Introduction
The genus Aporcella Andrássy, 2002 is a widely distributed dorylaimid (Dorylaimida) taxon, inhabiting soils of all zoogeographical realms/regions except Antarctica (Álvarez-Ortega et al., Reference Álvarez-Ortega, Subbotin and Peña-Santiago2013; Vazifeh et al., Reference Vazifeh, Niknam, Naghavi, Jabbari and Peña-Santiago2020). In Africa, it occurs in the southern part of the continent (Botswana and South Africa), where four species have so far been recorded: Aporcella adriaani (Botha & Heyns, Reference Botha and Heyns1990) Álvarez-Ortega, Subbotin & Peña-Santiago, 2013; Aporcella debruinae Álvarez-Ortega, Subbotin & Peña-Santiago, 2013; Aporcella parapapillata (Botha & Heyns, Reference Botha and Heyns1990) Andrássy, 2009; and Aporcella tropica (Jana & Baqri, 1981) Álvarez-Ortega, Subbotin & Peña-Santiago, 2013.
Since its original description by Andrássy (Reference Andrássy2002), the taxonomy of Aporcella has been subjected to a number of changes, including a new definition (Álvarez-Ortega et al., Reference Álvarez-Ortega, Subbotin and Peña-Santiago2013), the description of new species (Andrássy, Reference Andrássy2012; Álvarez-Ortega & Peña-Santiago, Reference Álvarez-Ortega and Peña-Santiago2016; Naghavi et al., Reference Naghavi, Niknam, Vazifeh and Peña-Santiago2019, Reference Naghavi, Niknam, Vazifeh and Peña-Santiago2020) and a recent update of its taxonomy (Vazifeh et al., Reference Vazifeh, Niknam, Naghavi, Jabbari and Peña-Santiago2020). Molecular analyses have repeatedly supported the monophyly of Aporcella. Surprisingly, the closer evolutionary relationship of Aporcella with non-aporcelaimid taxa (tylencholaims, discolaims) than with other aporcelaims has also been reported by various researchers (Álvarez-Ortega & Peña-Santiago, Reference Álvarez-Ortega and Peña-Santiago2016; Imran et al., Reference Imran, Abolafia, Peña-Santiago and Ahmad2019; Naghavi et al., Reference Naghavi, Niknam, Vazifeh and Peña-Santiago2019; Vazifeh et al., Reference Vazifeh, Niknam, Naghavi, Jabbari and Peña-Santiago2020).
From a 2016 survey of nematode fauna associated with watermelon in Nigeria, specimens of an Aporcella population that belong to an undescribed species were recovered. This would represent a novel geographical record for the genus, and, by means of its molecular analysis, offers the chance to confirm previous findings about its monophyly and phylogeny.
Materials and methods
Extraction and processing of nematodes
Nematode specimens were recovered from rhizosphere soil samples that were collected from watermelon fields in south-west Nigeria during 2016. Extraction of nematodes from soil followed a modified pie pan method (Coyne et al., Reference Coyne, Nicol and Claudius-Cole2007). Nematodes were fixed in a hot 4% formaldehyde solution for morphological observations (Nico et al., Reference Nico, Rapoport, Jiménez-Díaz and Castillo2002) and subsequently mounted in anhydrous glycerine as permanent slides (De Grisse, Reference De Grisse1963), while others were stored in dimethyl sulphoxide, disodium ethylenediamine tetra-acetic acid and saturated sodium chloride (abbreviated here as DESS) solution for molecular analysis. Morphological features of nematodes were observed, measured and photographed using an Eclipse 80i microscope (Nikon, Tokyo, Japan) equipped with differential interference contrast optics, a drawing tube (camera lucida) and a DS digital camera (Nikon, Tokyo, Japan).
Molecular identification
Fixed specimens in DESS were rinsed using double-distilled water, and two specimens were transferred to individual 1.5 ml tubes containing 15 μl nuclease-free water. DNA were extracted using chelex-100 by adding 20 μl of chelex (5% w/v) and 5 μl proteinase K to each tube, followed by incubation at 57°C for 120 min and at 95°C for 10 min (Rashidifard et al., Reference Rashidifard, Marais, Daneel, Mienie and Fourie2019).
