INTRODUCTION
In spring 2011, toxigenic Vibrio cholerae serogroup O75 caused an outbreak of vibriosis in the United States. The subsequent investigation determined that oysters harvested in Apalachicola Bay, located off the Florida Panhandle, were the source [Reference Onifade1]. Infections caused by this pathogen and similar toxigenic V. cholerae serogroups that cause vibriosis are rare. Little is known about them or the epidemiology of the illness they cause.
These pathogens are among ~200 known serogroups of V. cholerae [Reference Tobin-D'Angelo2] and are referred to as toxigenic V. cholerae non-O1, non-O139. This unusual name distinguishes them from toxigenic strains of V. cholerae serogroups O1 and O139, which cause cholera and have caused the death of millions of people worldwide. Like the strains that cause cholera, toxigenic V. cholerae non-O1, non-O139 produce cholera toxin, cause diarrhoea, and are transmitted through contaminated seafood or water, but they have not caused epidemics and, by definition, have not caused cholera [Reference Tobin-D'Angelo2–Reference Dalsgaard4]. Nevertheless, these pathogens can cause severe illness and small outbreaks, and thus warrant investigation.
We studied the clinical, epidemiological, and microbiological features of toxigenic V. cholerae non-O1, non-O139 infections detected in the United States from 1984 to 2014 and describe them here. We also discuss the public health implications of our findings.
METHODS
Toxigenic V. cholerae non-O1, non-O139 infections are reported to the United States Centers for Disease Control and Prevention (CDC) as vibriosis cases via the Cholera and Other Vibrio Illness Surveillance (COVIS) system. COVIS was launched in 1988 by the Gulf Coast states of Alabama, Florida, Louisiana, and Texas, the United States Food and Drug Administration, and CDC. COVIS also incorporates data available before 1988. By 2005, state and local health departments from nearly every state were submitting reports of laboratory-confirmed cases to COVIS. In 2007, vibriosis became a nationally notifiable condition. Reports to COVIS contain demographic and clinical information on the patient; the Vibrio species isolated; exposures, such as seafood consumption and water contact; and domestic and international travel that occurred within 7 days of symptom onset [Reference Newton5].
CDC's Enteric Diseases Laboratory Branch requests that local and state health departments submit all V. cholerae isolates they receive. The laboratory determines whether the strains are toxigenic by using polymerase chain reaction, testing specifically for the ctxA gene (cholera toxin) and the species-specific genes toxR and ompW [Reference Ghosh6, Reference Nandi7]. The laboratory tests each isolate for agglutination in specific antisera for V. cholerae serogroups O1, O75, O139, and O141, which are the most common toxigenic serogroups detected in the United States. If the isolate is not one of these four serogroups, it is untypable with available antisera and is recorded as non-O1, non-O139. The isolates are also subtyped by using pulsed-field gel electrophoresis (PFGE) with two restriction enzymes (SfiI and NotI) [Reference Tobin-D'Angelo2]. The laboratory uses either agar disk diffusion or broth microdilution to test for antimicrobial susceptibility, methods standardized by the Clinical and Laboratory Standards Institute (CLSI) [8]. CLSI interpretive criteria for V. cholerae susceptibility are available only for ampicillin, azithromycin, chloramphenicol, sulfisoxazole, and trimethoprim-sulfamethoxazole, although other drugs of medical importance were also tested.
Our analysis covers all reports to COVIS of toxigenic V. cholerae non-O1, non-O139 infections from 1984 to 2014. We describe and summarize the cases collectively and by serogroup. When calculating exposure proportions, we exclude patients whose exposure status was unknown. We also map infections to the state that probably was the source of the exposure by using patient's travel history, number and type of exposures, and available seafood traceback information.
