Introduction
Wetland ecosystems harbour a wide variety of species including different parasites that often go undetected. This can negatively affect the viability of wildlife populations. The detection of parasites and pathogens is important for the identification of endemic as well as exotic diseases, in order to know the host population's health status, the pressures they face and the actions that can be developed for their conservation and management (Lebarbenchon et al., Reference Lebarbenchon, Brown, Poulin, Gauthier-Clerc and Thomas2008). Wild birds are important natural reservoirs and potential dispersers of a wide variety of parasites (Kruse et al., Reference Kruse, Kirkemo and Handeland2004; Chang et al., Reference Chang, Eden, Hall, Shi, Rose and Holmes2020). In particular, migratory birds represent a basic mechanism in the emergence of new sources of infection at great distances from their original areas (or endemic areas) (Koprivnikar and Leung, Reference Koprivnikar and Leung2015).
Among avian endoparasites, haemosporidians (Apicomplexa, Haemosporida) are protozoans that infect vertebrate blood cells and are transmitted by vectors (haematophagous dipterans). A great diversity of haemosporidian species were reported in birds, represented in 3 genera: Haemoproteus (containing 2 subgenera Haemoproteus and Parahaemoproteus), Leucocytozoon and Plasmodium, the aetiological agent of avian malaria (Bell et al., Reference Bell, González-Acuña and Tkach2020). These parasites are globally distributed in most bird families, and the rates of infection and prevalence are variable depending on the bird order (Quillfeldt et al., Reference Quillfeldt, Arriero, Martínez, Masello and Merino2011). Although it is assumed that avian haemosporidian infections are relatively benign in wild populations, the study of these parasites is important because they can affect host fitness, in some cases cause severe pathology (tissue necrosis, haemorrhages and anaemia) and usually mortality is difficult to detect (Valkiūnas, Reference Valkiūnas2005; Palinauskas et al., Reference Palinauskas, Iezhova, Križanauskienė, Markovets, Bensch and Valkiūnas2013; Groff et al., Reference Groff, Lorenz, Crespo, Iezhova, Valkiūnas and Sehgal2019).
The avifauna of South America supports a high diversity of haemosporidians, whose distribution is strongly associated with its complex biogeography (Fecchio et al., Reference Fecchio, Bell, Pinheiro, Cueto, Gorosit, Lutz, Gaiotti, Paiva, França, Toledo-Lima, Tolentino, Pinho, Tkach, Fontana, Grande, Santillán, Caparroz, Roos, Bessa, Nogueira, Moura, Nolasco, Comiche, Kirchgatter, Guimarães, Dispoto, Marini, Weckstein, Batalha-Filho and Collins2019). However, most of the studies focused on passerine hosts, reflecting the need for sampling and describing avian haemosporidian parasites in non-passerine hosts (Bell et al., Reference Bell, González-Acuña and Tkach2020).
Other blood parasites of concern in birds are piroplasmids of the genus Babesia, a protozoan parasite causing babesiosis, an emerging and potentially zoonotic disease transmitted by ticks. Symptoms may include anaemia, leucocytosis and depressed liver function (Ebani and Mancianti, Reference Ebani and Mancianti2021). By now, only the species Babesia shortii is known to cause pathogenicity in birds (Khan et al., Reference Khan, Provencher, Forbes, Mallory, Lebarbenchon and McCoy2019). Currently, studies that focus on avian Babesia from seabirds in South America are scarce (Quillfeldt et al., Reference Quillfeldt, Martinez, Bugoni, Mancini and Merino2014).
