INTRODUCTION
Plague is a life-threatening disease caused by the bacterium Yersinia pestis. It has been responsible for over 100 million deaths during three pandemics. Clinical manifestations of plague include bubonic, septicaemic, and pneumonic forms of disease. Pneumonic plague, the most serious form, arises though direct inhalation of bacteria into the lungs (primary pneumonic plague) or through haematogenous spread of bacteria following percutaneous exposure (secondary pneumonic plague). Unlike other clinical forms, pneumonic plague is transmissible from person-to-person and therefore poses a special threat to public health [Reference Kool1].
Found naturally in many parts of the world, Y. pestis is considered a prime candidate for use as a bioweapon [Reference Khan, Morse and Lillibridge2, Reference Rotz3]. Aerosol release in urban centres could lead to large outbreaks of primary pneumonic plague with waves of secondary transmission [4]. In the USA, considerable resources have been devoted to preparing for a plague bioweapon attack, including emergency response exercises and stockpiling of antimicrobials for treatment, post-exposure, and pre-exposure prophylaxis [5, Reference Inglesby, Grossman and O'Toole6]. To be most effective, however, these countermeasures require a better understanding of primary pneumonic plague transmission dynamics.
Models of infectious disease transmission commonly utilize the basic reproductive number, R 0. This value represents the expected number of secondary infections following introduction of a single infected individual into a completely susceptible population [Reference Anderson and May7, Reference Rothman and Greenland8]. Alternately, R C is defined as the average number of secondary cases generated by a single infectious case after control measures have been implemented [Reference Bauch9]. This value indicates whether an outbreak will be self-sustaining under defined levels of control. When control measures are effective, R C<R 0, and for an epidemic to die out, R C must be <1 [Reference Bauch9, Reference Lipsitch10].
While R values provide an important measure of overall transmissibility, considerable variation can exist in the number of transmissions or secondary infections per individual. Recent studies have underscored the importance of this heterogeneity on transmission dynamics [Reference Bauch9–Reference Riley12]. For example, for SARS and other diseases, there are rare individuals, so-called ‘superspreaders’, who for immunological or social reasons generate an especially large number of transmission events. Superspreading events (SSEs), especially those that occur early in an outbreak, can markedly affect the course of an epidemic [Reference Lloyd-Smith11–Reference Shen14].
Previously published models suggest that primary pneumonic plague has an R 0 >1, meaning that an epidemic would be likely to spread until intervention [Reference Gani and Leach15, Reference Nishiura16]. However, these models have several limitations with respect to outbreak modelling and bioweapon event planning. First, they rely on data from large publicized outbreaks where multiple generations of transmission were observed, overlooking data on numerous isolated, unpublished cases where plague was not transmitted. This potentially biases estimates of R 0 upwards and affects selection of the underlying secondary transmission model. In addition, these models use data from developing countries where public health infrastructure and social networks differ from those in the USA and other industrialized nations.
We reviewed demographic and other data for all cases of primary pneumonic plague occurring in the USA since the disease was first introduced to North America in 1900. We used data from cases occurring in the pre-antibiotic era (1900–1943) to estimate R 0 and R C, and to model individual variability in secondary transmissions and the potential for pneumonic plague SSEs. These results were used to evaluate transmission potential, effectiveness of non-pharmaceutical control measures, and the effect of extreme events on the epidemic potential of this disease.
METHODS
We reviewed surveillance data on all confirmed, probable, and suspect cases of plague reported to the US Public Health Service from 1900 to 2009. We identified all cases of primary pneumonic plague and compared demographic features, source of infection, mortality rates, and frequency of secondary transmission for cases occurring in the pre-antibiotic era (1900–1943) with those in the antibiotic era (1944–2009). To assess transmission dynamics in the absence of pharmacological interventions, we estimated R 0, R C, and variability in secondary transmission in individuals using only data from the pre-antibiotic era.