RESULTS
From 1984 to 2014, of ~13 000 vibriosis cases reported to COVIS, 52 cases were caused by toxigenic V. cholerae non-O1, non-O139. Thirty of the 52 infections were with V. cholerae serogroup O75, and 21 were with serogroup O141. One isolate was untypable. Patients with O75 and O141 infections were similar: about half were female (50% for O75, 43% for O141), and the median age was in the 40s. The untypable isolate was from a 38-year-old woman (Table 1).
Table 1. Toxigenic V. cholerae non-O1, non-O139 infections, Cholera and Other Vibrio Illness Surveillance (COVIS) system, United States, 1984–2014*
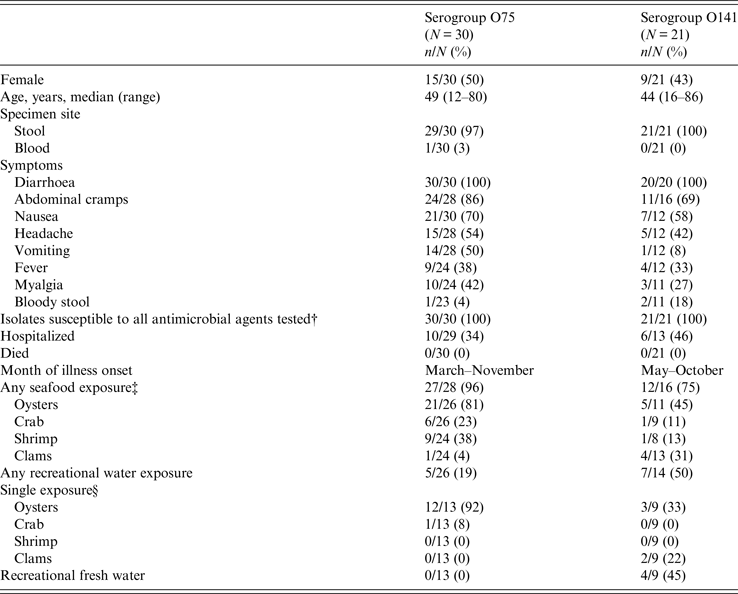
* In addition, a 38-year-old woman from Texas had an untypable infection. She experienced diarrhoea, abdominal cramps, nausea, headache, and vomiting. The isolate was susceptible to all antimicrobials tested. She was not hospitalized and survived her illness. She did not report seafood exposure or out-of-state travel. She ingested river water while tubing down the Comal River near Austin, Texas, in the week before her illness began.
† Clinical and Laboratory Standards Institute interpretive criteria for V. cholerae susceptibility include ampicillin, azithromycin, chloramphenicol, sulfisoxazole, and trimethoprim-sulfamethoxazole. Other drugs of medical importance also were tested.
‡ Nineteen of the patients infected with O75 ate raw oysters, while five of the O141 patients ate raw oysters. Two of the patients infected with O141 ate raw clams.
§ Eleven patients infected with O75 reported eating raw oysters. Three of the O141 patients reported eating raw oysters and two reported eating raw clams.
Patients resided in 21 states, with Florida (n = 8), Louisiana (n = 8), Georgia (n = 5), and Alabama (n = 4) being the most common (Fig. 1). Serogroup O75 infection only occurred in residents of the Eastern United States. Most (23/30, 77%) patients infected with serogroup O75 lived in southern coastal states and, based on their reported travel history, were probably infected there. Three patients travelled to a southern coastal state during the 7 days preceding symptom onset. Patients infected with serogroup O141 were from states across the country. Five of these patients travelled domestically during 7 days preceding symptom onset; three visited a state in the South and two a state in the Northeast. One patient travelled internationally.

Fig. 1. State of residence and probable origin of exposure for (a) serogroup O75 infections (state of residence for 30 persons and probable origin of exposure for 12 persons) and (b) serogroup O141 infections (state of residence for 21 persons and probable origin of exposure for eight persons), United States, 1984–2014. ![]() , No cases;
, No cases; ![]() , state of residence;
, state of residence; ![]() , seafood exposure (n);
, seafood exposure (n); ![]() , freshwater exposure (n).
