Introduction
The acanthocephalan genus Corynosoma includes 20 species from the gastrointestinal tract of pinnipeds (Laskowski & Zdzitowiecki Reference Laskowski, Zdzitowiecki, Klimpel, Kuhn and Mehlhorn2017), thirteen of which parasitize seals and fur seals (Hernández-Orts et al. Reference Hernández-Orts, Smales, Pinacho-Pinacho, García-Varela and Presswell2017; Laskowski & Zdzitowiecki Reference Laskowski, Zdzitowiecki, Klimpel, Kuhn and Mehlhorn2017). Some of these species have been described in Antarctic and sub-Antarctic pinnipeds, e.g., C. hamanni (Linstow, Reference Linstow1892), C. bullosum (Linstow, Reference Linstow1892), C. australe Johnston, Reference Johnston1937 , C. arctocephali Zdzitowiecki, Reference Zdzitowiecki1984b , C. hannae Zdzitowiecki, 1984, C. pseudohamanni Zdzitowiecki, 1984, C. evae Zdzitowiecki, 1984, and C. gibsoni Zdzitowiecki, 1986 (Table 1). In particular, the southern elephant seal, Mirounga leonina, has been reported as the only definitive host for C. bullosum. A questionable record suggests that the northern elephant seal, M. angustrirostris, may be another definitive host (Laskowski & Zdzitowiecki Reference Laskowski, Zdzitowiecki, Klimpel, Kuhn and Mehlhorn2017). In addition, larval stages of C. bullosum have been found in the sperm whale, probably as a non-definitive host (Laskowski & Zdzitowiecki Reference Laskowski, Zdzitowiecki, Klimpel, Kuhn and Mehlhorn2017). Larval specimens have also been found in the intestine of seabirds, such as the cormorant and the gentoo penguin (Laskowski & Zdzitowiecki Reference Laskowski, Zdzitowiecki, Klimpel, Kuhn and Mehlhorn2017).
Table 1. Different Corynosoma species described in Antarctic and sub-Antarctic pinnipeds

* Non definitive host; (?) Questionable report.
The acanthocephalan C. bullosum was first described by Linstow in 1892 (Linstow Reference Linstow1892), based on observations from elephant seals in South Georgia. Several later findings and reports are mentioned below. The different available descriptions for the species present several discrepancies in measurements, principally in total body length, the number of rows of hooks, and the number of hooks per row (Johnston & Edmonds Reference Johnston and Edmonds1953; Edmonds Reference Edmonds1954; Zdzitowiecki Reference Zdzitowiecki1986a; Laskowski & Zdzitowiecki Reference Laskowski, Zdzitowiecki, Klimpel, Kuhn and Mehlhorn2017). The report by Johnston and Edmonds (Reference Johnston and Edmonds1953) briefly described the species based on a single female specimen found in the intestine of an elephant seal on the Campbell and Auckland Islands. From an Antarctic expedition in 1949, Edmonds (Reference Edmonds1954) obtained several acanthocephalans in the Heard Islands that allowed him to provide several measurements. Later, in a morphological description carried out by Zdzitowiecki (Reference Zdzitowiecki1986a), measurements of 20 females and 20 males were taken, which were recovered from the intestine of three elephant seals from the South Shetland Islands.
At present, the contribution of Laskowski and Zdzitowiecki (Reference Laskowski, Zdzitowiecki, Klimpel, Kuhn and Mehlhorn2017) constitutes the most complete study of the morphology of Corynosoma species from Antarctica and the sub-Antarctic region. They also provide a list of definitive and non-definitive hosts, as well as paratenic and intermediate hosts. The phylogenetic analysis of the genera Corynosoma based on internal transcribed spacer (ITS) and 5.8s rRNA sequences supported the monophyly of the family and some of their genera (García-Varela et al. Reference García-Varela, Pérez-Ponce de León, Aznar and Nadler2013). However, considering the discrepancies in the morphological descriptions and the absence of a correlation between molecular and morphological data, the present report provides a morphological and molecular update of C. bullosum.
Materials and methods
Specimen collection
During the reproductive seasons of 2016/2017 and 2017/2018, 30 specimens of acanthocephalans were collected from faecal samples from 41 southern elephant seals, Mirounga leonina Linnaeus, 1758 (Pinnipedia, Phocidae). The fieldwork was conducted in the Antarctic Specially Protected Area ASPA N°132, “Península Potter”, near the Argentinean Scientific Base “Dr. Alejandro Carlini”, 25 de Mayo/King George Island (62°14’S 51 58°40’W) (Figure 1). Faecal samples were obtained directly from the ground, very close to the animals. To avoid contamination, we collected only the top of the faeces. Animals showed no signs of illness at the time of collection, and all faeces samples were fresh. The samples were frozen at -20 °C and then examined under a stereomicroscope. Parasites were collected, placed in distilled water, and repeatedly washed; subsequently, they were individually preserved in 96% ethanol. We also checked faecal samples from eight Weddell seals (Leptonychotes weddellii) and one crabeater seal (Lobodon carcinophaga), but we did not find any acanthocephalan.