DNA amplification was carried out on a Vacutec thermocycler (www.vacutec.co.za) using the primers: small subunit (SSU) F04 (5′-GCTTGTCTCAAAGATTAAGCC-3′), and SSU R26 (5′-CATTCTTGGCAAATGCTTTCG-3′) (Blaxter et al., Reference Blaxter, De Ley and Garey1998) for SSU ribosomal DNA (rDNA), as well as D2A (5′-ACAAGTACCGTGAGGGAAAGTTG-3′) and D3B (5′-TCGGAAGGAACCAGCTACTA-3′) (Subbotin et al., Reference Subbotin, Sturhan, Chizhov, Vovlas and Baldwin2006) for D2–D3 large subunit (LSU) rDNA. The polymerase chain reaction (PCR) amplification process was followed as reported by Rashidifard et al. (Reference Rashidifard, Bello, Fourie, Coyne and Peña-Santiago2020). Four microliters of PCR products were loaded on 1% agarose gel and visualized by an ultraviolet transilluminator to check the quality of DNA. The DNA was sequenced in both forward and reverse directions by the Inqaba Biotec™ genomic company (South Africa; www.inqaba-southafrica.co.za).
Phylogenetic analyses
The new SSU and LSU sequences obtained for the new species were compared to other available sequences in the GenBank using the BLASTN search tool. One data set was prepared for each of the SSU and LSU regions and taxa selection was carried out according to Rashidifard et al. (Reference Rashidifard, Bello, Fourie, Coyne and Peña-Santiago2020). The available sequences as well as outgroups in the SSU data set were aligned using the MUSCLE alignment tool (Edgar, Reference Edgar2004) in Geneious Prime® 2020.2.3 (www.geneious.com), while the LSU data set was aligned using the E-INS-I algorithm in the MAFF alignment tool (Katoh & Standley, Reference Katoh and Standley2013). According to the jModeTest 2.1.10 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012), the Hasegawa–Kishino–Yano with a gamma distribution (HKY + G) was the best substitution model for the SSU dataset. However, the General Time Reversible with proportion of invariable sites and a gamma distribution (GTR + I + G) was selected as the most appropriate model for the LSU dataset.
Bayesian inference was conducted in MrBayes v3.1.2 (Ronquist & Huelsenbeck, Reference Ronquist and Huelsenbeck2003) implemented in Geneious Prime® 2020.2.3 (www.geneious.com). The chain was running 3 × 106 generations for both the SSU and LSU data sets, and after discarding the 25% burn-in samples, the remaining samples were used for further analyses. The Markov chain Monte Carlo (MCMC) algorithm was implemented to calculate the posterior probabilities (PB) for each of the phylogenetic trees (Larget & Simon, Reference Larget and Simon1999) using the 50% majority rule. The mononchid taxa were used as outgroups for both the SSU and LSU phylogenies.
Results
Aporcella femina sp. n.
Material examined
Ten females in variable but generally good state of preservation (figs 1–3).