The order Charadriiformes contains 19 families and 384 species (del Hoyo et al., Reference del Hoyo, Elliott and Sargatal1996), most of them being seabirds and shorebirds. They have a cosmopolitan distribution and are usually associated with seas, rivers and wetlands (del Hoyo et al., Reference del Hoyo, Elliott and Sargatal1996). Among Charadriiformes seabirds, representatives of the family Laridae (gulls, terns and skimmers) are widely distributed. They are mostly coastal; some are pelagic or can inhabit inland environments (Winkler et al., Reference Winkler, Billerman, Lovette, Billerman, Keeney, Rodewald and Schulenberg TS2020). Within this family, the most studied species in the Northern Hemisphere is the common tern (Sterna hirundo) (Burger and Gochfeld, Reference Burger and Gochfeld1991). Common terns breed in most of Europe, Asia and North America, and undertake long-distance migratory movements to the Southern Hemisphere during the non-breeding season. They have extremely large population sizes, and are listed as Least Concern (IUCN, 2019). However, they are vulnerable to habitat deterioration and loss (del Hoyo et al., Reference del Hoyo, Elliott and Sargatal1996). There is less information available about this species in its wintering grounds in the Western Hemisphere (South America). The southernmost sightings were made in Argentina (Mauco et al., Reference Mauco, Favero and Bó2001; Yorio, Reference Yorio2005), mostly on the Atlantic coast of Buenos Aires, which constitutes the largest roosting area of the common tern in South America (30 000 birds recorded at Punta Rasa) (Hays et al., Reference Hays, Di Costanzo, Cormons, Antas, do Nascimento, do Nascimento and Bremer1997; Mauco et al., Reference Mauco, Favero and Bó2001).
The MalAvi database contains only 47 genetic haemosporidian lineages recovered from wild Charadriiformes (MalAvi database, accessed on 16 January 2023), and there is currently 1 record (i.e. Leucocytozoon sp.) in common terns (Włodarczyk et al., Reference Włodarczyk, Bouwhuis, Bichet, Podlaszczuk, Chyb, Indykiewicz, Dulisz, Betleja, Janiszewski and Minias2022). This paucity of information is probably due to the apparent absence or scarcity of blood parasites in certain groups of birds such as seabirds and shorebirds (Quillfeldt et al., Reference Quillfeldt, Martínez, Hennicke, Ludynia, Gladbach, Masello, Riou and Merino2010; Soares et al., Reference Soares, Escudero, Penha and Ricklefs2016). Published information shows that of 113 seabird species studied, in only 27% haematozoan infections were found. Specifically, in the case of the family Laridae, 59% of the species studied were infected with at least 1 haemoparasitic species, mostly by Haemoproteus (Quillfeldt et al., Reference Quillfeldt, Arriero, Martínez, Masello and Merino2011). There are some previous studies focusing on haemosporidia in charadriiforms from Argentina. In shorebirds for example, 2 studies reported negative results (D'Amico et al., Reference D'amico, Bertellotti, Baker and Diaz2007, Reference D'Amico, Bertellotti, Baker, Junior and González2008) and only 1 reported 1% of prevalence (Soares et al., Reference Soares, Escudero, Penha and Ricklefs2016). In the case of larids, the only positives have been found in Malvinas Islands from the dolphin gull (Larus scoresbii) (Quillfeldt et al., Reference Quillfeldt, Martínez, Hennicke, Ludynia, Gladbach, Masello, Riou and Merino2010); the other study has shown negative results (Jovani et al., Reference Jovani, Tella, Forero, Bertellotti, Blanco, Ceballos and Donázar2001).
Migratory movements can expose host populations to novel parasites as well as introduce parasites into new geographic areas (Koprivnikar and Leung, Reference Koprivnikar and Leung2015). Therefore, it is of interest to explore parasitic species in migratory birds in South America, to increase their detection and to contribute to the knowledge of their diversity, with the ultimate goal of determining parasite–host distribution patterns. The aims of the present study were (i) to assess the prevalence of the Haemosporida (Plasmodium, Haemoproteus and Leucocytozoon) as well as Babesia spp. in common terns from Punta Rasa, their most important wintering site in Argentina, (ii) to identify the genetic diversity of haematozoa parasites and (iii) to compare the genetic relationships among haematozoa haplotypes in Charadriiformes from South America to those identified in previous investigations.
Materials and methods
Study area and sample collection
Fieldwork was conducted during the non-breeding season between December 2020 and April 2021, in Punta Rasa, on the Atlantic coast of Argentina (Fig. 1). Punta Rasa is located in Samborombon Bay in the Río de La Plata estuary (36°17′22.8″ S; 56°46′52.9″ W) and is used by several shorebirds (e.g. sandpipers, plovers and oystercatchers) and marine birds (e.g. skimmers, terns and gulls) as roosting and feeding areas during the non-breeding season (Mauco et al., Reference Mauco, Favero and Bó2001). Birds were caught in January 2020 and February and March of 2021 using canon nets (permit number DI-2019-241). Blood samples of maximum 75 μL were collected by brachial venepuncture and stored on blotting paper (Biodynamics SRL, Buenos Aires, Argentina). Blood samples on the blotting paper were stored in the dark and at room temperature in separate tubes to avoid cross-contamination until DNA isolation. For each bird, 2 thin smears on microscope slides were made. Blood films were air dried immediately in the field, and then fixed with absolute methanol and stained with Giemsa in a working solution prepared with phosphate buffer pH 7.0 (ratio 1:5) for 30 min in the laboratory.

