonella control on farms and across the whole production pyramid, which had a long-term beneficial effect on the Salmonella situation in these species.
Within Great Britain, the early described epidemic multi-resistant Salmonella Typhimurium phage types, such as S. Typhimurium DT29, 204, 193 and 204c, were mainly confined to cattle. It was the emergence of the multi-resistant S. Typhimurium DT104, which caused a lot of interest, as it was seen in several livestock species as well as in people at the same time [Reference Threlfall21]. Of interest is also to mention how effectively these clones spread across the world, with reports from several European countries, North America, the Middle East and Asia. Genetic analysis demonstrated that in multi-resistant S. Typhimurium DT104 of resistance type ACSSuT, antibiotic resistance to these compounds is chromosomally encoded [Reference Threlfall23]. Later, in particular, following the licensing of enrofloxacin for veterinary use, isolates of multi-resistant DT104 with decreased susceptibility to ciprofloxacin were seen. In our study, the number of S. Typhimurium isolates from turkeys, pigs and cattle which showed resistance to nalidixic acid increased markedly from 1993/1994 onwards, which coincides temporally with the introduction of enrofloxacin for veterinary use.
However, the exchanges of multi-resistant S. Typhimurium DT104 between animal species and man remain a matter of debate. Mather and co-workers claim that the bacterium and its resistance genes were largely maintained separately within human and domestic animal populations and that there was limited transmission [Reference Mather13], while Leekitcharoenphon and co-workers came to a different conclusion [Reference Leekitcharoenphon24]. They analysed over 300 isolates of MDR DT104 from 21 countries and came to the conclusion that several host switches occurred between different animal species, from animals to humans and also likely from humans to animals.
The emergence of multiple clones of S. Typhimurium-like strains lacking expression of the second flagellar antigen (monophasic Salmonella Typhimurium) has been reported worldwide [25, Reference Moreno Switt26], and has been observed in all four livestock species described in the current study. However, numbers began to increase earlier in cattle and pigs (from 2007 onwards) compared to chickens and turkeys, where the first isolates were reported in 2010 and 2011, respectively. A possible explanation for this could be that monophasic strains of S. Typhimurium were introduced to pigs and cattle first, but that only after a gradual build-up of environmental contamination which also affected feed, infection was introduced into other animal sectors, which generally operate to higher biosecurity measures compared to the pig and cattle sectors. It is not clear whether the few isolates from the 1990s were genuine monophasic strains, and further molecular analysis on these isolates may answer that question.
Animal isolates of S. Enteritidis and S. Typhimurium in Great Britain are currently still phage typed; this has proven to be a valuable tool for source attribution and outbreak investigations, and to compare human and animal isolates. Phage typing has also been a valuable tool to identify possible multi-country outbreaks without the need for sophisticated molecular methods. However, phage typing has been phased out in many countries already and is likely to end in Great Britain within the next few years due to the cessation of supply of typing phages. This means that alternative methods for rapid comparison of isolates from human, veterinary and other sources and between countries will need to be agreed and established. Due to EU legislation demanding to type of Salmonella isolates from chickens and turkeys under the statutory surveillance programmes according to the Kauffman–White–Le Minor scheme, a complete change of veterinary diagnostic laboratories to molecular serotyping/genotyping methods is going to be a challenging task in the near future.
Antimicrobial resistance
Different clones and phage types of S. Typhimurium isolates show individual, usually typical resistance patterns, and the dominance of a particular phage type/clone can therefore significantly bias the overall resistance data at a certain time. DT104, for example, is usually penta-resistant (ampicillin, chloramphenicol, streptomycin, sulphonamide compounds and tetracycline), with only 15% of isolates from all livestock species reported as fully sensitive in 2014. Undefined phage type U288, the most common phage type seen in pigs, is usually resistant to ampicillin, chloramphenicol, streptomycin, sulphonamide compounds, tetracycline and sulphamethoxazole/trimethoprim, and in 2014, none of the U288 isolates from pigs were fully sensitive [27]. Therefore, since U288 was the most common phage type in pigs, the overall percentage of fully sensitive pig isolates was lower compared to the other species.
A particular feature of S. Typhimurium isolates from turkeys has always been the high proportion of isolates resistant to nalidixic acid, which is not observed in any of the other species. This is most likely due to a relatively high historic usage of fluoroquinolones in turkeys compared with other species, in particular when compared to pigs, where fluoroquinolone treatment in drinking water (or feed) is not used.
Resistance to apramycin, which is increasingly being used to treat diarrhoea in pigs, was mainly seen in pigs (with the exception of two years for chickens, one year for cattle and one year for turkeys). The situation was very similar for gentamicin, and the most likely explanation for this phenomenon is cross-resistance to both apramycin and gentamicin, conferred by the enzyme AAC(3)IV, which is predominantly responsible for resistance. However, both apramycin and gentamicin resistance in pigs was lower in pigs recently than in the previous years (35% and 34% in 2012 but 0% and 6% in 2014).
Resistance to nalidixic acid has been traditionally low in pigs (10% or less per year), since fluoroquinolones are not commonly administered as an oral antimicrobial, and only slightly higher in cattle (up to 16% of isolates per year). In chickens, resistance to nalidixic acid was 0% in eight of the 13 calendar years, but reached 58% in 2005 and 29% in 2006. In turkeys, where oral fluoroquinolones are commonly used as therapeutic antibiotics, resistance has been considerably higher in most years, with up to 84% of isolates being resistant in 2005, 71% in 2004 and 63% in 2003. However, in some years, as little as 0% resistance was reported, such as in 2009. A higher resistance of Salmonella isolates from poultry, in particular from turkeys, against quinolones compared to cattle and pigs was also observed in Germany [Reference Malorny28].
Acknowledgements
This work was supported by the Department for Environment, Food and Rural Affairs under project codes FZ2000 and FZ2026.