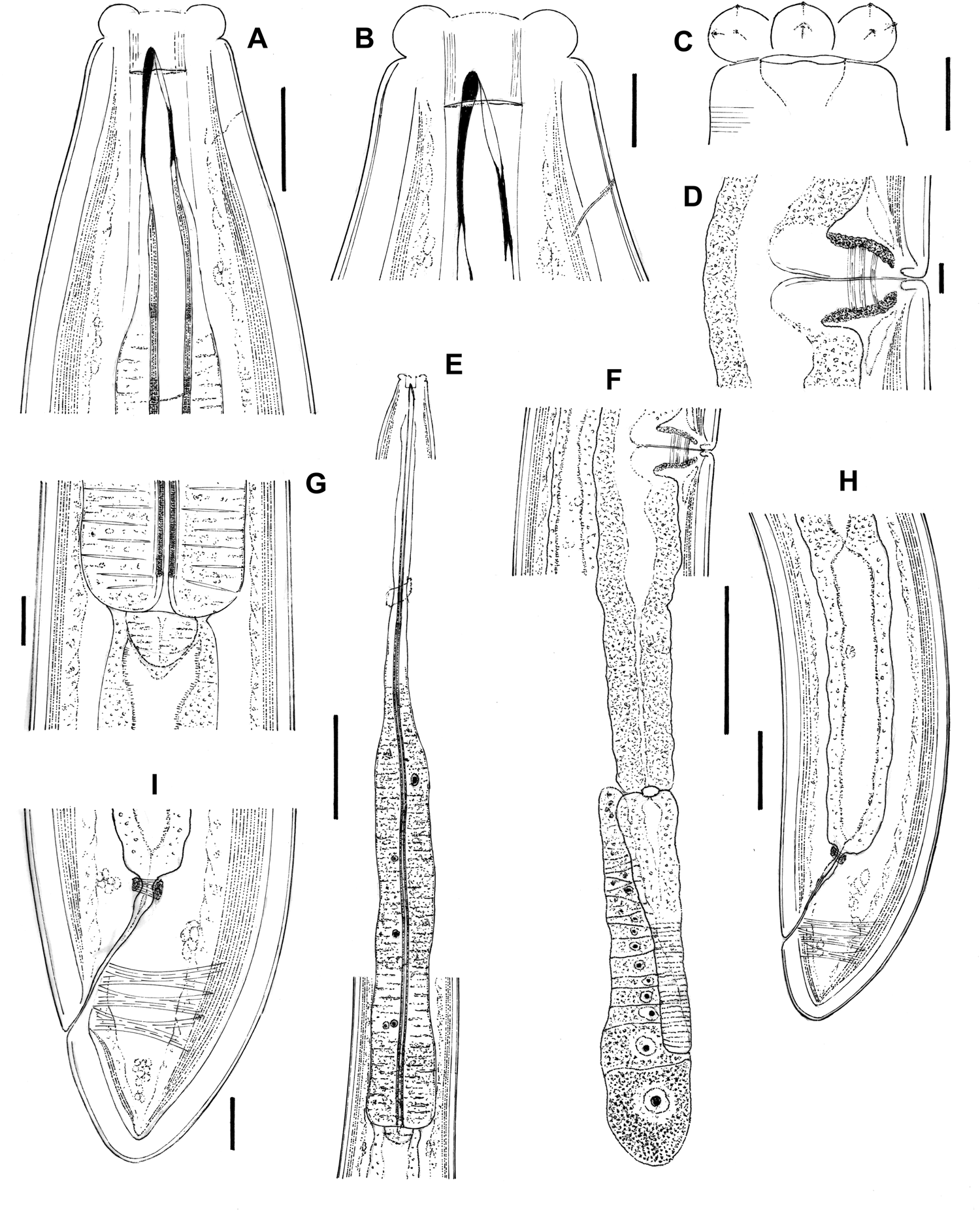
Fig. 1. Aporcella femina sp. n. (female): (a, b) anterior region in lateral median view; (c) anterior region in lateral surface view; (d) vagina region; (e) neck region; (f) posterior genital branch; (g) pharyngo–intestinal junction; (h) posterior body region; (i) caudal region. Scale bars: (a, g, i) 20 μm; (b–d) 10 μm; (e, f) 100 μm; (h) 50 μm.

Fig. 2. Aporcella femina sp. n. (female: light microscopy): (a–c) anterior region in lateral median view; (d) cuticle; (e) anterior region in lateral surface view; (f) vagina region; (g) posterior body region; (h) pharyngo–intestinal junction; (i) neck region; (j) posterior genital branch; (k) caudal region. Scale bars: (a, h, k) 20 μm; (b–f) 10 μm; (g) 50 μm; (i, j) 100 μm.
