Introduction
Wood, as a natural high-performance material, has served human needs for generations in structural applications. Advances have been made in the manufacture of high-strength lightweight products from nanocellulose and production of bioethanol [Reference Rajinipriya1–Reference Duque3] from wood and other lignocellulosic materials. Wood is also an important renewable resource in the design and construction of intelligent devices, with a wide range of functionalities and applications in biomedicine, energy storage, electronics, and as sensors in filtration and absorption devices [Reference Farid4]. If protected from the environment, wooden products can last hundreds of years. However, microorganisms can deteriorate wooden products, particularly in outdoor environments where the products can be subjected to several deteriorating factors including UV radiation. Among these, fungi are known to decay wood severely by degrading wood cell wall polymers (cellulose, hemicellulose, lignin) to obtain nutrients for their growth and function. More recently, evidence suggests that bacteria can also decay wood. Although bacteria do not decay wood as rapidly as fungi, they are adapted to degrade cell wall polymers under conditions (high moisture, toxic preservatives, and extractives in wood) considered extreme for wood decay fungi that cause the most severe damage. While microbial decay of wood, such as conversion of cell wall polymers in wood residues after tree harvest, is essential from ecological and environmental perspectives (for example, carbon cycling), each year decay brings enormous economic losses resulting from shortened life of wooden products. Therefore, wood protection based on the fundamental knowledge of microbial decay processes is of increasing importance, with advances made in the development of more efficient and targeted wood protection technologies.
Microscopy plays an important role in research on wood biodegradation. While it is possible to recognize the patterns of fungal wood decay using light microscopy (LM), bacterial decay patterns are clearly revealed only when transmission electron microscopy (TEM) is applied. Because of its ability to resolve fine details of cell structures, TEM is an invaluable tool in clearly revealing the micromorphological patterns of cell walls undergoing bacterial degradation. Patterns produced by bacteria are unique and very different from fungal decay patterns. TEM also reveals the spatial relationship of bacteria with degrading cell wall regions, clearly resolving the micromorphological features associated with cell wall degradation by bacteria [Reference Blanchette, Rowell and Barbour5–Reference Singh9]. The high definition achieved using TEM makes it possible to unambiguously identify bacterial decay, even in wooden objects exposed to natural environments, where mixed fungal and bacterial attack are often present in the same wood tissues [Reference Singh9] and regions of a cell wall [Reference Singh9,Reference Nagashima10]. This was not achievable in earlier LM-based studies because of lack of clarity due to greater thickness of sections when compared to ultrathin TEM sections and inadequate resolution of LM. Inability to clearly resolve cell wall degradation patterns in decaying tissues at the LM level, although bacterial presence could be detected, led to the belief that bacteria could not degrade lignocellulosic cell walls [Reference Singh and Butcher6,Reference Daniel, Nilsson, Bruce and Palfreyman7,Reference Schmidt and Liese11,Reference Clausen12].
Following TEM confirmation, bacterial degradation of wood was placed into two distinct categories, tunneling and erosion, based on the micromorphological patterns produced. Scanning electron microscope (SEM) alone, or in combination with TEM, is useful in characterizing bacterial erosion of cell walls that involves degradation progressing from exposed faces of the cell wall [Reference Holt13–Reference Singh16]. However, TEM is the high-resolution tool of choice for precisely understanding the nature of cell wall degradation by tunneling bacteria (TB) [Reference Nilsson and Daniel17–Reference Singh20]. This review concerns bacterial tunneling of cell walls using TEM to clearly resolve a unique, intriguing architecture of tunnels. TEM serves as an important diagnostic feature for detecting this type of bacterial attack on wood subjected to a wide range of conditions and environments.
Wood Structure and Composition
To understand wood decay involving bacterial tunneling of cell walls and the relationship of cell wall structure to tunneling bacterial degradation, it is important to understand the structure of wood, particularly cell wall ultrastructure and composition. The structure and composition of wood is briefly reviewed here; more detailed descriptions can be found in publications, including that by Schmitt et al. [Reference Schmitt, Asiegbu and Kovalchuk21]. Wood consists of cells with a cell wall around a lumen, where much of the water is stored in living trees. After tree harvest, the sawn timber is appropriately dried to remove water prior to being placed in service in the form of a specific product. However, wood products placed in service outdoors are exposed to moisture and other factors. Particularly hazardous are the conditions where products are in contact with soil. With moisture absorption by wood over time, wood-destructive microorganisms can invade.