We calculated R 0 and R C as maximum-likelihood estimates (MLEs) for discrete probability distributions (Poisson, geometric, negative binomial) based upon number of secondary transmissions per primary pneumonic plague case. The non-parametric bootstrap method was used to compute 95% confidence intervals (CIs). Selection among the candidate models was made using weighted Akaike's Information Criterion (AIC) and a goodness-of-fit (GOF) test based on likelihood-ratio (or G 2) parameter estimates. To estimate R 0, we included only those primary pneumonic plague cases that occurred (i.e. died or recovered) before public health interventions were implemented. To provide a conservative estimate of R C, we included data from cases occurring both before and after interventions were implemented. We assumed, in all instances, that infected persons could only transmit Y. pestis while symptomatic.
To estimate variability in secondary transmission among individuals, we used a negative binomial model to calculate the dispersion parameter, k 0, and assumed the variance-to-mean ratio to be 1+R 0/k 0. From this expression, the smaller the k 0, the greater the variation expected in secondary transmissions among infected individuals. The dispersion parameter was estimated by applying the method of maximum-likelihood to the reciprocal value (α=1/k 0) in the negative binomial likelihood expression. Using the non-parametric bootstrap method, we computed the 95% bias-corrected accelerated confidence interval for k 0 based on 10 000 resamples. We performed the Potthoff–Whittinghill ‘index of dispersion’ test to investigate whether this dataset deviates significantly from one describing a disease with homogeneous transmission probabilities, as estimated under Poisson distribution [Reference Potthoff and Whittinghill17].
We relied upon criteria proposed by Lloyd-Smith and colleagues to define a SSE for primary pneumonic plague in the US population. We used ![]() 0, as computed above, to construct a fitted Poisson distribution, a probability distribution for secondary transmissions that neglects individual variation. We then defined a SSE as an event in which an infected individual transmits plague to more people than is expected at the 99th percentile of this fitted Poisson distribution. We calculated the expected frequency of SSEs as the proportion of cases under a negative binomial distribution having mean
0, as computed above, to construct a fitted Poisson distribution, a probability distribution for secondary transmissions that neglects individual variation. We then defined a SSE as an event in which an infected individual transmits plague to more people than is expected at the 99th percentile of this fitted Poisson distribution. We calculated the expected frequency of SSEs as the proportion of cases under a negative binomial distribution having mean ![]() 0 and dispersion parameter
0 and dispersion parameter ![]() 0 that is ⩾99th percentile of the fitted Poisson distribution described above [Reference Lloyd-Smith11].
0 that is ⩾99th percentile of the fitted Poisson distribution described above [Reference Lloyd-Smith11].
We simulated a branching process under the fitted negative binomial model to investigate outbreak extinction probabilities according to transmission generation. Under this method, we considered that a single case initiated the outbreak by infecting a Poisson random number of individuals, considered generation 1; the mean of this Poisson distribution was drawn from a gamma distribution with shape=![]() 0 and scale=
0 and scale=![]() 0/
0/![]() 0. Individuals in generation 1 then infected a Poisson random number of individuals, generation 2, with individual means drawn from the same gamma distribution. The process continued in this manner until a generation with zero infected individuals was reached. For computational reasons, an outbreak simulation was truncated at the first of 40 generations or total outbreak size 7500 individuals, and this outcome was recorded. We also evaluated the conditional probabilities of extinction for each generation, given or conditional upon only those outbreaks not extinct in the previous generation. For this analysis, we simulated 50 000 outbreaks and also stratified these by number of transmissions made by the initial case (i.e. SSE vs. non-SSE).
0. Individuals in generation 1 then infected a Poisson random number of individuals, generation 2, with individual means drawn from the same gamma distribution. The process continued in this manner until a generation with zero infected individuals was reached. For computational reasons, an outbreak simulation was truncated at the first of 40 generations or total outbreak size 7500 individuals, and this outcome was recorded. We also evaluated the conditional probabilities of extinction for each generation, given or conditional upon only those outbreaks not extinct in the previous generation. For this analysis, we simulated 50 000 outbreaks and also stratified these by number of transmissions made by the initial case (i.e. SSE vs. non-SSE).