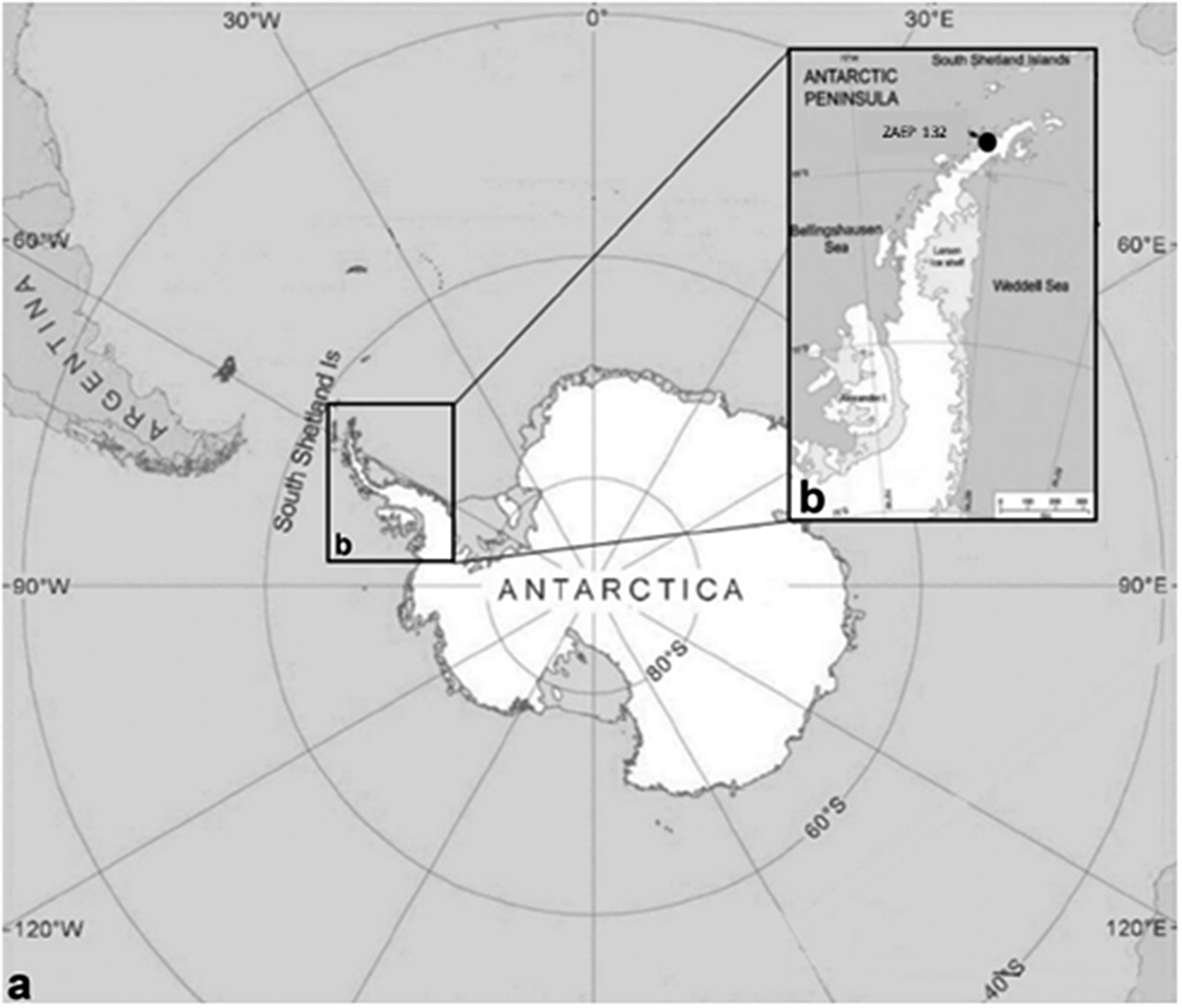
Figure 1. a. General view of Antarctica where specimens of the acanthocephalan Corynosoma bullosum were collected from faeces of the southern elephant seal; b. Antarctic Specially Protected Area (ASPA) N° 132, in “Península Potter”, South Shetland Islands, Antarctica.
Morphological study
Some selected specimens (10 females and 10 males in good condition) were cleared in lactic acid and studied under a light microscope (Leica DM 2500) for morphological description. Measurements were taken using the Leica Application Suite microscope software. For scanning electron microscopy (SEM), a female and a male were gradually dehydrated in an ethanol series, immersed in hexamethyldisilazane for 15 min, and air-dried for 10 min. Photomicrographs of a female and a male were obtained with a Jeol JSM-6460LV SEM operating at 15 kV (Figure 2). All the measurements are in millimetres unless otherwise stated (Table 2). The number of hooks per row was determined in all the specimens by counting in four neighbouring rows, following Zdzitowiecki (Reference Zdzitowiecki1986a). Vouchers were deposited at the Parasitological Collection of the Instituto de Biología de Organismos Marinos, Puerto Madryn, Chubut province, Argentina (CNP-Par 197 and CNP-Par 198).