Wood is placed into two broad categories: softwood and hardwood, based on cell types and composition of the cell wall. Softwoods consist largely of one cell type (tracheid) (Figure 1), whereas several types of cells are present in hardwoods. The cell wall characteristics described here relate to softwoods, which have been studied more extensively for bacterial tunneling degradation, although many of the features are also common to hardwoods. Depending on the season when tracheids are produced, their size and cell wall thickness differ. Tracheids produced in the spring (springwood or earlywood) are large in diameter and consist of thin cell walls. In comparison, those produced in the summer (summerwood or latewood) are distinctly smaller in diameter and consist of considerably thicker cell walls. It is not surprising that latewood cell walls, with a more abundant supply of nutrients, are the preferred target for TB, although the cell wall of earlywood tracheids is sufficiently thick for these bacteria to tunnel through. Structurally, tracheid cell walls consist of a compound middle lamella (ML) and a secondary wall that is differentiated into three layers (S1, S2, and S3). ML is the first structure to form and functions as a glue between cells that are derived from the wood-forming meristem destined to mature into wood cells. Subsequently, the secondary wall layers form, with S1, S2, and S3 deposited sequentially. Although this organization of the cell wall is resolvable in high-magnification images obtained using LM and confocal laser scanning microscopes, TEM provides superior textural information [Reference Schmitt, Asiegbu and Kovalchuk21–Reference Singh23] (Figure 2).
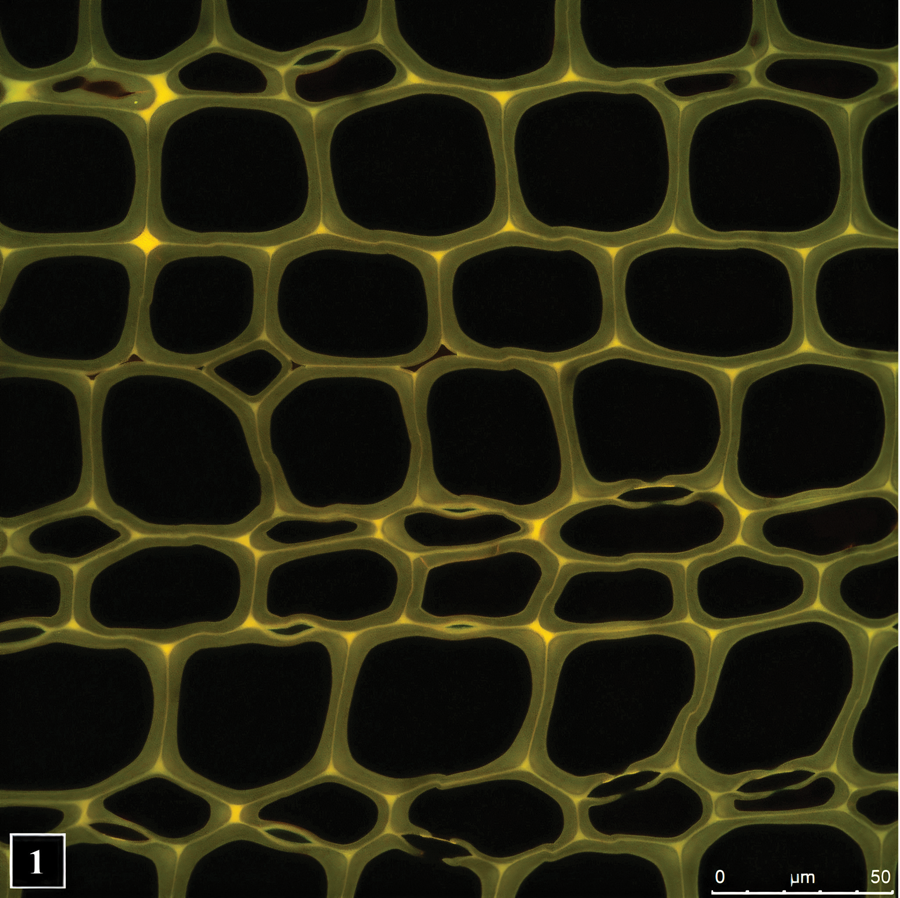
Figure 1: Transverse section of a pine wood showing lignin autofluorescence. Fluorescence intensity is greatest in the middle lamella, the cell wall region common to neighboring cells. The image was captured using confocal fluorescence microscopy. Micrograph courtesy of Dr. Lloyd Donaldson.