Fig. 1. Common tern sampling location in Argentina. Arrows in orange indicate the migratory route of the species in South America.
Blood parasite detection
Haemosporidians
The presence or absence of avian haemosporidians was determined through nested polymerase chain reaction (PCR) through amplification of the mitochondrial cytochrome b (cytb) gene. Total DNA was extracted using the ammonium-acetate DNA precipitation protocol (Martínez et al., Reference Martinez, Martinez-de La Puente, Herrero, Del Cerro, Lobato, Aguilar, Vasquez and Merino2009). DNA was obtained from 58 of the 60 birds sampled. NanoDrop2000c UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, USA) was used to measure the DNA concentration after extraction. If the DNA concentration was higher than 80 ng μL−1, samples were diluted to 20 ng μL−1. In addition, we tested the quality of the DNA by performing the PCR sexing, resulting in 97% of the samples successfully sexed. Molecular sexing was performed following the protocol of Quintana et al. (Reference Quintana, López and Somoza2008). An initial PCR was run with the primer pair HaemNFI/HaemNR3 [see Hellgren et al. (Reference Hellgren, Waldenström and Bensch2004) for cycling conditions]. An aliquot of this PCR product was subsequently used as template DNA for the second PCR with specific primer pairs HaemF/HaemR2 for Haemoproteus and Plasmodium cytb gene amplification (Bensch et al., Reference Bensch, Stjernman, Hasselquist, Ostman, Hansson, Westerdahl and Pinheiro2000), and HaemFL/HaemR2L for Leucocytozoon cytb amplification (Hellgren et al., Reference Hellgren, Waldenström and Bensch2004). The PCRs with HaemF/HaemR2 and HaemFL/HaemR2L consisted of 25 μL reaction volumes containing 4 μL DNA template, 1.65 μL of each primer (10 mm), 12.5 μL of DreamTaq Master-Mix (Thermo Fisher Scientific, Waltham, USA) and 5.2 μL of nuclease-free water. Samples of different species of Columbiformes with known infections have been used as positive controls in each PCR run (Schumm et al., Reference Schumm, Bakaloudis, Barboutis, Cecere, Eraud, Fischer, Hering, Hillerich, Lormeé, Mader, Masello, Metzger, Rocha, Spina and Quillfeldt2021). Negative controls [template replaced with double-distilled water (ddH2O)] were included in runs to check for possible contamination. Then, PCR products of the samples were visualized using QIAxcel Advanced (Qiagen, Hombrechtikon, Switzerland) high-resolution capillary gel electrophoresis. A positive PCR result was interpreted as an infected bird, and positive samples were Sanger sequenced bidirectionally by Microsynth-Seqlab (Sequence Laboratories Goettingen GmbH, Göttingen, Germany). For the new lineages, PCR and sequencing were performed twice to verify the results. The new cytb sequences were deposited in GenBank under the corresponding accession numbers.
To confirm the presence or absence of intracellular parasite gametocytes, smears (n = 120) were double-blind scanned by microscopic examination at ×1000 magnification using a light microscope (PrimoStar Zeiss, Göttingen, Germany).
Piroplasms
Detection of piroplasms was performed by nested PCR on the 18S rRNA gene following the protocol previously developed for the detection of Babesia in yellow-legged gull's (Larus michahellis) blood (Bonsergent et al., Reference Bonsergent, Vittecoq, Leray, Burkart, McCoy and Malandrin2022). Primers 18SBp_fw and 18SBp_rev and BAB-GF2 and BAB-GR2 were used in 2 different nested PCRs to amplify the expected fragments of 1529 and 560 bp, respectively, after a primary amplification with CRYPTOF and CRYPTOR primers (Malandrin et al., Reference Malandrin, Jouglin, Sun, Brisseau and Chauvin2010). A positive control was included in each run to ensure the PCR had worked properly. A Babesia infecting human, Babesia sp. FR1, was used in the first PCR as a PCR success control. The use of genomic DNA from a Babesia species not infective for birds allows the identification of eventual sample contaminations from the positive control, after sequencing. Babesia sp. YLG genomic DNA was used as a positive control in the second PCR. Negative controls (template replaced with ddH2O) were included in runs to check for possible PCR contamination.