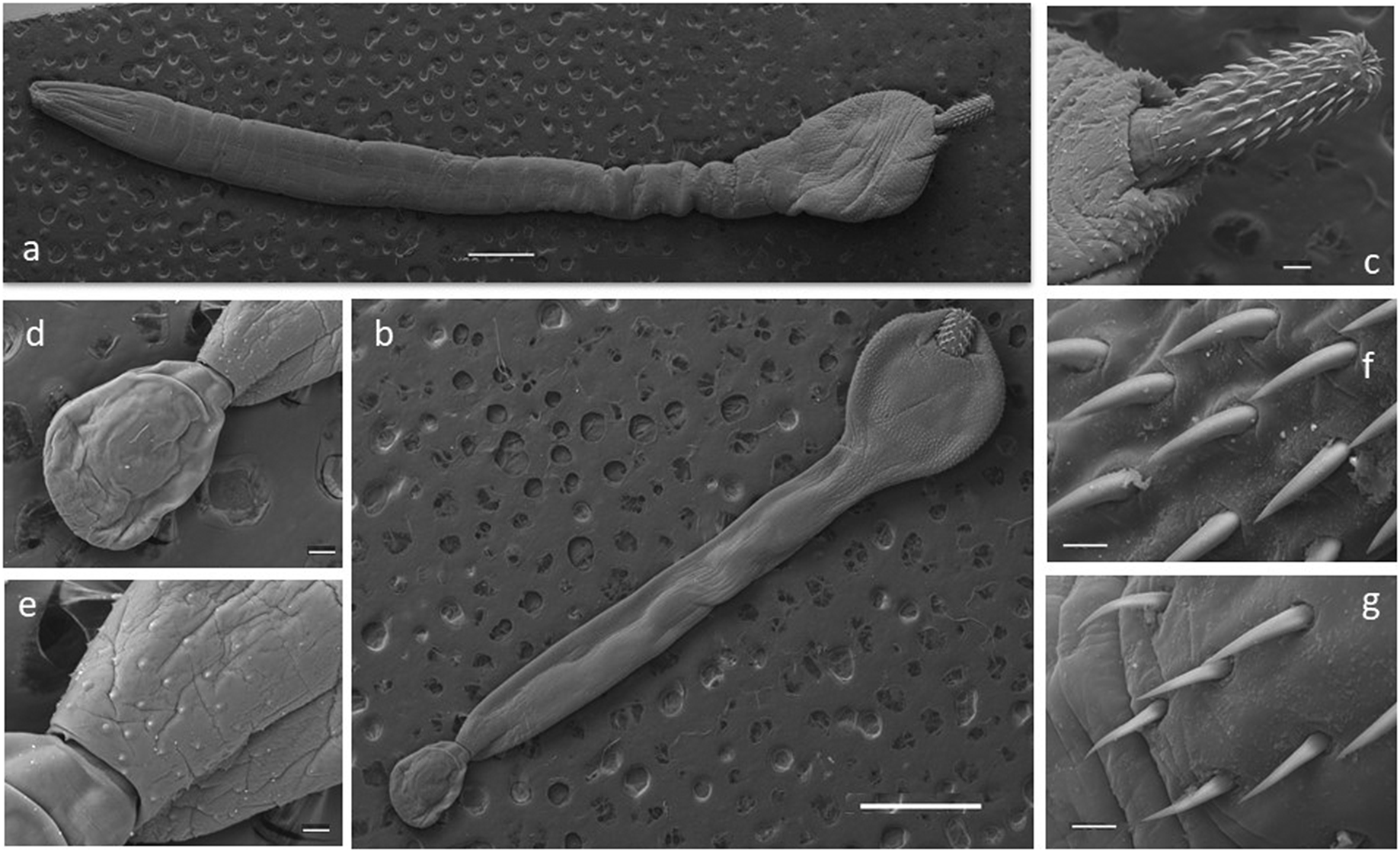
Figure 2. Scanning electron microscopy (SEM) photographs of specimens of the acanthocephalan Corynosoma bullosum from the southern elephant seal. a. adult female; b. adult male; c. neck and proboscis detail; d. everted copulatory bursa; e. genital spines surrounding the genital pore; f. apical hooks; g. basal hooks. Scale bars: a–b 1mm; c, d, e 100μm; f–g 20μm.
Table 2. Comparative measurements of Corynosoma bullosum from southern elephant seal; measures in millimetres, mean and range in parentheses

* The author did not differ by sex.