Figure 2: Transverse section through a cell corner region (CC) between pine wood cells, imaged using TEM. ML, middle lamella; S1, S2, S3, secondary wall layers. The arrowheads point to warty protuberances. Scale bar = 1 μm. Micrograph courtesy of Professor Jong Sik Kim.
The secondary cell wall is composed of three main polymers: cellulose, hemicellulose, and lignin. Among these, the concentration of cellulose is greater than the latter two, which are present in similar amounts [Reference Schmitt, Asiegbu and Kovalchuk21]. The chains of cellulose molecules in the cell wall are organized in the form of stiff nano cables, referred to as microfibrils [Reference Donaldson22], which are responsible for much of the strength of the cell wall [Reference Reza24,Reference Reza25]. Hemicellulose and lignin are matrix polymers. Together with cellulose they form a complex interconnecting network in the cell wall. Collectively, these polymers provide strength and stability to wood in standing trees, as well as wood products made after tree harvest. Microfibrils in the S2 layer, the thickest part of the cell wall, are oriented along the length of cells and perpendicular to this in the S1 and S3 layers [Reference Schmitt, Asiegbu and Kovalchuk21,Reference Donaldson22]. From the perspective of wood biodegradation, the orientation of microfibrils, and the amount, type, and distribution of cell wall polymers, particularly lignin, are determining factors for some wood decay microorganisms, but not for others. For example, cavities produced in the cell wall by soft rot fungi and channels formed by cell wall eroding bacteria are always aligned with microfibrils [Reference Daniel and Nilsson15,Reference Khalili26,Reference Anagnost27], reflecting a highly specific mechanism employed by these organisms to attack cellulose. Microfibril orientation does not appear to be a limiting factor for tunneling bacteria. Similarly, the concentration of lignin, the most recalcitrant cell wall polymer, is a limiting factor for certain wood decay organisms, but not for others [Reference Singh9].
TB and Tunneling of Wood Cell Walls
TEM has revealed that TB are Gram-negative type. It must be emphasized that it has not been possible thus far to obtain pure cultures of TB, as the standard protocols for culturing bacteria do not work for TB. Here, involvement of microbiologists with expertise in culturing bacteria might help, but most microbiologists are not familiar with bacterial wood degradation studies, as most work has been undertaken by wood scientists and published in wood-related journals. The classification of bacteria as Gram-negative and Gram-positive is based on differences in cell wall composition and ultrastructure as revealed by TEM. The cell wall of TB is thin and membranous [Reference Singh9], an ultrastructural feature typical for Gram-negative bacteria, and one that imparts flexibility with respect to shape changes during cell wall degradation. SEM observations have shown TB to be non-flagellate (devoid of surface appendages that function in swimming motility) rods, a feature also observed using TEM [Reference Singh and Butcher6,Reference Daniel18].
TEM has played a key role in understanding bacterial cell entry and the tunneling process. The initial process leading to entry into wood tissues involves surface colonization in natural environments of appropriate moisture and temperature. Although this aspect has not been investigated for TB, considerable information is available from other systems involving bacterial colonization of solid surfaces [Reference Dang and Lovell28]. The biofilm that develops contains microbial communities and an extracellular slime matrix [Reference Donlan29]. The microorganisms within the biofilm communicate chemically with other biofilm bacteria. Wood decay microorganisms take the lead and penetrate wood tissues from the biofilm coated surface, initially decomposing non-lignified tissues (parenchyma) rich in readily metabolizable storage nutrients. Then they make their way into tracheids that form the bulk of softwood.