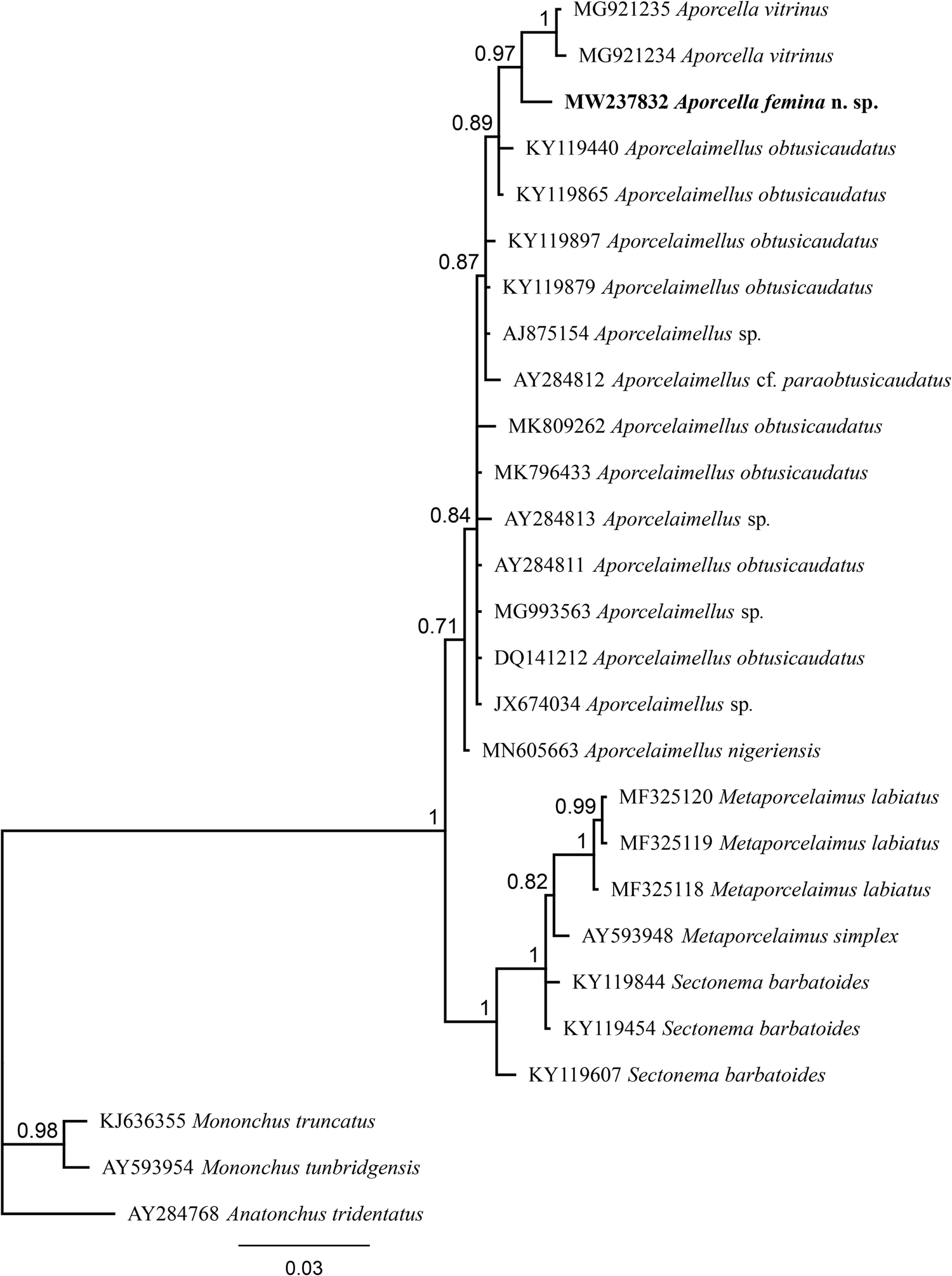
Fig. 3. Bayesian tree with 50% majority rule of Aporcella femina n. sp. from Nigeria using partial SSU rDNA sequences under the HKY + G model. The sequence of new species is indicated by bold font. Scale bar shows the number of substitution per site.
Measurements
All measurements are presented in table 1.
Table 1. Morphometrics of Aporcella femina sp. n. from Nigeria and two other African species of the genus.