RESULTS
A total of 1001 (985 confirmed, 12 probable, 4 suspect) plague cases were reported in the USA from 1900 to 2009, including 74 cases of primary pneumonic plague (Table 1). Sixty (81%) of 74 cases occurred in the pre-antibiotic era, of which 38 (63%) occurred before implementation of routine public health interventions during an outbreak (e.g. quarantine). Fourteen cases occurred in the antibiotic era, with antimicrobials used to treat all but the most recent illness [Reference Wong18]. As shown in Table 1, during the pre-antibiotic era, 83% of cases were reportedly acquired through human contact. Of those, 86% were associated with three epidemics (San Francisco 1904, Oakland 1919 and Los Angeles 1924). During the antibiotic era, the principle route of transmission has been through animal contact (64%), while laboratory exposures accounted for another 21% of all cases. In the pre-antibiotic era, 92% of all cases reported were fatal, whereas only 36% of cases have died since.
Table 1. Demographic, transmission, and outcome information for all primary pneumonic plague cases reported to occur in the USA during two time periods: 1900–1943 (pre-antibiotic era) and 1944–2009 (antibiotic era)
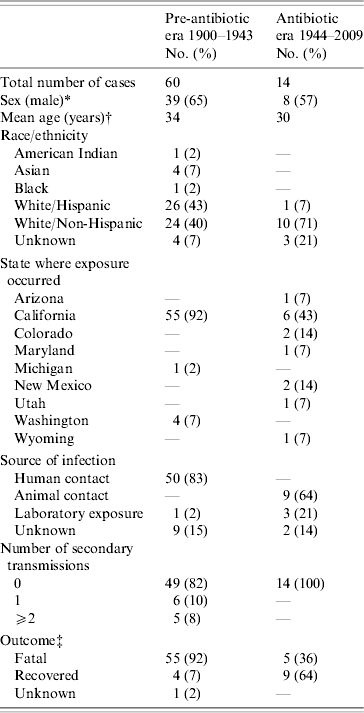
* No significant difference observed for sex (χ2 test, P=0·6) in the pre-antibiotic vs. antibiotic eras.
† No significant difference (t test, P=0·44) between cases in the pre-antibiotic vs. antibiotic eras.
‡ Significant difference observed (χ2 test, P<0·001) for outcome in cases in the pre-antibiotic vs. antibiotic eras.
The number of secondary transmissions resulting from individual primary pneumonic plague cases has also changed markedly between the pre-antibiotic and antibiotic eras (Table 1). In the pre-antibiotic era, 11 (18%) of those affected by pneumonic plague went on to transmit Y. pestis to at least one individual. In total, five people (8%) transmitted Y. pestis to at least two additional contacts. In the San Francisco outbreak of 1904, available evidence suggests that a 14-year-old female transmitted plague to three of her family members. An alternate explanation is that she infected her father (aged 54 years), who went on to transmit plague to the other two family members [Reference Blue19]. In the Oakland outbreak of 1919, one 32-year-old male transmitted plague to five individuals; one of these (a 31-year-old male) went on to transmit to another four individuals, another 31-year-old male transmitted to another two individuals [Reference Kellogg20]. The most extreme event affected an entire Los Angeles neighbourhood in 1924. In this case, one 37-year-old male was probably responsible for transmission of pneumonic plague to 26 individuals [Reference Dickie21, Reference Link22]. No primary or secondary pneumonic plague cases were reported between 1924 and 1944. No secondary transmissions have been reported in the USA since the 1924 outbreak.