Phylogenetic analyses
The forward and reverse sequences of the cytb fragment were assembled and trimmed in CLC Main Workbench 7.6.4 (CLC Bio, Qiagen, Denmark). To identify haemosporidian lineages, an online BLAST (National Center for Biotechnology Information) using BLASTN 2.3.0+ (Zhang et al., Reference Zhang, Schwartz, Wagner and Miller2000) was performed in the MalAvi database (Bensch et al., Reference Bensch, Hellgren and Pérez-Tris2009). The resulting reference sequences were aligned with our sequences and trimmed to MalAvi sequence length (479 bp) using the program BioEdit (Hall, Reference Hall1999). Sequences were considered as distinct lineages if they differ by 1 or more nucleotides. Lineages with no database records in MalAvi were considered as novel, and new MalAvi names were assigned.
Constructions of lineage networks for Plasmodium, using the median-joining network method were performed with PopART 1.7 (Bandelt et al., Reference Bandelt, Forster and Röhl1999; Leigh and Bryant, Reference Leigh and Bryant2015) (Table 1).
Table 1. Taxa included in the median-joining network of mitochondrial cytb gene lineages of Plasmodium spp.

Results
Of the 58 samples, we obtained quality DNA in 56 samples. After PCR assay, we found 2 positive samples (3.6%), and there were no double infections. Only Plasmodium infections were detected, and no Haemoproteus- or Leucocytozoon-positive samples were found. We obtained sequences of 501 and 499 bp for the positive samples. Then sequences were assembled and edited, resulting in 479 bp fragments.
We identified 2 new Plasmodium lineages, named STEHIR01 (accession number ON872158) and STEHIR02 (accession number ON872159), which differ in 1 and 2 nucleotides from their closest MalAvi match PARUS67, respectively (Fig. 2). BLASTN analysis from MalAvi database revealed an identity of 99% with the Plasmodium lineages TURDUS1, BT7 (representing the morphospecies Plasmodium circumflexum), SFC6, PARUS67 and FALTIN14. The 2 new lineages appear phylogenetically clustered with Plasmodium lineages isolated from other bird orders such as Passeriformes, Charadriiformes, Strigiformes, Falconiformes and Anseriformes (Fig. 2). Despite positive PCR, we did not detect haemoparasites in the corresponding blood smears.
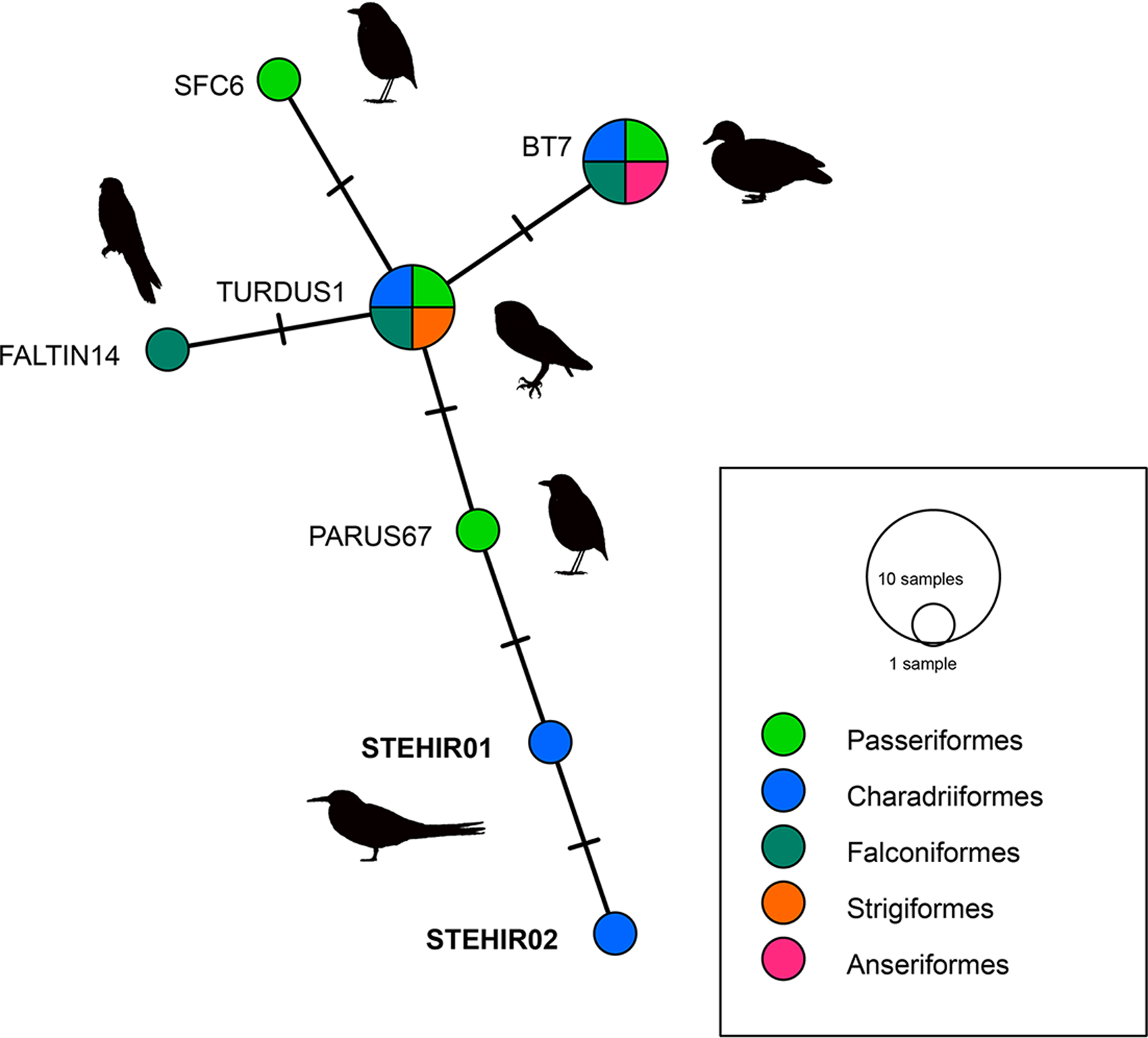
