To the Editor—The critical threat of carbapenemase-producing (CP) Enterobacteriaceae has garnered significant attention.Reference Logan and Weinstein 1 However, the hazards posed by CP Pseudomonas aeruginosa and CP Acinetobacter baumannii have been underestimated.Reference Gniadek, Carroll and Simner 2 The transmission of mobile genetic elements containing carbapenemase-encoding genes is not species specific. Early and accurate detection of CP P. aeruginosa and CP A. baumannii is necessary to prevent the propagation of carbapenemases across all gram-negative organisms in healthcare settings.
Both P. aeruginosa and A. baumannii pose challenges for carbapenemase detection, but for different reasons. Although accurate and cost-effective phenotypic assays are available to detect CP P. aeruginosa, carbapenem nonsusceptibility among P. aeruginosa isolates in the United States are predominantly mediated by non-carbapenemase (non-CP) mediated mechanisms (eg, the loss of OprD porin expression and/or upregulation of MexAB-OprM efflux pumps).Reference Gniadek, Carroll and Simner 2 , Reference Simner, Opene, Chambers, Naumann, Carroll and Tamma 3 Clinical microbiology laboratories may not see the resources needed to identify carbapenemase detection in carbapenem-nonsusceptible P. aeruginosa as a “high return on investment.” Although carbapenemase production is the primary resistance mechanism among carbapenem nonsusceptible A. baumannii in the United States, commonly employed methods for carbapenemase detection in Enterobacteriaceae and P. aeruginosa are limited in their ability to detect carbapenemases in A. baumannii. Reference Simner, Opene, Chambers, Naumann, Carroll and Tamma 3 We sought to determine whether a carbapenem minimum inhibitory concentration (MIC) cutoff value exists to accurately distinguish CP and non-CP P. aeruginosa and A. baumannii. Identification of such a cutoff value could overcome challenges posed by phenotypic carbapenemase detection methods for these 2 species while eliminating the need to place all people with carbapenem-nonsusceptible isolates on contact precautions.Reference Tamma, Huang, Opene and Simner 4
We included 199 carbapenem-nonsusceptible P. aeruginosa and A. baumannii isolates: 111 were obtained from the Centers for Disease Control and Prevention and Food and Drug Administration Antimicrobial Resistance Isolate Bank (CDC-FDA) and 88 consecutive clinical isolates were obtained from the Johns Hopkins Hospital (JHH). CDC-FDA isolates had been previously molecularly characterized to identify β-lactamase genes using whole-genome sequencing and/or polymerase chain reaction (PCR). The Phoenix Automated System (Becton Dickinson Diagnostics, Sparks, MD) was used for antimicrobial susceptibility testing (AST) for the JHH isolates and carbapenem AST results were confirmed using the ETEST method (bioMérieux, Marcy-l'Étoile, France). Carbapenem nonsusceptibility was defined as meropenem or imipenem MIC of ≥4 mcg/mL. The β-lactamase genes in the JHH isolates were identified using the Check-MDR CT103XL kit microarray-based assay (Check-Points, Wageningen, Netherlands).
Receiver operating characteristic (ROC) curves were generated using various carbapenem MICs to determine the optimal MIC for the detection of CP isolates. The discriminatory power was evaluated using the area under the ROC curve (AUC), with an AUC value of 0.5 indicating no discriminative ability and an AUC value >0.8 indicating good-to-excellent prediction. Sensitivities and specificities were calculated at various carbapenem MIC values. It was determined a priori that sensitivities would be more relevant than specificities in establishing MIC cutoff values. It was more important to have sensitivity approaching 100% so that all or most CP-Pseudomonas and Acinetobacter could be detected, at the expense of specificity. Analyses were performed using the R statistical package (R Foundation for Statistical Computing, Vienna, Austria).
Of 118 P. aeruginosa isolates, 23 (19%) were CP producers. Ambler Class B carbapenemases were identified in 74% of the CP P. aeruginosa isolates; the carbapenemase gene most frequently detected was bla VIM. Of 81 A. baumannii isolates, 50 (62%) were CP producers. With the exception of 3 isolates that produced NDM-1, all A. baumannii isolates produced at least 1 acquired OXA-type carbapenemase.
