With the rapid growth of aquaculture, demand for aquafeed with less fishmeal (FM) has increased because of the cost of this protein source and its limited supply( Reference Naylor, Hardy and Bureau 1 ). During the past decade, considerable progress has been made towards replacing portions of FM in aquafeeds with alternative protein sources( Reference Naylor, Hardy and Bureau 1 , Reference Hardy 2 ). Currently, FM is becoming a minor protein source in the feed for omnivorous species. However, it continues to be the primary protein source in aquafeed for marine species and other species during the fry or fingerling stage( Reference Hardy 2 ). The oversubstitution of FM has generally led to reduced growth performance in marine carnivorous species( Reference Naylor, Hardy and Bureau 1 , Reference Hardy 2 ). The inferior performance of non-FM protein sources (plant proteins, in particular) has been attributed to nutritional limitations, such as the imbalanced amino acid profiles, presence of anti-nutritional compounds and other factors( Reference Hardy 2 ). However, little is known regarding the nutrient-sensing and metabolic changes after FM replacement in aquatic animals.
Postprandial responses represent a critical step towards defining the utilisation efficiency of dietary protein sources( Reference Boirie, Dangin and Gachon 3 , Reference Bos, Metges and Gaudichon 4 ). Feeding-induced stimulation of anabolic protein synthesis depends on postprandial repletion of amino acid pools in plasma and other tissues( Reference Frühbeck 5 ). Mediated by amino acid transporters, amino acid availability in turn mediates the activation of nutrient-sensing cascades, including target of rapamycin (TOR) and amino acid response (AAR) pathways, both of which control protein synthesis and downstream metabolism( Reference Kimball and Jefferson 6 ). The activated TOR signalling pathway promotes the translation of many anabolic enzymes and other proteins involved in diverse cellular functions( Reference Ma and Blenis 7 – Reference Jewell, Russell and Guan 9 ). However, any individual amino acid limitation can activate the AAR pathway, which triggers global protein synthesis repression and induces translation of rate-limiting enzymes related to amino acid and lipid metabolism( Reference Kilberg, Pan and Chen 10 , Reference Guo and Cavener 11 ). The counter-regulatory mechanisms of amino acid sensing exist to coordinate the action of TOR and AAR pathways and their downstream effects on translation, which provide the molecular basis for nutritional responses( Reference Yan and Lamb 12 ).
Most nutrient-sensing studies in fish focus on cellular responses in vitro ( Reference Dai, Panserat and Plagnes-Juan 13 – Reference Lansard, Panserat and Plagnes-Juan 15 ). However, such information may not provide important explanations concerning the physiological responses generated by a certain protein source in animals. We hypothesised that dietary FM replacement would probably change the postprandial nutritional response of fish, which, in turn, would have an impact on their metabolism and phenotypic performance. To test this hypothesis, an economically valuable marine carnivorous fish species, turbot (Scophthalmus maximus L.), was chosen as the model species because of its high dietary protein requirement( Reference Lee, Cho and Park 16 ). Fish were fed either an FM diet or partial FM replacement by soyabean meal with or without essential amino acid (EAA) supplementation diets, which represented the most frequently used strategy for improving the performance of non-FM proteins( Reference Gatlin, Barrows and Brown 17 ). After the feeding trial, a comprehensive characterisation of the postprandial dynamics of the expression of amino acid transporters, free amino acid pools, the activation of nutrient-sensing molecules and the regulated expression of key metabolic enzymes was evaluated. Our results could contribute to mechanistic explanations on the performance changes after FM replacement in mariculture.
Methods
Diets
The ingredients and composition of the experimental diets are shown in Table 1. Their amino acid profiles are shown in online Supplementary Table S1. Turbot were fed with three isonitrogenous (50·1 % crude protein) and isoenergetic (20·8 kJ/g) diets comprising different protein sources (Table 1): 60 % FM and 33 % FM+34·6 % soyabean meal (45 % FM replaced with soyabean meal) incorporated (SMI) without or with (SMI diet with dietary EAA supplementation (SMI+AA)) amino acids supplemented to match the EAA profile of the FM diet. The ingredients were ground into a fine powder through a 320-μm mesh. All of the ingredients were thoroughly mixed and extruded as pellets, dried at 45°C for 12 h, sieved and refrigerated at −20°C before feeding.