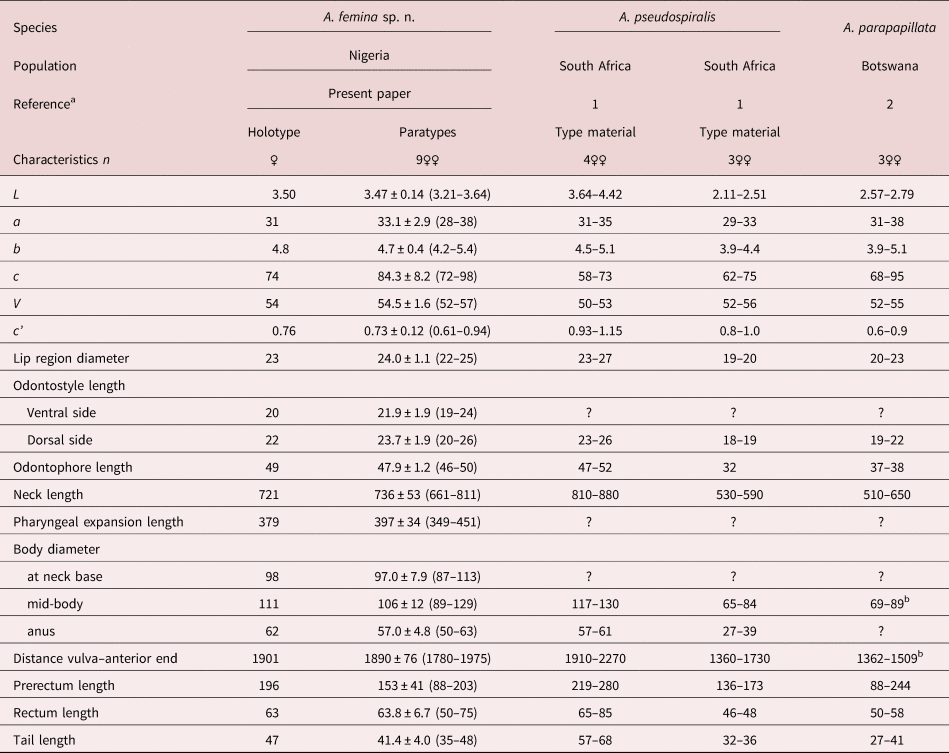
Measurements in μm except L in mm, and in the form average ± standard deviation (range).
a 1: Botha & Heyns (Reference Botha and Heyns1990); 2: De Bruin & Heyns (Reference De Bruin and Heyns1992).
b Calculated from other morphometrics.
Description
Female. Moderately to slender (a = 28–38) nematodes of large size, 3.21–3.64 mm long. Body cylindrical, tapering towards both ends but substantially towards the anterior region, while the tail is short and rounded. Upon fixation, habitus regularly curved ventrad, C- to G-shaped. Cuticle two-layered, very thick throughout the entire body, 4–7 μm in anterior region, 7.5–9.5 μm at mid-body and 7.5–11.5 μm on tail, consisting of a thin outer layer with nearly smooth surface, and a thicker inner layer that displays a fine but distinct transverse striation (fig. 2g). Lateral chords narrow, 6–17 μm wide or 6–19% of mid-body diameter. Ventral and dorsal pores restricted to cervical region, two or three in number at each side at odontostyle–odontophore level; lateral pores small. Lip region offset by a distinct constriction, 3.3–3.8 times wider than high and less than one-third (22–29%) of body diameter at neck base; lips separate, with moderately protruding papillae. Amphidial fovea stirrup-shaped, its opening 9–10.5 μm, less than one-half (38–46%) of lip region diameter. Cheilostom broad, 10.5–14 μm long, with thick walls. Odontostyle strong, 3.8–4.3 times longer than wide, barely shorter (0.9–1.0 times) than lip region diameter and 0.58–0.73% of total body length, with aperture 13–17 μm long or c. two-thirds (58–76%) of total length. Guiding ring simple but distinct, plicate. Odontophore linear, rod-like, 1.9–2.4 times longer than odontostyle. Pharynx entirely muscular, enlarging very gradually, with its basal expansion 5.8–9.0 times as long as wide, 3.1–4.5 times the body diameter at neck base and occupying 