We explored the possibility of an AUC that maximized both sensitivity and specificity for the detection of CP and non-CP P. aeruginosa and A. baumannii. The meropenem ROC curves were relatively poor at distinguishing CP and non-CP P. aeruginosa and A. baumannii, with AUCs of 0.66 (95% confidence interval [CI], 0.57–0.75) and 0.55 (95% CI, 0.49–0.62), respectively. For imipenem, the AUC of the ROC curve was 0.86 (95% CI, 0.74–0.97) for P. aeruginosa. An imipenem MIC of 64 mcg/mL had the greatest overall sensitivity and specificity (82% and 97%, respectively) for distinguishing CP and non-CP P. aeruginosa. However, an imipenem MIC value of 64 mcg/mL would fail to detect an unacceptably large portion of CP P. aeruginosa. The imipenem ROC curve was poor at distinguishing CP and non-CP A. baumannii (AUC, 0.61; 95% CI, 0.51–0.72).
We refocused our efforts to identify the meropenem and imipenem MIC values that maximized sensitivity, at the sake of specificity, of CP nonfermenters (Table 1). A meropenem MIC of 8 mcg/mL yielded a sensitivity of 98% for CP P. aeruginosa. An imipenem MIC of 8 mcg/mL detected 100% of CP Pseudomonas. For Acinetobacter, a meropenem MIC of 8 mcg/mL detected 100% of CP Acinetobacter and an imipenem of 8 mcg/mL detected 98% of CP Acinetobacter.
TABLE 1 Sensitivities and Specificities Distinguishing Carbapenemase-Producing and Non-carbapenemase-Producing Carbapenem Nonsusceptible Pseudomonas and Acinetobacter Isolates
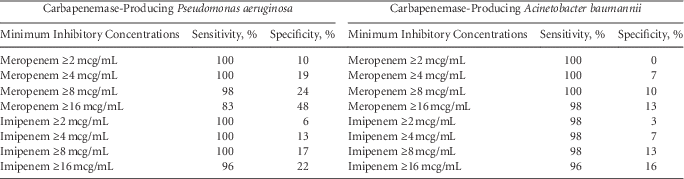
Our findings suggest that a meropenem or imipenem MIC cutoff value of 8 mcg/mL could detect ≥98% of CP P. aeruginosa and CP A. baumannii. The specificities associated with these values, however, would be poor. We prioritized sensitivity over specificity because failing to recognize the presence of carbapenemase-producing organisms in healthcare settings could have unfortunate infection control implications. An MIC cutoff value above the carbapenem susceptibility breakpoint would reduce the proportion of patients placed on contact precautions.
Mobile genetic elements containing carbapenemase genes can spread rapidly in healthcare settings, between both glucose-fermenting (e.g., Enterobacteriaceae) and nonfermenting organisms.Reference Logan and Weinstein 1 , Reference Gniadek, Carroll and Simner 2 Identifying carbapenem cutoff values highly sensitive for detecting carbapenemase production can support enhanced infection control practices for patients harboring CP organisms, potentially averting outbreaks.
The isolates provided by the CDC-FDA bank purposefully contain an overrepresentation of carbapenemase producers to allow for diverse resistance mechanisms to be evaluated. The inclusion of the CDC-FDA isolates improved the accuracy of our sensitivity estimates; however, the prevalence of CP isolates in our cohort should not be extrapolated to general US prevalence estimates. Because only US isolates were included in our cohort, our results may not be generalizable to carbapenem-nonsusceptible isolates from other parts of the world.
Our findings suggest that meropenem or imipenem MIC cutoff values of 8 mcg/mL have sensitivities approaching 100% for the detection of CP P. aeruginosa and CP A. baumannii. Carbapenem susceptibility patterns and resistance mechanisms for nonfermenters are anticipated to change over time, and appropriate MIC cutoff values need to be reviewed periodically to remain accurate.
ACKNOWLEDGMENTS
Financial support: This work was supported by funding from The National Institutes of Health (grant no. K23-AI127935 awarded to P.D.T. and grant no. R21-AI130608 awarded to P.J.S.). Potential conflicts of interest: All authors report no conflicts of interest relevant to this article.