Under all models evaluated (Poisson, geometric, negative binomial), R 0 was estimated to be 1·18. As expected, implementation of standard, non-pharmaceutical control measures lowered the transmission potential, providing an estimate of ![]() C=0·76. For both the R 0 and R C estimates, the negative binomial model, having individual secondary transmission probabilities that are gamma-distributed with means R 0 or R C, and dispersion parameters, k 0 and k C, provided the best fit for the pre-antibiotic era data, according to the weighted AIC values. However, the P value (0·012) for goodness-of-fit (Table 2) indicated that the negative binomial distribution (R 0: 95% CI 0·37–3·82) still did not adequately fit these datasets. Observed proportions and fitted distributions for the number of secondary transmissions per case of primary pneumonic plague for the three models are displayed in Figure 1.
C=0·76. For both the R 0 and R C estimates, the negative binomial model, having individual secondary transmission probabilities that are gamma-distributed with means R 0 or R C, and dispersion parameters, k 0 and k C, provided the best fit for the pre-antibiotic era data, according to the weighted AIC values. However, the P value (0·012) for goodness-of-fit (Table 2) indicated that the negative binomial distribution (R 0: 95% CI 0·37–3·82) still did not adequately fit these datasets. Observed proportions and fitted distributions for the number of secondary transmissions per case of primary pneumonic plague for the three models are displayed in Figure 1.
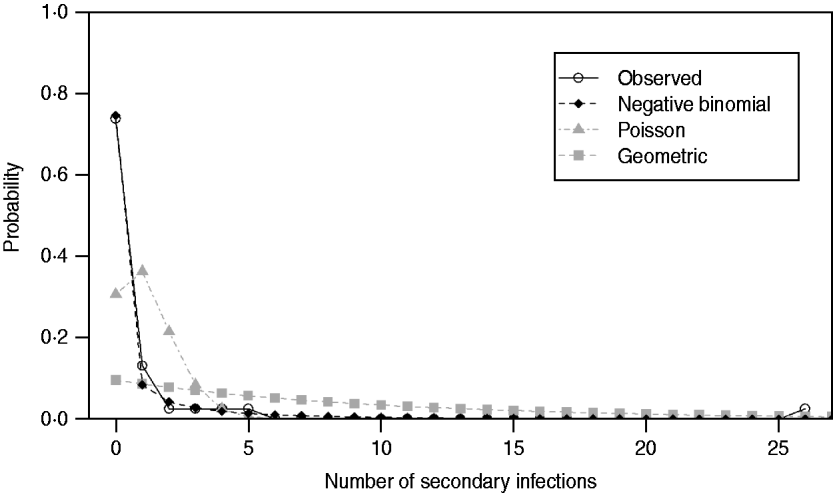
Fig. 1. Observed proportions and fitted probability distributions for the number of secondary transmissions per case of primary pneumonic plague in the USA.
Table 2. Model fit results for three candidate models used to estimate the basic reproductive number, R0, for primary pneumonic plague in the USA
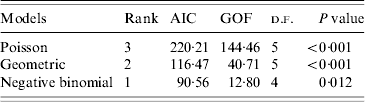
AIC, Akaike's Information Criterion; GOF, goodness of fit.
The dispersion parameter, k 0, was estimated at 0·126 (95% CI 0·033–0·360). The P value for the Potthoff–Whittinghill test was <0·0001, indicating that it is unlikely that the observed high degree of variance arose by chance from a Poisson distribution.
In this analysis, a 99th percentile SSE for pneumonic plague in the USA was determined to be any event where at least four people were infected by a single case. Moreover, it was estimated that 10% of all primary pneumonic plague cases occurring in the USA would lead to SSEs. We identified three possible SSEs in this historical dataset. Two SSEs occurred as part of the Oakland outbreak of 1919 (described above) [Reference Kellogg20]. The third event, where plague was transmitted by one person to 26 individuals, was part of the Los Angeles outbreak of 1924 [Reference Dickie21, Reference Link22]. This single event had a very strong impact on the overall distribution chosen to represent secondary transmission probabilities, as well as the corresponding R and k values. Exclusion of this extreme value gives us an estimate for ![]() 0 of 0·51.
0 of 0·51.