52–56% of total neck length; pharyngeal gland nuclei located as follows (n = 1): DO = 47–52, DN = 48–56, DO-DN = 3–5, S1N1 = 61–66, S1N2 = 73–75, S2N = 85–88. Cardia short and rounded conoid, 21–32 × 20–34 μm, surrounded by intestinal tissue that forms a short extension projection into the intestinal lumen. Genital system didelphic–amphidelphic, with well-developed genital branches, the anterior 346–565 μm or 11–15% of body length, the posterior 381–638 μm or 12–18% of body length. Ovaries comparatively small, mostly not reaching the oviduct–uterus junction, the anterior 103–259 μm and posterior 106–231 μm long. Anterior oviduct 127–200 μm or 1.4–1.8 body diameters long, posterior oviduct 133–203 μm or 1.4–1.9 body diameters long, both consisting of a slender distal portion made of prismatic cells and a distinct pars dilatata with perceptible lumen. Oviduct and uterus separated by a distinct sphincter. Anterior uterus 191–325 μm or 1.9–3.1 body diameters long, posterior uterus 204–350 μm or 2.1–3.3 body diameters long, often appearing as a simple, tube-like structure, but with some (well-preserved) specimens having a longer uterus, which enables the appreciation of a wider proximal uterine region and a more slender distal region, both of about similar length. No uterine egg observed. Vagina extending inwards 40–54 μm, occupying c. one-half (41–45%) of body diameter: pars proximalis 28–41 × 19–28 μm, with distally convergent walls and surrounded by a relatively weak musculature; pars distalis well-developed, 10–14 μm long; pars refringens absent. Vulva a slightly posterior transverse slit. Prerectum 2.4–4.0, rectum 0.8–1.5 times the anal body diameter long. Tail short, convex conoid, its ventral side weakly straighter than the dorsal side; caudal pores two pairs, close together, at the posterior half of tail, one lateral, another sublateral.
Male. Unknown. Females do not contain sperm cells.
