Article contents
End-to-End Deep Learning-Based Cells Detection in Microscopic Leucorrhea Images
Published online by Cambridge University Press: 02 March 2022
Abstract
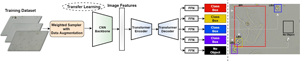
Vaginitis is a prevalent gynecologic disease that threatens millions of women’s health. Although microscopic examination of vaginal discharge is an effective method to identify vaginal infections, manual analysis of microscopic leucorrhea images is extremely time-consuming and labor-intensive. To automate the detection and identification of visible components in microscopic leucorrhea images for early-stage diagnosis of vaginitis, we propose a novel end-to-end deep learning-based cells detection framework using attention-based detection with transformers (DETR) architecture. The transfer learning was applied to speed up the network convergence while maintaining the lowest annotation cost. To address the issue of detection performance degradation caused by class imbalance, the weighted sampler with on-the-fly data augmentation module was integrated into the detection pipeline. Additionally, the multi-head attention mechanism and the bipartite matching loss system of the DETR model perform well in identifying partially overlapping cells in real-time. With our proposed method, the pipeline achieved a mean average precision (mAP) of 86.00% and the average precision (AP) of epithelium, leukocyte, pyocyte, mildew, and erythrocyte was 96.76, 83.50, 74.20, 89.66, and 88.80%, respectively. The average test time for a microscopic leucorrhea image is approximately 72.3 ms. Currently, this cell detection method represents state-of-the-art performance.
Keywords
- Type
- Biological Applications
- Information
- Copyright
- Copyright © The Author(s), 2022. Published by Cambridge University Press on behalf of the Microscopy Society of America
References
- 2
- Cited by