Molecular study
Genetic comparison and phylogenetic analyses were based on a fragment of 18S rDNA sequences. Sequences of two individuals of Coryonsoma bullosum from the southern elephant seal, Mirounga leonina, from Península Potter, Antarctic Peninsula, were analysed. The samples were digested overnight at 55 °C, and genomic DNA from individual worms was isolated using Qiagen DNeasy tissue kit (Qiagen Inc., Valencia, California, USA) according to the manufacturer’s instructions. Polymerase chain reaction (PCR) was performed in 30 μL volumes containing 2 x red PCR premix (Ampliqon, Odense, Denmark), 20 pmol of each primer, and 3 μL of extracted DNA. A 222 bp fragment of the partial 18S was amplified using the primers MGF (5′-GATCGGGGAGGTAGTGACG-3′) and MGR (5′-ACCCACCGAATCAAGAAAGAG-3′). PCR conditions for the 18S rDNA gene were amplified following the protocol of Rodríguez et al. (Reference Rodríguez, Amin, Heckmann, Sharifdini and D’Elía2022). PCR products were analysed on 1.5% agarose gel and visualized with a UV transilluminator. Later, the PCR products were sequenced in both directions using the same PCR primers with ABI 3130 sequencer. Amplicons were sequenced using an external sequencing service (Macrogen, Inc., Seoul, South Korea). Finally, all DNA sequences were edited using CodonCode (CodonCode Aligner, Dedham, Massachusetts) and deposited in GenBank (OQ192986-OQ192987).
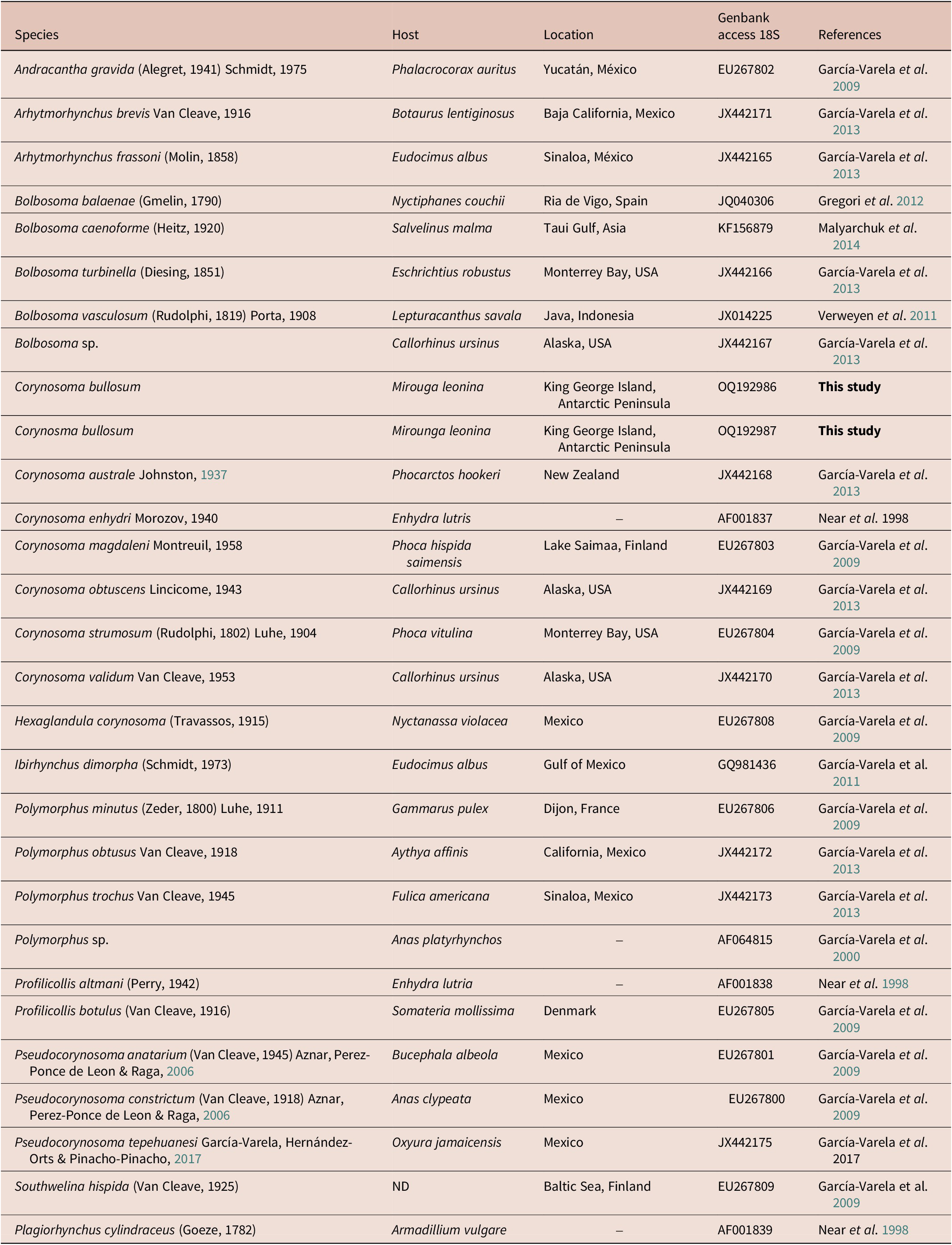
The new partial 18S rDNA gene was integrated into a matrix with one representative of each genus of the family Polymorphidae. A total of 26 belonging to the family Polymorphidae was downloaded from GenBank (Table 4) and analysed together with the two new sequences of C. bullosum. Last, one sequence of Plagiorhynchus cilindraceus (Plagiorhynchidae family) was used as an outgroup (Table 4). The final analysed matrix had a total of 29 sequences.