Bacterial attachment to and penetration into the cell wall. The processes of attachment to the exposed face of the cell wall (the S3 layer which faces the lumen) (Figure 2), and subsequent initial penetration of the cell wall by TB, have been reviewed [Reference Singh and Butcher6] (Figure 3). It will suffice to say that TB attach themselves using the extracellular slime (likely a mucopolysaccharide in composition) that they abundantly produce throughout cell attachment, a mechanism for surface attachment by bacteria not equipped with motility appendages [Reference Kimkes and Heinemann30] and degradation processes. Despite loss of slime components that may occur in conventional sample preparation (for example, dehydration using acetone or alcohol), TEM images clearly show slime rings around the posterior of penetrating TB that connect to the exposed face of the S3 wall (Figure 2) and thus secure the bacterial position at the point of cell wall penetration. It is intriguing that TB assume a pear shape to achieve initial penetration into the cell wall, using the pointed front to gain entry [Reference Singh and Butcher6] (Figure 3). “Point” penetration may be a strategy to economically use resources such as enzymes, making a hole of minimal size that can allow bacterial entry. TB are capable of changing their shape (pleomorphism) and squeezing through a hole smaller than their cross-sectional diameter. However, information on the initial entry into the cell wall is available only from a few studies, and this fascinating aspect requires detailed studies using a variety of wood substrates varying in the thickness and composition of the S3 layer, particularly lignin (a highly recalcitrant polymer) concentration.

Figure 3: The diagram shows attachment of a tunneling bacterium to the exposed face of the cell wall (1) and stages of subsequent bacterial penetration into the cell wall (2,3). TB can move in varying directions (arrows) and produce tunnels in all cell wall regions. S1, S2, S3, secondary wall layers. The diagram is reproduced from Singh et al. (2019), IAWA Journal [Reference Singh9].
Cell wall degradation by tunneling. Once entry is completed, and when in contact with the overlying nutrient rich S2 layer, TB return to their usual form and develop microcolonies that tunnel in varying directions. This gives an impression of repeat tunnel branching radiating from a central point, reminiscent of tree branching. While this feature is clearly resolved with TEM (Figure 4), with LM it appears as fine branching striations that can be mistaken for other form(s) of microbial attack, particularly in natural environments where different types of microorganisms often co-exist in wood tissues, some producing similar decay patterns.

Figure 4: The TEM micrograph shows repeat branching of tunnels (star), radiating from a central point in a wood cell wall. TB, tunneling bacteria. Scale bar = 2 μm. The image is reproduced from Singh et al. (2019), IAWA Journal [Reference Singh9].
As mentioned earlier, microfibril angle does not appear to pose a restriction on TB movement within the cell wall, which allows them to optimize use of nutrients and take full advantage of the available resources. Wood cell walls are not homogenous in their composition, with lignin concentration being a limiting factor for some wood-degrading bacteria that degrade cell walls by way of erosion and soft rot fungi that produce cavities within the cell wall [Reference Daniel, Nilsson, Bruce and Palfreyman7,Reference Singh9]. However, TB do not appear to be constrained by lignin concentration, judging by the presence of tunnels in and across all cell wall layers and regions [Figure 5], including the ML, the most highly lignified cell wall region [Reference Schmitt, Asiegbu and Kovalchuk21].

Figure 5: TEM micrograph of heavily degraded wood cell walls filled with numerous TB (arrowhead) and tunnels (arrows). All cell wall regions, including the highly lignified middle lamellae (stars), are tunneled, and the direction of tunneling is variable. Scale bar = 8 μm. The image is reproduced from Singh et al. (1987), Journal of the Institute of Wood Science [Reference Singh19].
The intricate architecture of tunnels provides the most important diagnostic feature. Many early bacterial wood degradation studies were based on LM that could not clearly resolve the pattern of tunneling degradation, although the presence of bacteria in decaying wood was detected [Reference Schmidt and Liese11,Reference Clausen12]. However, use of TEM in subsequent studies provided a wealth of information on the pattern of cell wall degradation, revealing that degradation was caused by bacteria producing tunnels with a distinct and unique architecture. This formed the basis for diagnosing the type of bacterial tunneling degradation in wood exposed to both terrestrial and aquatic environments [Reference Blanchette, Rowell and Barbour5–Reference Singh and Kim8]. The intricate architecture of tunnels formed within wood cell walls stems from the presence of crescent-shaped periodic bands along the tunnel behind the bacteria (Figure 6). The crescent shape of tunnel bands conforms to the hemispherical form of the posterior of the TB present at the leading edge of the tunnel, with band concavity facing the direction in which they move to reveal the direction of tunneling (Figure 6).