, freshwater exposure (n).
Hospitalization rates were somewhat lower for patients infected with O75 (34%) than for those with O141 (46%). Diarrhoea, abdominal cramps, nausea, and headache were the four most common symptoms reported by patients infected with either serogroup. Fourteen patients infected with O75 reported vomiting (50%), but only one infected with O141 reported this symptom (Table 1). No one died.
The illness onset date was reported for 48 patients. Serogroup O75 cases occurred both earlier and later in the year (March–November) than O141 cases (May–October).
Twenty-seven (96%) of 28 patients with O75 infection and 12 (75%) of 16 patients with O141 infection who provided a food history reported consuming seafood. Most patients with O75 infection ate oysters (21/30, 70%), primarily raw (19/21, 90%), and 12 of them reported no other seafood exposures (11 ate only raw oysters). Both oysters and clams were common seafood exposures for patients infected with serogroup O141. Some of these patients reported eating only raw oysters (3) or raw clams (2) and had no other suspect exposures (Table 1). Traceback information on the origin of the seafood was available for 12 patients infected with serogroup O75, all of whom ate oysters from the Florida Panhandle. Traceback information from four patients with O141 infection showed that two consumed oysters from Florida and two consumed clams from New Jersey (Fig. 1).
Half (7/14) of the patients infected with serogroup O141 reported recreational fresh- or saltwater exposure, but only 19% (5/26) of patients with O75 infections reported this. Recreational water was the most common exposure for persons with O141 infections, and four of these patients only reported freshwater exposure in lakes or rivers. Three of these patients reported that they did not consume seafood in the week prior to illness onset; it is not known if the fourth patient ate seafood. They were exposed in Arizona, Michigan, Missouri, and Texas. The patients exposed in Michigan and Missouri reported swimming and boating in Whitmore Lake and Big Piney River, respectively, while the patient from Texas reported swimming in the Guadalupe River. The patient from Arizona drank water from the San Carlos Lake, which had been closed due to elevated algae levels; this is the patient whose seafood exposure is unknown. The patient with the untypable isolate ingested water while tubing on the Comal River near Austin, Texas, and she reported that she did not consume seafood. In contrast, no patients with O75 infection reported recreational water exposure as a sole exposure (Table 1, Fig. 1).
There were seven PFGE pattern combinations (SfiI and NotI) in the O75 isolates and 13 in the O141 isolates. All isolates were susceptible to antimicrobial agents tested.
DISCUSSION
The toxigenic V. cholerae non-O1, non-O139 serogroups described here are closely related pathogens that cause similar illness: most patients experienced diarrhoea, abdominal cramps, and nausea. These patients were also demographically similar. Nevertheless, there were notable differences in how and where the patients were infected.
Although seafood, specifically raw shellfish, was often reported and probably was the vehicle for many infections, recreational exposure to fresh, non-potable water in rivers and lakes was the likely source for four O141 cases and the patient whose isolate was untypable. Freshwater is an unusual source for V. cholerae non-O1, non-O139 infection, although isolation has been reported from pond water samples in Austria [Reference Hirk9]. The culprit in the infections in the United States might have been copepods – small crustaceans present in fresh- and saltwater. V. cholerae attach to them to feed on the chitin in their exoskeletons, and the bacteria can be present in large numbers [Reference Huq10–Reference Heidelberg, Heidelberg and Colwell12]. The patients could have ingested one or more copepods when they swallowed the water, which would explain how they received an infectious dose.
The geographical distribution of exposures reflects the source of each serogroup. Serogroup O75 infections were regionally focused in the Gulf Coast, whereas O141 infections occurred throughout much of the continental United States, inland and along the coast. The patient with the untypable isolate was exposed in a river in the interior of Texas. Previous studies have identified V. cholerae non-O1, non-O139 serogroups in similar inland and coastal environments in other continents, including Asia, Europe, and South America [Reference Dalsgaard4, Reference Hirk9, Reference Diep13].