Sequences were aligned using MAFFT v.7 software (Katoh & Standley Reference Katoh and Standley2013), allowing the program to choose the alignment strategy (L-ins-i). To select the best-fitting model of molecular evolution, we used the proposed model tool in the program IQ-Tree v1.6.12 (Kalyaanamoorthy et al. Reference Kalyaanamoorthy, Minh, Wong, von Haeseler and Jermiin2017), which selected TVMe+I+G4 for 18S. We used a maximum-likelihood approach to obtain the best tree using the program IQ-Tree v1.6.12 (Trifinopoulus et al. 2016). We carried out 15 independent runs to explore the tree space, changing the value of the strength of the perturbation (-pers) parameter. For it, we conducted the following analyses: Five runs modifying the strength of the perturbation parameter from 0.3 (default value) to 0.5; five runs using the default value (0.5) for the strength of the perturbation parameter; five runs modifying the strength of the perturbation parameter from 0.5 (default value) to 0.7. In all cases, the number of unsuccessful interactions to the stop parameter (-nstop) value was changed from 100 (default value) to 1000. The tree with the highest likelihood score was chosen (18S = log-likelihood: -6366.004, -pers 0.7, and -nstop 1000). Support for the nodes was evaluated using two approaches: the aBayes test (Anisimova et al. Reference Anisimova, Gil, Dufayard, Dessimoz and Gascuel2011) and the ultrafast bootstrap procedure using 1000 replicates (Hoang et al. Reference Hoang, Chernomor, von Haeseler, Minh and Vinh2018). Finally, observed genetic p-distances (p) between haplotype and sample pairs were calculated in MEGA 7 (Tamura et al. Reference Tamura, Stecher, Peterson, Filipski and Kumar2013).
Results
In Table 2 we summarise the comparative measurements of females and males of the acanthocephalan Corynosoma bullosum from the southern elephant seal, reported by different authors and observed in the present data.
Morphological data of Corynosoma bullosum (Linstow, Reference Linstow1892)
The presence of 16–18 rows of hooks with 11–15 hooks each (8–12 anterior and 3–4 rootless basal hooks) are the diagnosis characteristics for C. bullosum. Specimens are white, great in size, and with evident sexual dimorphism. Females are larger than males. The hind-trunk is cylindrical and longer than the fore-trunk. The trunk expands anteriorly forming a disc; spines spread ventrally. Genital spines surround the genital pore in both sexes. Lemnisci are flat and equal in size, shorter than the proboscis receptacle. Hooks’ measurements are given in Table 3. In females, the fore-trunk constitutes 20–30% of the trunk length. The somatic armature covers 30–40% of the trunk length on the ventral side. Genital pore is in terminal position. In males, the fore-trunk constitutes 25–35% of the trunk length. The somatic armature covers 35–50% of the trunk length on the ventral side.
Table 3. Dimensions, mean (min–max), of C. bullosum proboscis hooks, based on four neighbouring rows of hooks
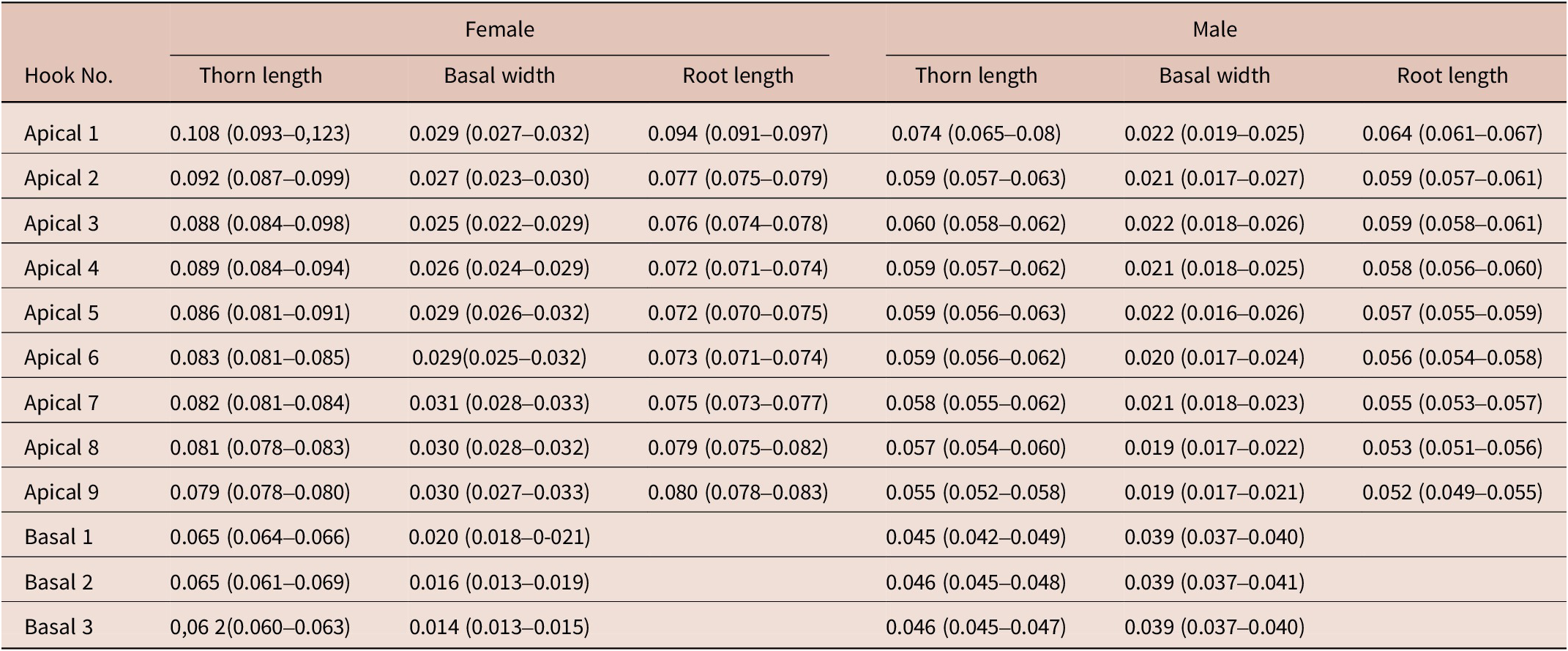
Table 4. Species of acanthocephalans, their hosts, locations, and GenBank accession number of the sequences used in the phylogenetic analysis. (–) = Not indicated
Molecular results