Type locality and habitat
South-west Nigeria, Odeda (7°10′59.999″N, 3°26′42.01″E), an agrarian locality with a history of cassava (Manihot esculenta Crantz) and vegetable production; specimens were collected from a field cultivated with watermelon (Citrullus lanatus) on sandy-loam soil (sand = 66%, silt = 13%, clay = 21%, organic matter = 11.18%, pH 6.22).
Molecular characterization
After sequencing and editing, two totally (100%) identical sequences per each locus (SSU and LSU) were obtained; therefore, only one sequence was used for analysis and deposited into the NCBI GenBank: a 745 bp-long LSU sequence (accession number MW237838) and another 855 bp-long SSU sequence (accession number MW237832). The evolutionary relationships of the new species are presented in figs 3 and 4.
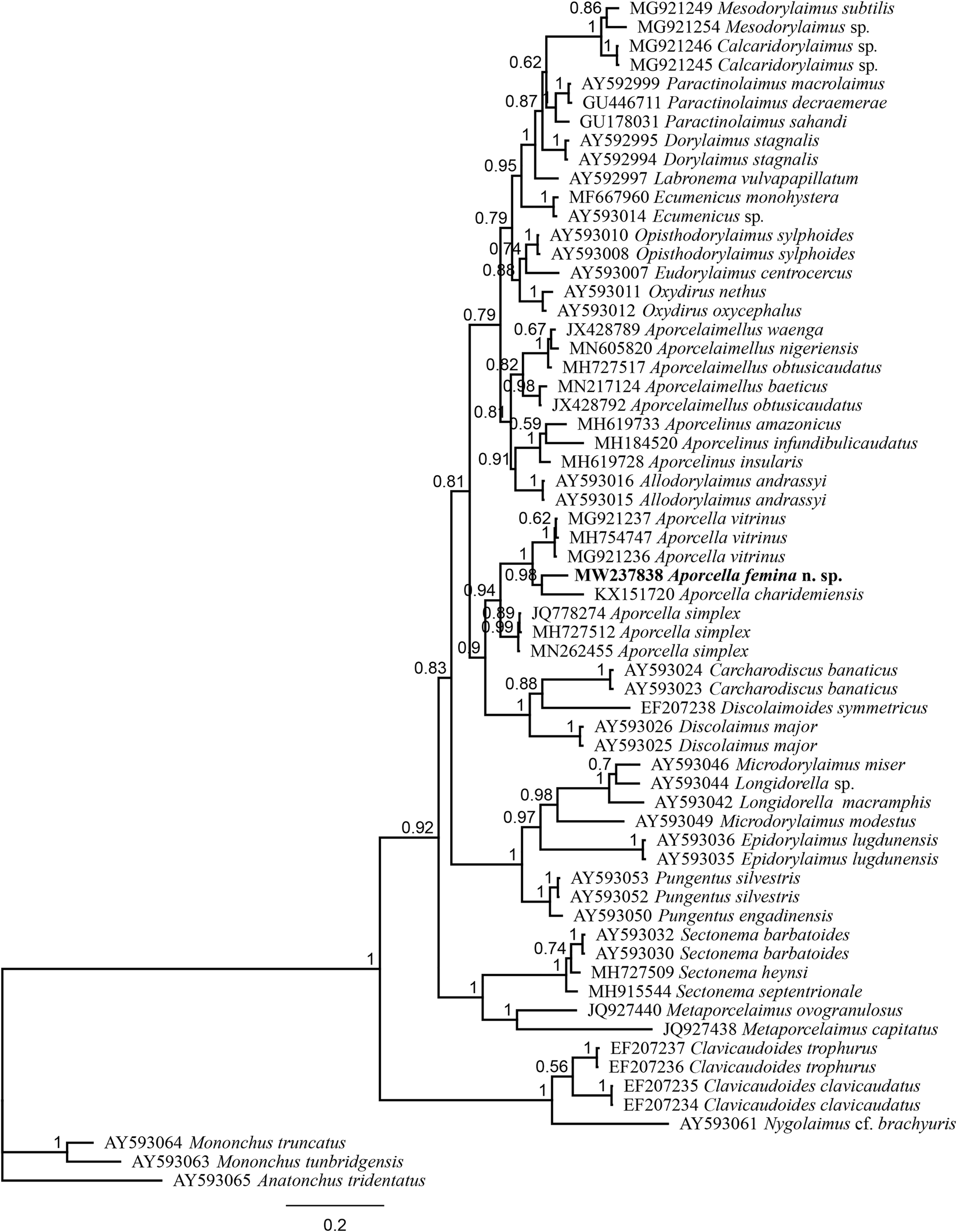
Fig. 4. Bayesian tree with 50% majority rule of Aporcella femina n. sp. from Nigeria using partial LSU (D2–D3) rDNA sequences under the GTR + I + G model. The sequence of new species is indicated by bold font. Scale bar shows the number of substitutions per site.
Type material
Female holotype and nine female paratypes, deposited in the nematode collection of the University of Jaén, Spain.
Etymology
The specific epithet is a Latin term meaning woman or female, and refers to the existence of only females in this species.
Diagnosis and relationships
The new species is characterized by its 3.21–3.64 mm-long body, inner cuticle layer with fine but distinct transverse striation, lip region offset by deep constriction and 22–25 μm broad, odontostyle 20–26 μm at its dorsal side and 19–24 μm at its ventral side, neck 661–811 μm long, pharyngeal expansion 349–451 μm long or 52–56% of total neck length, female genital system didelphic–amphidelphic, uterus 191–350 μm or 1.9–3.3 body diameters long, V = 52–57, tail short and convex conoid (35–48 μm, c = 72–98, c′ = 0.7–0.9) and males absent.