Figure 6: The TEM micrograph shows a tunnel (T) containing a tunneling bacterium (star) and prominent crescent-shaped slime bands (arrowhead). Scale bar = 2 μm. The image is reproduced from Singh (1989), IAWA Bulletin [Reference Singh20].
How does this peculiar architecture develop during tunneling? Bacteria lacking flagella generally move on solid or semi-solid surfaces with the help of extracellular slime they produce, which is composed of polysaccharides or mucopolysaccharides that play an important role in microbial function and survival [Reference Costa31]. In regard to bacterial tunneling, thus far only static images using TEM and SEM have been acquired, but hypotheses have been advanced suggesting that as they degrade the cell wall TB continuously produce slime and are encased in a slime tube. Slime aided bacterial movement (bacterial gliding) has been the focus of many studies in recent years that suggest the mechanism is likely to involve interaction between bacteria, the slime, and the substrate surface. This mechanism may also be operative for TB, the difference being that, as the tunnel architecture suggests, TB do not continually move but stop in places, perhaps to replenish cell wall degrading enzymes. Where TB stop, the slime mass becomes more dense around the posterior, producing a banded appearance of the tunnel (Figure 6). Microbial slime is known to be a highly hydrated polymer [Reference Costa31] and due to dehydration during sample preparation is likely to become compacted in regions where TB stop. Consequently, there is greater accumulation of the slime visible as bands under TEM. Elsewhere in the slime tube (inter-band regions), the slime is distributed thinly as fibrils [Reference Singh and Butcher6] (Figure 6). Although the compact appearance of tunnel bands is likely an artifact introduced during TEM sample preparation, the presence of discrete periodic bands in the tunnel has consistently served to diagnose the presence of bacterial tunneling. This is also true in situations involving mixed microbial attack in wood involving fungi and bacteria [Reference Singh9,Reference Nagashima10,Reference Santhakumaran and Singh35] (Figure 7) and in extensively degraded cell walls where only fragmented tunnels remained [Reference Singh36]. Diagnosing the presence of TB, particularly in wood products placed in service after treatment with such toxic chemicals as copper-chrome-arsenate (CCA), has been of paramount importance in engineering preservative formulations to protect wood against fungal and bacterial attack, particularly involving TB that are highly resistant to toxic metals.
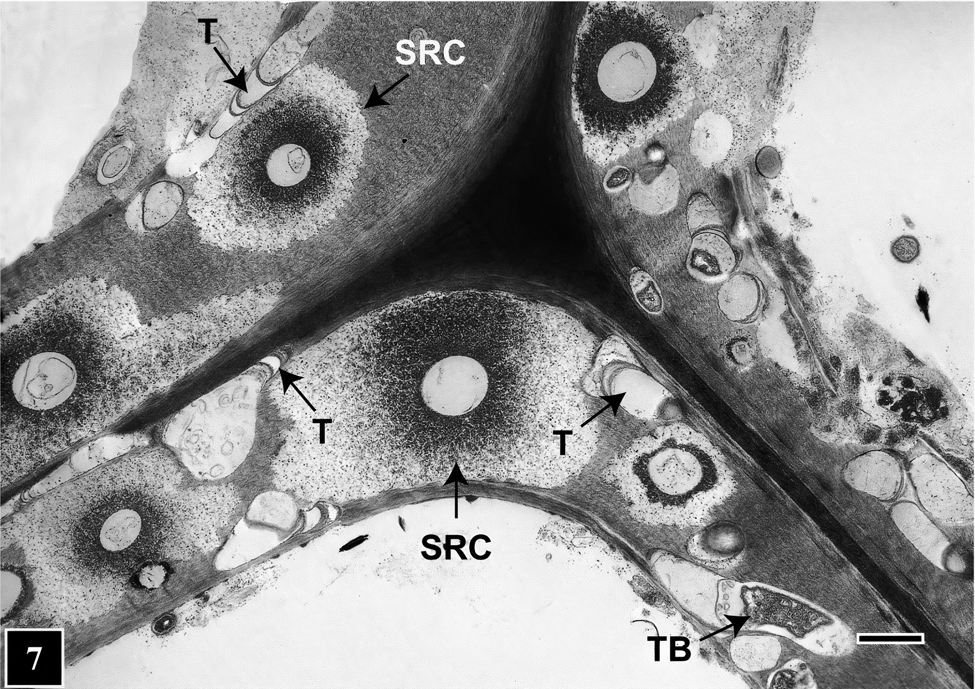
Figure 7: TEM micrograph of a transverse section through a corner region between neighboring cells. The image shows mixed microbial attack on the cell wall involving TB and soft rot fungi. Tunnels (T) are present in cell wall regions not occupied by soft rot cavities (SRC). Scale bar = 2 μm. The image is reproduced from Singh et al. (2019), IAWA Journal [Reference Singh9].