Fig. 2. Median-joining network of mitochondrial cytb gene lineages of Plasmodium spp. (the 2 lineages found in the present study and the first 5 reference lineages from MalAvi, all of 479 bp, are presented in Table 1). Circles represent distinct genetic lineages, and the circle sizes are proportional to the lineage frequencies. One hatch mark represents 1 mutation. Lineage names are noted at the associated circles.
The amplification of partial 18S rRNA genes using BAB-GF2 and BAB-GR2 and 18SBp_fw and 18SBp_rev primers resulted in amplicons of non-expected sizes, most probably corresponding to non-specific primer hybridization. Sequencing and BLASTN analyses were performed on, respectively, 3 and 2 of the amplicons and confirmed non-specific amplifications. Neither Babesia sp. YLG nor other Babesia/Theileria piroplasms were detected in common terns' blood samples of this study.
Discussion
Members of the Laridae family have been reported as carriers of haemoparasites in South America, such as the black skimmer (Rynchops niger) and the brown noddy (Anous stolidus) from Brazil, the swallow-tailed gull (Creagrus furcatus) from Galapagos, Ecuador and the dolphin gull from Malvinas Islands, Argentina (Quillfeldt et al., Reference Quillfeldt, Martínez, Hennicke, Ludynia, Gladbach, Masello, Riou and Merino2010, Reference Quillfeldt, Martinez, Bugoni, Mancini and Merino2014; Levin et al., Reference Levin, Valkiūnas, Santiago-Alarcon, Cruz, Iezhova, O'Brien, Hailer, Dearborn, Schreiber, Fleischer, Ricklefs and Parker2011; Roos et al., Reference Roos, Belo, Silveira and Braga2015). However, these lineages from Laridae are not genetically close to the new lineages found in the present study (see Supplementary information Figs S1 and S2). In the common tern there are only 2 previous studies on the subject from the Americas. One was performed at the breeding sites in the northeastern USA showing the absence of haemoparasites. This was only carried out by screening blood smears (Fiorello et al., Reference Fiorello, Nisbet, Hatch, Corsiglia and Pokras2009). The second one is a checklist of avian blood parasites from North America reporting 0% prevalence in the common tern by blood smears screening (Greiner et al., Reference Greiner, Bennett, White and Coombs1975). Recently, haemosporidians of the genus Leucocytozoon have been reported in common terns from Europe, but with low prevalence (<0.5%), and no Plasmodium positives were found (Włodarczyk et al., Reference Włodarczyk, Bouwhuis, Bichet, Podlaszczuk, Chyb, Indykiewicz, Dulisz, Betleja, Janiszewski and Minias2022).