Table 1 Formulations of experimental diets
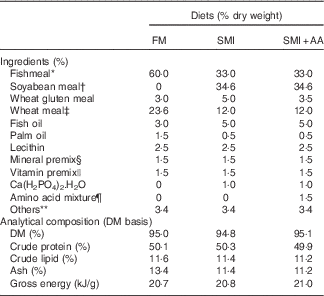
FM, fishmeal; SMI, soyabean meal-incorporated diet; SMI+AA, SMI diet with dietary essential amino acid supplementation.
* FM: steam-dried FM (Copeinca Group), with crude protein: 70·10 %, crude lipid: 7·58 %.
† Soyabean meal: dehulled, solvent-extracted soyabean meal (Great Seven Bio-tech), with crude protein: 54·61 %, crude lipid: 2·33 %.
‡ Wheat meal and wheat gluten meal act as a carbohydrate source and as a filler (Great Seven Bio-tech).
§ Mineral premix (mg/kg diet): CoCl2.6H2O (1 %), 50; CuSO4.5H2O (25 %), 10; FeSO4.H2O (30 %), 80; ZnSO4.H2O (34·50 %), 50; MnSO4.H2O (31·80 %), 45; MgSO4.7H2O (15 %), 1200; sodium selenite (1 %), 20; calcium iodine (1 %) 60; zeolite, 11 470.
|| Vitamin premix (mg/kg diet): thiamin (98 %), 25; riboflavin (80 %), 45; pyridoxine-HCl (99 %), 20; vitamin B12 (1 %), 10; vitamin K3 (51 %), 10; inositol (98 %), 800; pantothenic acid (98 %), 60; niacin acid (99 %), 200; folic acid (98 %), 20; biotin (2 %), 60; retinyl acetate (15 %), 32; cholecalciferol (1·25 %), 5; α-tocopherol (50 %), 240; ascorbic acid (35 %), 2000; antioxidants (oxygen ling grams, 100 %), 3; rice husk powder (100 %), 11 470.
¶ Amino acid mixture: l-lysine (coated amino acid, 60 %) 0·6 %, l-methionine (coated amino acid obtained, 90 %) 0·32 %, l-isoleucine (crystalline amino acid, 99·4 %) 0·12 %, l-leucine (crystalline amino acid, 99·4) 0·13 %, l-threonine (crystalline amino acid, 99·9 %) 0·13 %, l-valine (crystalline amino acid, 99·1 %) 0·16 %. Coated amino acid was obtained from Beijing XingHuo Yuan Science and Technology Co. Ltd and crystalline amino acid was obtained from Jizhou City Huayang Chemical Co. Ltd.
** Others: beer yeast 2 %, choline chloride 0·25 %, mould inhibitor 0·1 %, antioxidant 0·05 %, attractant (betaine–dimethylpropiothetin–glycine–alanine–inosine 5'-phosphate=4:2:2:1:1) 1·0 %.
Feeding trial and sampling
All experimental protocols were approved by the Animal Care Committee of Ocean University of China. Juvenile turbot were obtained from Haiyang fish farm (Haiyang, China). During the acclimatisation period, fish were fed a commercial diet (Great Seven Bio-tech) twice per day, for 2 weeks. To start the experiment, the juvenile turbot with an initial weight of 9·19 (sem 0·01) g were randomly distributed into tanks filled with 500 litres of seawater, with forty fish in each tank. Diets were randomly allocated in triplicate to the tanks. Fish were fed twice per day, until apparent satiation, for 30 d. During the experimental period, the water temperature was 16–20°C. The uneaten feed was collected 1 h after each meal, dried to a constant weight and weighed to allow calculation of food intake.