Using a branching process, the initial case was a SSE for 5049 (10·1%) of 50 000 simulated outbreaks having a probability of secondary transmission randomly selected from a negative binomial distribution with ![]() 0=1·18 and
0=1·18 and ![]() 0=0·126. Overall, 73·6% (3715 simulations) of outbreaks begun by a SSE reached extinction, and 99·9% of outbreaks begun by a non-SSE reached extinction (before 40 generations). Of those outbreaks that reached extinction, in Figure 2 a, we demonstrate that extinction given introduction by a single individual is nearly 80% in the first generation of transmission. In subsequent generations, the probability of extinction approaches zero, owing to the relatively few outbreaks persisting past the fourth generation and the continued potential for SSEs. As shown by the conditional probability figures at bottom, the probability of extinction (past the fourth generation) ranges from 20% to 30%. In Figure 2 b, which displays extinction probabilities given disease introduction by a SSE, the probability of extinction in the early generations is much lower in generation 1 than in Figure 2 a, although the conditional probability of extinction past the fourth generation is very similar. The results in Figure 2 c, which displays extinction probabilities given introduction by a single non-SSE individual, are similar to those in Figure 2 a, except for a slightly higher probability of extinction in the early generations of transmission. This similarity is due to predominance of non-SSE individuals among pneumonic plague cases.
0=0·126. Overall, 73·6% (3715 simulations) of outbreaks begun by a SSE reached extinction, and 99·9% of outbreaks begun by a non-SSE reached extinction (before 40 generations). Of those outbreaks that reached extinction, in Figure 2 a, we demonstrate that extinction given introduction by a single individual is nearly 80% in the first generation of transmission. In subsequent generations, the probability of extinction approaches zero, owing to the relatively few outbreaks persisting past the fourth generation and the continued potential for SSEs. As shown by the conditional probability figures at bottom, the probability of extinction (past the fourth generation) ranges from 20% to 30%. In Figure 2 b, which displays extinction probabilities given disease introduction by a SSE, the probability of extinction in the early generations is much lower in generation 1 than in Figure 2 a, although the conditional probability of extinction past the fourth generation is very similar. The results in Figure 2 c, which displays extinction probabilities given introduction by a single non-SSE individual, are similar to those in Figure 2 a, except for a slightly higher probability of extinction in the early generations of transmission. This similarity is due to predominance of non-SSE individuals among pneumonic plague cases.