What cell wall components can TB degrade? Many microorganisms, such as rumen bacteria [Reference Wang and McAllister32], produce enzymes that can degrade cellulose and hemicellulose in plant cell walls. Wood cell walls also contain lignin, which is highly resistant to degradation by microorganisms. However, wood-degrading microorganisms can uncouple the protective lignin from the cellulose-hemicellulose complex, which allows their enzymes to gain access to these more readily degradable polymers. The exact mechanism used by wood-degrading bacteria (erosion and tunneling) is not known. Chemical analysis using a range of techniques has indicated that erosion bacteria can at best only slightly modify lignin. Similar analytical work has not been carried out for TB, so the interpretation of whether TB can degrade lignin is based on ultrathin sections stained with potassium permanganate, a reagent known to contrast lignin in plant and wood cell walls [Reference Hepler33,Reference Bland34]. In most lignocellulosic substrates, tunnels appear empty of contents except for the presence of slime as discrete bands, as tunnel lining, and as diffuse fibrils. Additionally, tunnels are found in the most highly lignified cell wall regions, such as the ML [Reference Singh and Butcher6,Reference Daniel18] (Figure 5). The extensively degraded cell wall regions appear empty except for the presence of disintegrating tunnels and scavenging bacteria [Reference Singh and Butcher6] (Figure 5), which rely on simple leftover sugars for their nutrition. Based on these TEM observations, it has been proposed that TB have the capacity to degrade all wood cell wall components. But the question remains as to what extent lignin is degraded.
Enzymes, Slime
Wood decay microorganisms adapt specific strategies to gain access to readily convertible cell wall polymers for their nutrition. The microorganisms for which enzymes have been well-characterized and the mechanism of cell wall degradation proposed are white rot and brown rot fungi. For example, brown rot fungi deploy small molecular substances, including hydroxyl radicals [Reference Arantes, Goodell and Schultz37], to loosen the compact wood cell wall (intermolecular pore size around 2 nm) for their rather large size enzymes to gain access to cellulose and hemicellulose, the main nutrient source for these fungi. TEM has served as a powerful tool for immunolocalization of fungal enzymes to understand their spatial relationship to the substrate [Reference Daniel38]. However, for wood-degrading bacteria, including TB, nothing is known about the enzymes/radicals they deploy to degrade wood cell walls. Since TB are surrounded by an extracellular slime, and thus their outer membrane cannot make direct contact with wood cell wall polymers, the enzymes are likely to be extracellular. Apparently, the slime facilitates in channelizing enzymes to the substrate, and thus loss of this precious bacterial product is prevented or minimized. A similar mechanism may be operative for slime-mediated nutrient sequestration by TB.
Why Tunneling?
Bacterial tunneling can be compared with larval tunneling mechanisms adapted by some well-known wood-eating organisms, such as ship worms and some beetles, in terms of safety and performance efficiency. Securing shelter from predation can be considered an important purpose for TB. Bacterial predation (grazing) by other types of organisms, such as protozoa, has been known for many years [Reference Clarholm39]. TB live in habitats such as soils where protozoa are also common. Protozoa have been found in extensively decaying wood, allowing them to penetrate via gaps and voids formed within the disintegrating tissues. However, protozoa cannot dig into intact lignified cell walls, and thus TB are safe in the tunnels they form and continue to be safe as they move into cell wall regions well away from the site inhabited by protozoa.