Despite these differences, interventions that prevent most enteric Vibrio infections also probably would prevent many infections from these toxigenic serogroups. Two interventions have been used in the past: providing safer food by using techniques that reduce or prevent bacterial growth during and shortly after harvest and that reduce pathogen loads prior to sale and consumption, and educating the public about the risks of consuming raw shellfish, primarily in the form of placards or other printed warnings in restaurants. Little evidence exists, however, that this form of consumer education is an effective means of public health messaging [Reference Mouzin14, Reference Vugia15]. Unless more effective consumer education methods are developed, public health interventions should focus less on encouraging consumers not to eat raw shellfish and more on making food safe.
For more than a decade, an array of proven techniques have been available to prevent Vibrio growth and reduce pathogen loads in oysters, the principal food vehicle for V. cholerae non-O1, non-O139 as well as other pathogenic Vibrio species. The most effective of these techniques is post-harvest processing, which includes individual quick freezing, heat-cool pasteurization, and high hydrostatic pressurization [Reference Vugia15, 16]. In 2003, California enacted regulation requiring raw oysters harvested in the Gulf of Mexico between April and October – when pathogen loads are highest – to undergo post-harvest processing to reduce contamination by V. vulnificus, a highly virulent species of the bacteria. A recent study demonstrated that this regulation has been highly effective, yielding a significant and sustained reduction in V. vulnificus cases in California since its enactment [Reference Vugia15]. Other studies have shown that these techniques also reduce loads of V. parahaemolyticus – another pathogenic Vibrio species often detected in oysters – to levels safe for raw consumption [Reference DePaola17–Reference Ye19]. No studies have specifically assessed the effectiveness of these techniques on toxigenic V. cholerae non-O1, non-O139, but given their biological similarity to V. vulnificus and V. parahaemolyticus [Reference Thompson, Iida and Swings20], the interventions might result in an adequate reduction in bacterial load.
Some states might choose to follow California's example, but other states, such as those with many small oyster farms for which setting up post-harvest processing would be too expensive, might consider limiting their requirements to minimizing pathogen growth after harvest. These interventions include icing oysters within 1 h of harvest and maintaining a storage temperature below 10 °C, the temperature threshold for limiting Vibrio growth [21]. These interventions have been shown to inhibit bacterial growth occurring after oysters are removed from their beds [Reference Lydon, Farrell-Evans and Jones22, Reference Melody23], but they do not reduce bacterial loads already present in the shellfish [Reference Melody23]. Therefore, when dangerous levels of pathogen are present prior to harvest, which is more likely when water temperatures exceed 10 °C, these interventions would not make oysters safe for raw consumption, leaving consumers at risk. Water temperatures in the Gulf of Mexico, one of the principal sources of oysters in the United States, often exceed this threshold, with average temperatures sometimes reaching 30 °C during the summer months [24]. Post-harvest processing techniques are the only interventions currently available that can ensure the safety of oysters harvested in these conditions that are intended to be consumed raw.
While the economic burden of public health interventions should be assessed and taken into consideration when determining the most appropriate course of action, government and industry have an obligation to continue to develop food safety practices and regulations that aim to ensure safer shellfish, which can be risky when eaten raw. Together, they can work to reduce foodborne Vibrio infections – including those caused by toxigenic V. cholerae non-O1, non-O139.
ACKNOWLEDGEMENTS
The authors thank Luis Luque, Christina Scheel, and Sue Swensen for their assistance with the preparation of this manuscript.
This research received no specific grant from any funding agency, commercial or not-for-profit sectors. The conclusions, findings, and opinions expressed by the authors do not necessarily reflect the official position of the United States Department of Health and Human Services or the Centers for Disease Control and Prevention.
DECLARATION OF INTEREST
None.