In accordance with previous seabird studies (e.g. Fiorello et al., Reference Fiorello, Nisbet, Hatch, Corsiglia and Pokras2009; Quillfeldt et al., Reference Quillfeldt, Arriero, Martínez, Masello and Merino2011; Campioni et al., Reference Campioni, Martínez-de la Puente, Figuerola, Granadeiro, Silva and Catry2018; Ilahiane et al., Reference Ilahiane, De Pascalis, Pisu, Pala, Ferrario, Cucco, Rubolini, Cacere and Pellegrino2022; Roldán-Zurabián et al., Reference Roldán-Zurabián, José Ruiz-López, de la Puente, Figuerola, Drummond and Ancona2022) haemosporidian prevalence in the common tern from the Argentinean coast was low. Avian Plasmodium species are distributed worldwide except in Antarctica, but only 3 species, Plasmodium relictum, Plasmodium reticulum and Plasmodium matutinum, have been detected in wild seabirds, the former being the most common species in birds (Quillfeldt et al., Reference Quillfeldt, Arriero, Martínez, Masello and Merino2011; Włodarczyk et al., Reference Włodarczyk, Bouwhuis, Bichet, Podlaszczuk, Chyb, Indykiewicz, Dulisz, Betleja, Janiszewski and Minias2022). In our study, the new Plasmodium lineages differ in 1 and 2 nucleotides from their closest matching lineage PARUS67 that was found in the great tit (Parus major) and the marsh tit (Parus palustris) (Dubiec et al., Reference Dubiec, Podmokła, Zagalska-Neubauer, Drobniak, Arct, Gustafsson and Cichoń2016; Ellis et al., Reference Ellis, Huang, Westerdahl, Jönsson, Hasselquist, Neto, Nilsson, Nilsson, Hegemann, Hellgren and Bensch2020). This can indicate a recent divergence during a host switching process. At the same time, as we were not able to observe gametocytes in the blood smears, this finding could represent an abortive malaria infection. This happens when a parasite invades a host, but cannot complete its full life cycle and be transmitted to the vector as gametocytes are absent in the blood (Palinauskas et al., Reference Palinauskas, Žiegytė, Iezhova, Ilgūnas, Bernotienė and Valkiūnas2016).
Other shorebirds and seabirds were analysed for haemoparasites in the Argentinean coast. For example, Jovani et al. (Reference Jovani, Tella, Forero, Bertellotti, Blanco, Ceballos and Donázar2001) analysed blood smears of 560 birds from 13 avian species, and no haemoparasites were detected. Martinez-Abrain et al. (Reference Martínez-Abraín, Esparza and Oro2004) discussed different reasons for the apparent lack or lower presence of blood parasites in some avian species. Despite the more common explanation being the absence or scarcity of parasite vectors in some habitats, there are contradictions which show that it is not a universal explanation, and that other factors (e.g. immunological capabilities of the host, absence of the right host–parasite–vector assemblage) could be operating (Martinez-Abrain et al., Reference Martínez-Abraín, Esparza and Oro2004). Methodological weaknesses also could be influencing. According to our results and those from previous authors (e.g. Quillfeldt et al., Reference Quillfeldt, Martínez, Hennicke, Ludynia, Gladbach, Masello, Riou and Merino2010), PCR analyses are needed to avoid false negatives. This is because PCR screening techniques are reliable for detecting low intensity infections and are able to identify infections with low levels of parasitaemia that are not visible on smears (Durrant et al., Reference Durrant, Beadell, Ishtiaq, Graves, Olson, Gering, Peirce, Milensky, Schmidt, Gebhard and Fleischer2006).
It is difficult to establish where birds could have been infected, since they are inter-continental migrants, and avian Plasmodium parasites are found worldwide. Therefore, addressing transmission in the avian malaria system turns out to be challenging. Also, in the North–South American migratory route, migratory and resident host species share the same habitats, therefore Plasmodium spp. lineages can switch easily between a wide range of hosts (Ricklefs et al., Reference Ricklefs, Medeiros, Ellis, Svensson-Coelho, Blake, Loiselle, Soares, Fecchio, Outlaw, Marra, Latta, Valkiūnas, Hellgren and Bensch2017).