Conservation of enzymes may be another reason why TB degrade cell walls by tunneling. Enzyme production in living cells is an energy-demanding process, therefore it is in the best interest of wood-degrading microorganisms to minimize loss of enzymes during wood degradation. Tunnel formation enables a close spatial relationship with cell wall polymers via the encasing slime tube enabling enzymes to hit the target efficiently. Furthermore, the ability of TB to move in all directions within the cell wall unaffected by microfibril orientation, a limiting factor for some wood-degrading microorganisms, enables them to optimize use of the available nutrient resource in situations involving mixed microbial attacks on cell walls, common in natural environments.
For example, natural environments supporting adequate moisture and oxygen that favor TB growth and activity also suit soft rot fungi. These two types of organisms often co-exist within wood tissues and have been found as neighbors occupying the same regions of a cell wall (Figure 7). Judging by the extent of the cell wall area occupied in mixed soft rot attacks, soft rot fungi appear to be more aggressive [Reference Santhakumaran and Singh35]. TEM images show that TB cleverly avoid direct competition, as they tunnel available cell wall regions among cavities that soft rot fungi produce during cell wall degradation (Figure 7). This way, TB can squeeze through cell wall regions wide enough to tunnel through and thus use virtually all available nutrient resource, taking full advantage of the opportunity presented. In this quest, the ability of TB to move in all directions, making directional and shape changes as needed as well as small diameter tunnels, all have a role to play.
Biotechnological Potential
Heavy metal contamination in terrestrial and aquatic environments, which adversely affects the health of living organisms including humans, is of grave concern. Fortunately, dedicated attempts are being made to develop effective amelioration technologies, and the use of bacteria to remove toxic metals from such environments and convert them into non-toxic forms is proving to be an attractive way to combat the threat [Reference Sharma40]. TB are widely present in nature and can degrade wood substrates with which wood-degrading fungi have trouble. One example is treatment of wood products with highly toxic copper-chrome-arsenate (CCA) preservatives to extend their service life. TB can degrade naturally durable wood and timbers treated with CCA at concentrations high enough to discourage fungal attack. The exact mechanism of TB tolerance to CCA components is not known, but some studies have provided a clue. Again, TEM is an immensely useful tool in combination with energy dispersive X-ray spectroscopy (EDS), revealing that CCA components are trapped within the extracellular slime (biosorption) during bacterial tunneling [Reference Daniel18]. Thus, CCA components are prevented from reaching vital cell components. Whether TB have the capacity to also detoxify CCA components is not known, but if they can, opportunities exist for deploying TB for bioremediation of heavy metal contaminated environments, particularly taking advantage of available genetic engineering tools and technologies.
Future Perspectives
The stumbling block facing detailed fundamental understanding of the taxonomic affiliation and behavior of TB and the mechanism underlying cell wall degradation is the inability to isolate these bacteria in culture. Controlled experiments using pure cultures are needed to understand aspects as physiology, biochemistry, enzymology, and the factors affecting bacterial growth and function, including the mechanism of bacterial tolerance to toxic metals. It has been possible to reproduce the degradation pattern caused by TB in laboratory experiments using mixed bacterial cultures obtained from wood decaying in natural environments. However, in the meantime, microscopic characterization of the cell wall degradation process needs to be extended using advanced microscopy techniques, such as cryo-TEM [Reference Dohnalkova41]. Cryo-TEM should provide more precise information on the fine structure of tunnel slime, including banded formations, in their native or near-native state by eliminating dehydration-induced structural alterations common to conventional TEM preparations. The true porosity (interfibrillar spaces) of the slime can then be related to the size range of enzymes known from other well-characterized wood-degrading microbial systems to draw an inference that the enzymes produced by TB can traverse the slime and reach cell wall polymers. Determining the size and extent of porosity in the slime using nano-gold probes in conjunction with cryo-TEM may prove to be a useful complementary technique.
Confocal microscopy is a widely used tool in biological research for non-destructive imaging [Reference Price and Jerome42]. Live cell imaging can be performed using super-resolution confocal microscopy [Reference Wu43], in conjunction with fluorescent probes specific for Gram-negative bacterial cell wall components, to more clearly understand TB movement and the tunneling process. High-resolution confocal Raman microscopy with spectroscopic capabilities [Reference Antonio and Schultz44] can be used to understand to what extent TB can degrade highly recalcitrant lignin by mapping chemical distribution within tunnels, which should complement chemical analysis of TB-degraded wood using conventional advanced analytical tools.