Before the feeding trial, twenty fish from the same population were randomly collected for initial biochemical analysis. At the end of the feeding trial, fish were fasted for 48 h to obtain the basal levels of metabolites in fish( Reference Seiliez, Panserat and Lansard 18 ). After 24 h of fasting, fish in each tank were weighed and counted, and four fish per tank were randomly selected for biochemical analysis. At the end of the 48-h fasting period, the sampled fish were designated as the 0-h sample (fasted fish). The fish were then fed a single meal of their allocated diet until visible satiation. Samples were collected at 2-, 8- and 24-h intervals after refeeding. At each interval, six fish (two fish per tank) were randomly taken from every treatment (n 6). Fish were anaesthetised with benzocaine (30 mg/l) and killed by cervical section. The stomach and gut contents of each fish were checked to ensure that the fish had effectively consumed the diet. The liver, intestine and dorso-lateral white muscle were immediately dissected and frozen in liquid N2 and kept at −80°C. The other six fish in each treatment were selected, anaesthetised, visually checked as to whether or not the belly was bulging greatly because of a full diet in the digestive tract and blood was extracted from the caudal vein into heparin anti-coagulation tubes and centrifuged at 3000 g for 5 min. The recovered plasma was kept at −80°C until analysis.
Biochemical analysis
DM (105°C for 24 h), crude protein (N×6·25, Kjeltec nitrogen analyserTM 8400; FOSS), crude lipid (ether extraction, Soxhlet method) and ash (combustion in a muffle furnace at 550°C for 12 h) of the experimental diets and fish samples were analysed as described elsewhere( Reference Liu, He and Wang 19 ). The gross energies of feed and fish samples were measured using an adiabatic bomb calorimeter (C2000; Ika Werke). Amino acids in ingredients and diets were analysed using an amino acid analyzer (L-8900; Hitachi). Plasma glucose (GLU) and TAG concentrations were measured using commercial kits according to the manufacturer’s instructions (Sysmex).
Free amino acid analysis
The postprandial free amino acid concentrations in the plasma and muscle were analysed by an automated amino acid analyser (L-8900) with a lithium high-performance column( Reference Mente, Deguara and Santos 20 ). Briefly, plasma (400 µl) was deproteinised by mixing it with 1·2 ml of 10 % sulfosalicylic acid solution and incubating it at 4°C for 5 min. After centrifugation at 13 000 rpm for 15 min, 1 ml of supernatant was filtered through 0·22-µm filters for free amino acid measurement; in addition, white muscle samples (400 mg) were homogenised in 1·2 ml of 10 % sulfosalicylic acid solution. After centrifugation at 13 000 rpm for 15 min, the supernatant was filtered through 0·22-µm filters for free amino acid measurement.
Quantitative real-time PCR
Total RNA was extracted from intestine, liver or muscle (approximately 50 mg) using Trizol reagent (Invitrogen) according to the manufacturer’s recommendations, quantified by a Nanodrop 2000 spectrophotometer (Thermo) and electrophoresed on a 1·2 % denaturing agarose gel to test the integrity. complementary DNA synthesis and quantitative real-time PCR (qRT-PCR) reactions were conducted as described previously(
Reference Zuo, Ai and Mai
21
). Specific primer sequences of target genes for qRT-PCR are listed in Table 2. qRT-PCR analyses were focused on the postprandial kinetics of peptide and amino acid transporters, which were peptide transporter 1 (PepT1) mediating the uptake of essentially all dipeptides and tripeptides(
Reference Romano, Barca and Storelli
22
); B0-type amino acid transporter 1 (B
0
AT1) transporting all neutral amino acids; sodium-coupled neutral amino acid transporter 2 (SNAT2) preferring alanine and other small and polar neutral amino acids; and y+L-type amino acid transporter 1 (y
+
LAT1) mediating the transport of cationic amino acids(
Reference Bröer
23
). We also examined the gene expression of several key enzymes of hepatic metabolism, which were as follows: glucokinase (GK) and pyruvate kinase (PK) for glycolysis; fructose 1,6-bisphosphatase (FBPase) and glucose 6 phosphatase (G6Pase) for gluconeogenesis; transcription factor sterol regulatory element-binding protein 1 (SREBP1) and fatty acid synthase (FAS) for fatty acid synthesis; diacylglycerol O-acyltransferase homolog (DGAT)1 and DGAT2 for TAG synthesis; and carnitine palmitoyltransferase 1 isoforms A (CPT1A) and acyl-CoA oxidase 1 (ACOX1) for fatty acid oxidation(
Reference Dai, Panserat and Plagnes-Juan
13
,
Reference Cunha, Galante-Oliveira and Rocha
24
). Results were normalised to reference genes RNA polymerase II subunit D (RPSD) for intestine and muscle samples, and elongation factor 1α (EF1α) for liver samples. No expression changes of RPSD and EF1α were observed in the corresponding tissues among treatments (online Supplementary Fig. S1). The expression levels of target mRNA were calculated using the comparative cycle threshold (C
t
) values expressed as
![]() $$2^{{\left^( {^{{{\minus}\Delta \Delta C_{t} ) \right $$
. Transcription levels were normalised by the reference gene. Gene expression was represented as fold change to the control (T 0 h FM).
$$2^{{\left^( {^{{{\minus}\Delta \Delta C_{t} ) \right $$
. Transcription levels were normalised by the reference gene. Gene expression was represented as fold change to the control (T 0 h FM).