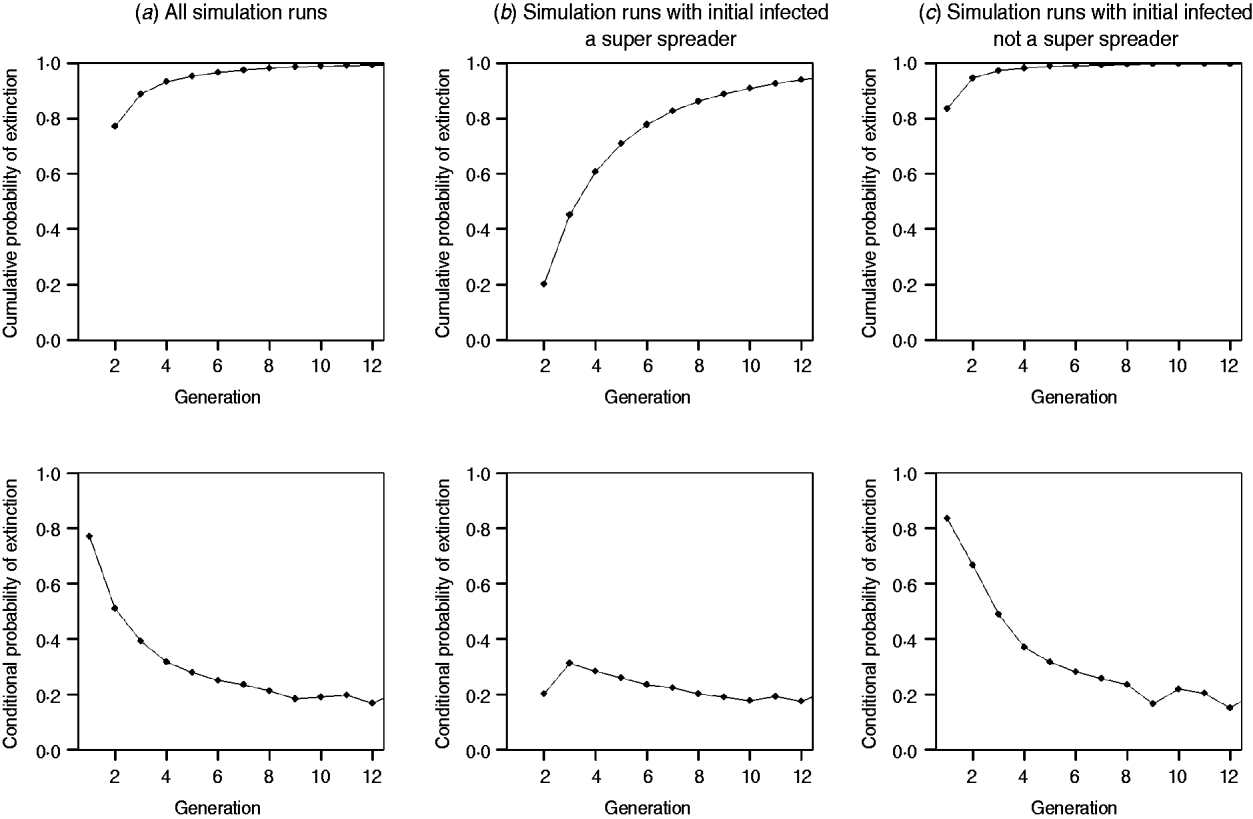
Fig. 2. The probability of extinction and conditional probability of extinction by generation of transmission for R 0=1·18, and k 0=0·126 (a) through random selection of a single introductory case according to the negative binomial distribution; (b) when introduction is by a single superspreading event (SSE) (⩾4 secondary transmissions); (c) when introduction is by a single non-SSE (<4 secondary transmissions).
DISCUSSION
Using models based on comprehensive surveillance data, we estimate that the R 0 for primary pneumonic plague in the USA is nearly equal to 1·0. This value is similar to a previous estimate; however, the underlying model for secondary transmission is substantially different, with important implications for transmission dynamics and response planning. In 2004, Gani & Leach [Reference Gani and Leach15] calculated R 0=1·3 for pneumonic plague based upon aggregated data from eight published outbreaks (n=74) occurring between 1907 and 1997 on four continents. More recently, Lloyd-Smith and colleagues [Reference Lloyd-Smith11] used the same dataset to evaluate individual variation on outbreak dynamics. Both groups found that the geometric model provided the best fit to the data, with the dispersion parameter, k 0, estimated at 1·37 (90% CI 0·88–3·53). In contrast, our data containing a larger number of non-transmitters had higher individual variability, best fit the negative binomial model, and yielded a k 0 of 0·126 (95% CI 0·052–0·339), tenfold lower [Reference Lloyd-Smith11, Reference Gani and Leach15].
Our results suggest a paradoxical aspect to pneumonic plague transmission. High variability in individual transmission potential, as reflected in the low k 0 value, indicates a possibility for SSEs characterized by sudden, explosive increases in cases. At the same time, the low R 0 value indicates that most outbreaks are unlikely to perpetuate for multiple generations and may be terminated with relatively modest interventions. In fact, all US outbreaks during the pre-antibiotic era, including one having two reported SSEs (Oakland 1919), were controlled quickly and effectively with routine measures, as demonstrated by an estimated value for R C of 0·76. Described in detail in the literature, these control measures included social-distancing, isolation, quarantine, enhanced surveillance, contact tracing, and simple barrier precautions [Reference Kellogg20–Reference Viseltear23].