Molecular analyses showed both phylogenetic trees gathered via maximum likelihood (ML) and Bayesian inference (BI) were mostly congruent (Figure 3). Sequences obtained from adults recovered from Southern elephant seals collected from the Antarctic Peninsula appear as sister relationships to C. australe, which was weakly supported in the BI analysis and moderately supported ML analysis (PP(Bayesian probability) = 0.36; BS(Bootstrap proportions)= 72). Haplotypes of C. bullosum show low genetic variation (average = 0.9%). The clade formed by C. bullosum and C. australe is sister to C. enhydri (PP = 1; BS = 98), and these clades differ on average by 0.9%. Meanwhile, the genus Bolbosoma is sister to this clade (PP = 1; BS = 97) and also appears as sister to the clade formed by all other species of Corynosoma in a clade well-supported only by the BI analysis (PP = 1; BS = 69; Figure 3). The average genetic p-distance between the clades of C. bullosum + C. australe, Bolbosoma, and another Corynosoma clade was 2.2% and 2.7%, respectively. This last clade is sister to the clade formed by the species Arhythmorhynchus brevis, Southwellina hispida, Hexaglandula corynosoma, and Ibirhynchus dimorpha, and it falls in a strongly supported clade only by the BI analysis and moderately supported by ML analysis (PP = 0.99; BS = 68). The average genetic p-distance between these clades was 5.8%. This latter clade is sister (PP = 1; BS = 100) to the clade formed by the genera Polymorphus and Pseudocorynosoma (PP = 1; BS = 84), who in turn is sister to Arhythmorhynchus frassoni species (PP = 0.97; BS = 73). The average genetic p-distance between the clade formed by C. bullosum + C. australe and Polymorphus trochus and Pseudocorynosoma and Arhythmorhynchus frassoni species was 7.9% and 9%, respectively. The other lineage is formed by Profilicollis and Polymorphus, which appears strongly supported (PP = 0.99; BS = 98; Figure 3). The average genetic p-distance between this later clade and the clade formed by C. bullosum + C. australe was 9.5%.

Figure 3. Phylogenetic relationships of haplotypes based on partial 18S rDNA gene sequences of specimens of the Polymorphidae family recovered in a Bayesian inference analysis. Numbers next to nodes refer to support values. Bayesian posterior probability values are shown left of the diagonal, and bootstrap proportions gathered in the maximum likelihood analysis (Ln = -6366.004) are shown right of the diagonal. GenBank accession numbers are included in the terminal labels.
Discussion
Our work provides a redescription of Corynosoma bullosum, including a comparison with existing measures and, for the first time, scanning electron microscope photographs and an assessment of its phylogenetic position based on 18S rDNA sequences. We considered that Zdzitowiecki (Zdzitowiecki Reference Zdzitowiecki1986a; Laskowski & Zdzitowiecki Reference Laskowski, Zdzitowiecki, Klimpel, Kuhn and Mehlhorn2017) provided valuable data on the morphology of C. bullosum, but that information is partial. On the other hand, molecular data presented by García-Varela and collaborators (Reference García-Varela, Aznar, Pérez-Ponce de León, Piñero and Laclette2005) include ITS and 5.8S rRNA sequences, but unfortunately, they are not comparable to previous assays. The use of 18S rDNA allowed us to assess the phylogenetic position of C. bullosum with other species of Corynosoma, allowing at the same time correlation of morphological with molecular data.