In having a comparatively long body (more than 3.0 mm long), Aporcella femina sp. n. resembles Aporcella magna Andrássy, 2012 and Aporcella pseudospiralis (Botha & Heyns, Reference Botha and Heyns1990) Andrássy, 2012, and significantly differs from its other congeners (body 3.21–3.64 vs up to 3.08 mm long) (see compendium by Álvarez-Ortega et al., Reference Álvarez-Ortega, Subbotin and Peña-Santiago2013 and Vazifeh et al., Reference Vazifeh, Niknam, Naghavi, Jabbari and Peña-Santiago2020). It can be distinguished from A. magna, a species occurring in Chile, in its smaller general size: body 3.21–3.64 versus 4.48–5.00 mm long, n = 5; neck 661–811 versus 888–1020 μm long; tail 35–48 versus 50–60 μm. It is also smaller than A. pseudospiralis, which is only known to occur in South Africa, with a smaller general size (body 3.21–3.64 vs 3.64–4.42 mm long, n = 4; neck 661–811 vs 810–880 μm long), shorter prerectum (122–203 vs 219–280 μm) and female tail (35–48 vs 57–68 μm, c = 72–98 vs 58–73, c′ = 0.61–0.94 vs 0.93–1.15) and male absent (vs present).
Among the small-sized Aporcella species (body up to 3.08 mm long), and due to the comparatively long odontostyle (>16 μm), short (c′ up to 1.1) and convex conoid tail, it is similar to Aporcella papillata (Bastian, 1865) Álvarez-Ortega, Subbotin & Peña-Santiago, 2013 and A. parapapillata (Botha & Heyns, Reference Botha and Heyns1990) Andrássy, 2009. The new species differs from A. papillata, currently recorded from UK only (Thorne & Swanger, Reference Thorne and Swanger1936; Álvarez-Ortega & Peña-Santiago, Reference Álvarez-Ortega and Peña-Santiago2010), in its larger general size (body 3.21–3.64 vs 2.43–3.08 mm long; neck 661–811 vs 510–650 μm long), longer uterus (191–350 vs 111–131 μm or 1.9–3.3 vs 1.3–1.5 body diameters long), more posterior vulva (V = 52–57 vs V = 45–50, n = 11) and male absent (vs present); and from A. parapapillata, recorded from South Africa (Botha & Heyns, Reference Botha and Heyns1990; De Bruin & Heyns, Reference De Bruin and Heyns1992), in its larger general size (body 3.21–3.64 vs 2.11–2.79 mm long; neck 661–811 vs 555–649 μm long), much longer odontophore (44–50 vs 26–38 μm, n = 12) and male absent (vs as frequent as female).
The new species described herein displays intermediate morphometrics between two South African species – namely, A. parapapillata and A. pseudospiralis – all of them described based on relatively few specimens. Aporcella femina sp. n. is very similar to A. pseudospiralis, but, as mentioned above, they differ in their overall size and especially in tail length. Therefore, the possibility of co-specificity could be raised, and the noted differences possibly due to geographical variations. The absence of molecular data for A. parapapillata and A. pseudospiralis currently prevents any molecular comparison, which, when available, would clarify any ambiguity. However, the available morphological information clearly distinguishes the three species from each other, supporting a separate status for them.
The evolutionary relations of the new species, as derived from the molecular analyses based on the partial sequence of SSU and LSU rDNA, are represented in figs 3 and 4. According to the SSU phylogeny, the new species is in a highly supported (BPP: 97%) sister relation with Aporcella vitrinus. Furthermore, Bayesian tree using partial LSU sequence shows that the new species forms part of a fully supported (BPP: 100%) subclade with Aporcella charidemiensis and A. vitrinus, the former being the closest relative, which is recorded from Spain. A highly supported (BPP: 94%) Aporcella clade is made of this subclade along with three Aporcella sequences belonging to A. simplex. Therefore, the present results confirm previous findings (for instance, see Naghavi et al., Reference Naghavi, Niknam, Vazifeh and Peña-Santiago2019; Vazifeh et al., Reference Vazifeh, Niknam, Naghavi, Jabbari and Peña-Santiago2020) about both the monophyly of Aporcella, and the belonging of Aporcella sequences to a large clade morphologically characterized by the absence of pars refringens vaginae, which also includes discolaims in the tree provided herein.
Financial support
One author (R.P.S.) is grateful for the financial support received from the scientific project (ref.: Action 1-PAIUJA 2019-2020: EIRNM02-2017), University of Jaén. Infrastructure and partial funding by the Nematology Unit at North-West University of South Africa is also acknowledged.
Conflicts of interest
None.
Author contributions
M.R. and T.T.B. contributed equally.