The k 0 and R 0 values estimated in this analysis for pneumonic plague are very similar to those reported for SARS [Reference Lloyd-Smith11]. Epidemics of SARS have been found contingent upon high variability in transmission and occurrence and timing of SSEs, and may therefore serve as a model for future pneumonic plague outbreaks. Other important factors for both diseases include spatial heterogeneity or social-networking [Reference Dye and Gay24]. Since 1924, plague in the USA has shifted from urban centres (e.g. San Francisco) to more sparsely populated areas of the Southwest. The lack of outbreaks since that time may be due partly to a lowered contact rate with susceptibles [Reference Bauch9, Reference Meyer25]. Inclusion of such social and spatial factors in future pneumonic plague transmission models may significantly improve model accuracy [Reference Davis26].
By using data from the pre-antibiotic era, our approach has the advantage of estimating the transmission potential of plague in a situation where antibiotic treatment or prophylaxis is not available or practical. This information may be particularly relevant to a bioweapon release of Y. pestis that has been engineered for antibiotic resistance. Nevertheless, our analysis has several limitations. First, although the negative binomial provided the best fit to our data, a mixed distribution model may be a more appropriate model for this highly skewed data. Second, the data used for this analysis were obtained from a retrospective review of surveillance data, public health reports and publications, and generally represent cases of naturally occurring plague. It is possible that these data may not represent the type of transmission that would occur following infection due to a bioweapon [Reference Inglesby27]. If the agent used is modified to be more virulent or infectious, the biological response could be different. In those cases, however, a more rapid and severe illness would probably also lead to faster deaths, and a shorter period of transmissibility (i.e. disease that is somewhat self-limiting). It is also possible that SSEs are associated with genetically distinct Y. pestis strains. However, in all of those outbreaks having SSEs, the majority of individuals failed to transmit, even after exposure to a SSE-transmitted strain. Moreover, all strains presently known to exist in the USA are of the same biovar [Reference Morelli28]. Other potential limitations of the data include a vague or unclear clinical picture (e.g. primary vs. secondary presentation) for some cases, and a poorly defined contact history for some of the earliest cases. However, nearly all records indicated a fairly short symptomatic period, which is most consistent with clinical presentation of primary pneumonic plague [29]. In addition, for those few cases where transmission history was less clear, the overall number of transmissions per outbreak did not change, and the R 0 estimate was unaffected.
Given the potential for explosive epidemics of pneumonic plague due to the occurrence of SSEs, an evaluation of efficacy of specific control measures is needed. Modelling efforts should focus on pneumonic plague exposures among hypothetical populations of varying sizes having multiple introductions, with a comparison of individual-level and population-wide controls. This may require construction of complex models used to evaluate variable contact rates and durations of infectiousness. An evaluation of cost, timing and the effect of combined control efforts would be most useful for policy and planning guidance [Reference Bauch9].
While it is likely that antimicrobial use could further blunt transmission of pneumonic plague during an outbreak, the logistical challenges associated with mass prophylaxis are not inconsequential, and could even be counterproductive if they detract from immediately available, non-pharmaceutical, interventions such as ‘social distancing’. As suggested by this analysis, the most effective intervention may involve rapid identification and treatment of ill persons, minimizing the potential for SSEs. It is critical that decision-makers understand the likely transmission dynamics of pneumonic plague, including both the potential for SSEs and the overall low risk of ongoing spread.
ACKNOWLEDGEMENTS
We thank the local, state, and federal public health practitioners who diligently collected and recorded all data used for this study. We also thank the following CDC staff: Kiersten Kugeler for general advice; Ken Gage for scientific guidance and manuscript review; and J. Erin Staples for her extensive review of literature and public documents to provide a more comprehensive record of human plague in the USA.
This work was funded by the Centers for Disease Control and Prevention. The views expressed in the publication are those of the authors and not necessarily those of the U.S. government.
DECLARATION OF INTEREST
None.