The type host for C. bullosum is the southern elephant seal, Mirounga leonina. However, it has been reported on other marine species of birds and mammals, which are considered non-definitive hosts (Edmonds Reference Edmonds1954; Zdzitowiecki Reference Zdzitowiecki1986b; Laskowski & Zdzitowieck Reference Laskowski, Zdzitowiecki, Klimpel, Kuhn and Mehlhorn2017). Besides C. bullosum, two other species of Corynosoma, C. australe (Sardella et al. Reference Sardella, Mattiucci, Timi, Bastida, Rodríguez and Nascetti2005; Hernández-Orts et al. Reference Hernández-Orts, Viola, García, Crespo, González, García-Varela and Kuchta2015) and C. cetaceum Johnston & Best, Reference Johnston and Best1942 (Silveira et al. Reference Silveira, Robaldo, Pinto-Colares, Bianchini, Muelbert, Martínez and Valente2014), have been found in southern elephant seals. However, we did not find any specimen of the latter species in our samples. Similarly, although it has been recorded mainly in Antarctic and sub-Antarctic regions, there are reports of immature specimens of C. bullosum in the sperm whale, in the north of Argentinean Patagonia (Zdzitowiecki Reference Zdzitowiecki1986a; Laskowski & Zdzitowiecki Reference Laskowski, Zdzitowiecki, Klimpel, Kuhn and Mehlhorn2017). In a pilot study with southern elephant seals in Patagonia, we analysed some samples with negative results for species of Corynosoma (Soto, unpublished data).
Available descriptions of C. bullosum differ among them, presenting variability in some measures. The first description by Linstow (Reference Linstow1892) mentions 25 rows of hooks with eight hooks per row. Later descriptions, including this study, report a mean of 16 rows of hooks with 12 hooks per row. However, the total body dimensions of females and males described by Linstow fall within the range reported by later authors (Edmonds Reference Edmonds1954; Zdzitowiecki Reference Zdzitowiecki1986a; present study). In the case of Edmonds (Reference Edmonds1954), the author provides a more detailed list of measures; however, for most of these measurements, the author did not differentiate between sexes. Finally, the description of Zdzitowiecki (Reference Zdzitowiecki1986a) is the most comprehensive and provided comparable information.
Several authors have discussed the difficulties in the morphological characterization of Corynosma spp. (Zdzitowiecki Reference Zdzitowiecki1984b; Stryukov Reference Stryukov2004; Sardella et al. Reference Sardella, Mattiucci, Timi, Bastida, Rodríguez and Nascetti2005; Hernández-Orts et al. Reference Hernández-Orts, Smales, Pinacho-Pinacho, García-Varela and Presswell2017, Reference Hernández-Orts, Montero, García, Crespo, Raga, García-Varela and Aznar2019). In this sense, Steinauer et al. (Reference Steinauer, Nickol and Ortí2007) argue that intraspecific variability is probably related to cryptic speciation or environmentally induced plasticity. For instance, C. australe presented intraspecific variation among different host species or even in the same host species (Hernandez-Orts et al. Reference Hernandez-Orts, Brandão, Georgieva, Raga, Crespo, Luque and Aznar2017). Our results seem to follow the same pattern. We considered the morphological differences between our samples and those from Linstow (Reference Linstow1892) as evidence of the intrinsic intraspecific variability of Corynosoma species.
Since our molecular and morphological data confirmed the identity of C. bullosum, it also opens a question regarding the taxonomic status of some acanthocephalan species. On the one hand, our molecular results corroborate that despite morphological variability between our samples, and with other Corynosoma species, C. bullosum showed low genetic variation. This low intraspecific genetic diversity could be explained by considering host behaviour. Acanthocephalan infects marine mammals and seabirds. These species usually exhibit a high vagility. Vagility contributes to the dispersal of parasites’ eggs, resulting in a genetic homogenisation of the population and, consequently, low genetic diversity (Goulding & Cohen Reference Goulding and Cohen2014; Rodríguez et al. Reference Rodríguez, D’Elía and Valdivia2017a; Reference Rodríguez, Diaz and D’Elía2017b; Lorenti et al. Reference Lorenti, Rodríguez, Cremonte, D’Elía and Diaz2018; Presswell et al. Reference Presswell, García-Varela and Smales2018; Santoro et al. Reference Santoro, Palomba, Gili, Marcer, Marchiori and Mattiucci2021). However, this may not be the only cause of the level of genetic diversity found. For example, one of the great challenges in acanthocephalan studies has been to collect parasites from various host species that make up their life cycles; among them are arthropods and paratenic hosts, fishes, birds, and marine mammals. Along with this, the wide distribution and movement/migration of many of them make the sampling process difficult (Verweyen et al. Reference Verweyen, Klimpel and Palm2011; Gregori et al. Reference Gregori, Aznar, Abollo, Roura, González and Pascual2012; Hernández-Orts et al. Reference Hernández-Orts, Smales, Pinacho-Pinacho, García-Varela and Presswell2017). In this context and especially for marine mammal parasites, among them Corynosoma species, we know that they are hard to sample. Therefore, we are aware and consider that although the present study is not yet comprehensive, an effort has been made to sample new specimens, including morphological assessment and additional molecular markers.