Many species of avian Plasmodium use Culicidae mosquitoes belonging to different genera for completing sporogony and transmission (Valkiunas and Iezhova, Reference Valkiūnas and Iezhova2018). Two hundred and twelve species of Culicidae mosquitoes have been recorded in Argentina. From these species, 39.75% are found in the centre of the country (Stein et al., Reference Stein, Rossi, Almirón, Berón, Campos, Gleiser, Díaz Nieto, Salomón and Schweigmann2016). This includes Buenos Aires province, where the present study was conducted. In 2010, 2 cases of lethal avian malaria were confirmed in Magellanic penguins (Spheniscus magellanicus). Birds were in permanent captivity in San Clemente del Tuyú (36°20′17″ S; 56°45′14″ W), the same area as the sampling site of the present study. Identifications were made by blood smears resulting in 3 species: Plasmodium (Haemamoeba) tejerai, Plasmodium (Huffia) sp. and Plasmodium (Novyella) sp. (Vanstreels et al., Reference Vanstreels, Capellino, Silveira, Braga, Rodríguez-Heredia, Loureiro and Catão-Dias2016). This indicates a chance for haemoparasitic infection at this site, but further investigations are needed in order to investigate the local transmission of avian blood parasites.
Several tick species have been proposed to transmit Babesia to different seabird species. They are particularly exposed to nidicolous vectors during the breeding season, given the abundance of hard or soft ticks in their colonies. In addition, it is not yet well understood whether Babesia species are highly host-specific (Bonsergent et al., Reference Bonsergent, Vittecoq, Leray, Burkart, McCoy and Malandrin2022). Although the common terns nest on the ground, in open areas with loose substrate and with scattered vegetation, which can increase the chances of encountering ticks, we do not have sufficient information to explain the absence of Babesia in the common terns tested. More studies of seabirds are required to understand the parasitic dynamics and transmission ecology of Babesia species.
In summary, we report for the first time Plasmodium from wild migratory seabirds from Argentina, the first report of a Plasmodium in the common tern, and the second report of Plasmodium from wild seabirds from the temperate region of South America (Quillfeldt et al., Reference Quillfeldt, Martínez, Hennicke, Ludynia, Gladbach, Masello, Riou and Merino2010). The results presented here show that in the southernmost part of South America the role of wild birds (especially non-passerines) in the spread of haemoparasites remains speculative, and may change over time. In this sense, this kind of studies requires urgent attention. Increasing surveillance will allow a better understanding of parasite transmission at sites frequented by birds.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182023000185
Data availability
The authors confirm that the data supporting the findings of this study are available within the article. Raw data are available from the corresponding and first author.
Acknowledgements
We thank all rangers from Reserva Natural Punta Rasa who assisted with fieldwork, and Sabine Wagner for her contribution in laboratory work. The authors would like to thank the anonymous reviewers as well as the editor, for the helpful comments on the manuscript.
Author's contribution
S. C. conceived and designed research. S. C., E. L., B. F., G. P., M. L. and G. C. carried out the samplings. S. C., Y. R. S., P. Q., C. B. and L. M. determined the parasites and analysed data. S. C., Y. R. S., P. Q., C. B., L. M. and J. I. D. contributed to the results and discussion as well as writing of both the draft and final versions of the manuscript. J. I. D. supervised the project. All authors read and approved the manuscript.
Financial support
We gratefully acknowledge the financial support for this research by the following agencies: Universidad Nacional de La Plata (J. I. D., grant number N859), Consejo Nacional de Investigaciones Científicas y Técnicas (J. I. D., grant number PIP-1787 and PUE CEPAVE), German Academic Exchange Services (DAAD to S. C.), Fondo para la Investigación Científica y Tecnológica (S. C., grant number PICT-2019-3115) and Organismo Provincial para el Desarrollo Sostenible (Ministerio de Ambiente, Buenos Aires province).
Conflict of interest
The authors declare that no conflicts of interest exist.
Ethical standards
All samples taken from birds were in compliance with all appropriate research permissions (permit number DI-2019-241) provided by Organismo Provincial para el Desarrollo Sostenible (Dirección de Áreas Protegidas, Ministerio de Ambiente, Buenos Aires province, Argentina).