Most studies have suggested that the classification of the Phylum Acanthocephala based only on morphological characters shows instability due to the conservative and simple morphology of the group (García-Varela & Pérez-Ponce de León Reference García-Varela and Pérez-Ponce de León2008; Presswell et al. Reference Presswell, García-Varela and Smales2018; Amin et al. Reference Amin, Rodríguez and Heckmann2019, Reference Amin, Rodríguez, Rubtsova, Heckmann, Peña, Castro, Rivera and D’Elía2022; Presswell et al. Reference Presswell, Bennett and Smales2020). Therefore, we are assuming that the combination of molecular and morphological tools is a proper approach to resolving taxonomic ambiguities. Our results with 18S rDNA showed that C. bullosum species fall in the same clade as C. australe; however, this relationship was weakly supported by both analyses. Likewise, the unique available sequence of C. bullosum was analysed by García-Varela et al. (Reference García-Varela, Aznar, Pérez-Ponce de León, Piñero and Laclette2005), who concluded that C. bullosum is a sister to C. australe (using ITS1&2 as well as 5.8S rRNA) and forms a monophyletic group. Also, some bootstrap resampling indicated that these relationships were strongly supported, while other relationships among Corynosoma species were inconsistent. However, they also yielded conflicting hypotheses. In light of this, other studies should tackle the relationships among species of the family Polymorphidae, which through various phylogenetic interpretations have shown that their relationships are still unstable; this suggests that the limits of the genera constituting this family warrant further evaluation. Overall, and for a better understanding of Polymorphidae diversity, we suggest expanding the identification of morphological characters to increase molecular markers for the same acanthocephalan species and incorporate ecological and biogeographical relationships of the phylum Acanthocephala.
Finally, it is worth mentioning that in previous studies, samples were obtained by necrosis or shooting. In the present work, we obtained parasites from faecal samples, which is a simple and non-invasive method. Even so, its quality is probably not optimal, which could have influenced the quantity and quality of DNA obtained. Regarding morphometric measurements, parasite quality was not altered due to faecal samples being frozen and carefully washed before taking measurements. As was reported by Rengifo-Herrera et al. (Reference Rengifo-Herrera, Ferre, Ortega-Mora, Rojo-Montejo, García-Moreno, García-Párraga and Pedraza-Díaz2005), we also found some helminth parasites. Remarkably, studies on host-parasite association studies in Antarctic phocids are still poor and scarce. In some cases, descriptions are incomplete, outdated, and/or differ over time. Considering the increasing impact of climate change in Antarctica, long-term analyses are required to understand the effect on the host-parasite dynamic. As such, the sampling technique used here may prove useful for geographically extensive and relatively fast parasite sampling.
Conclusions
We provided an updated morphological redescription for Corynosoma bullosum including electron microscopy photographs and molecular data. The resulting ML and BI trees for 18S rDNA showed a low level of congruence and supported that C. bullosum is sister to C. australe, and they fall in a separate clade from other Corynosoma species. Our results show that because the phylogenetic relationships among species of the family Polymorphidae still show instability, it is necessary to perform studies that combine morphological analyses and different molecular markers, especially of marine acanthocephalans.
Acknowledgments
We thank Alex González for his assistance with the laboratory work in Sistemática Lab from Universidad Austral de Chile. Further, we thank Dr. Guillermo D’Elía for his comments on the phylogenetic analysis; Mauricio Luquet (SEM unit, Aluminio Argentino) for help with SEM photographs; Martín Durante, Jorge Rossi, and Maximiliano Perier who were involved in fieldwork assistance at Península Potter. The study was financially and logistically supported by the Dirección Nacional del Antártico, Instituto Antártico Argentino. The permit for this work was granted by the Dirección Nacional del Antártico (Environmental Office). The authors also appreciate the restored Ministerio de Ciencia, Tecnologı́a e Innovación Productiva de la Nación for the promotion of the scientific Argentinean program and the support to public education. We also want to express our heartfelt gratitude and appreciation to all the healthcare workers and researchers working on the frontline against the pandemic for COVID-19. FS, MSL, JN, and FC are members of CONICET.
Financial support
This study was not grant funded.
Conflict of interest
The authors declare that they have no competing interests.
Ethical standard
Permission to work in the area was granted by Dirección Nacional del Antártico.