Table 2 Primer sequences used for real-time quantitative PCRFootnote *

* Abbreviations and GenBank accession nos: ACOX1, acyl-CoA oxidase 1, KC189925; CPT1A, carnitine palmitoyltransferase 1 isoforms A, KC189926; DGAT1, diacylglycerol O-acyltransferase homolog 1, KC189938; DGAT2, diacylglycerol O-acyltransferase homolog 1, KC189939; EF1α, elongation factor-1 α, AF467776.1; FAS, fatty acid synthase, KC189927; FBPase, fructose 1,6-bisphosphatase, KC184130; G6Pase, glucose 6 phosphatase, KC184131; GK, glucokinase, JX678944; PK, pyruvate kinase, DQ848903; RPSD, RNA polymerase II subunit D, DQ848899.1; partial sequences of some target genes in turbot were obtained through a degenerate PCR strategy in this study, including B 0 AT1, B0-type amino acid transporter 1; PepT1, peptide transporter 1; SNAT2, sodium-coupled neutral amino acid transporter 2; SREBP1, sterol regulatory element-binding protein 1; y + LAT1, y+L-type amino acid transporter 1.
† For the nucleotide sequences of genes B 0 AT1, PepT1, SNAT2, SREBP1 and y + LAT1, see the online Supplementary material.
Protein extraction and Western blot analysis
Tissues (approximately 40 mg) were homogenised with Glass Tenbroeck Tissue Grinders (Kimble Chase) on ice and lysed in radioimmunoprecipitation assay buffer (50 mm-Tris, 150 mm-NaCl, 0·5 % Nonidet P-40, 0·1 % SDS, 1 mm-EDTA, pH 7·5) with protease and phosphatase inhibitors (Roche). Lysates were centrifuged at 12 000 g for 20 min at 4°C. Protein concentrations were determined using an Enhanced BCA Protein Assay Kit (Beyotime Biotechnology) with bovine serum albumin as standard. Lysates (10 µg of total protein/lane) were separated by SDS-PAGE and transferred to 0·45-µm polyvinylidene fluoride membranes (Millipore) for Western blot analysis. Primary antibodies against phospho-Akt (Ser473) (Cat. no. 9271), Akt (Cat. no. 9272), phospho-TOR (Ser2448) (Cat. no. 2971), TOR (Cat. no. 2972), phospho-S6 (Ser235/236) (Cat. no. 4856), S6 (Cat. no. 2217), phospho-4E-BP1 (Thr37/46) (Cat. no. 9459), 4E-BP1 (Cat. no. 9452), phospho-eIF2α (Ser51) (Cat. no. 3597) and eIF2α (Cat. no. 9722) were purchased from Cell Signaling Technology Inc. Anti-ATF4 antibody (Cat. no. sc-200) was purchased from Santa Cruz Biotechnology Inc. All antigenic regions of these antibodies have been reported as conserved in turbot( Reference Wang, He and Mai 25 ), and also successfully used in rainbow trout in previous studies( Reference Seiliez, Panserat and Lansard 18 , Reference Dai, Panserat and Mennigen 26 ). Next, membranes were incubated with HRP-labelled goat anti-rabbit IgG (H+L) second antibody (Beyotime Biotechnology), and developed using a Beyo ECL Plus kit (Beyotime Biotechnology). The density of protein bands was quantified using the NIH Image 1.63 software (n 6).
Statistical analysis
The SPSS 16.0 software was used for all statistical analysis. The data of growth performance were subjected to one-way ANOVA, followed by Tukey’s multiple range tests. The data for the time course and different diets were analysed by two-way ANOVA, testing the main effects of time (T) and diet (D), and their full-factorial interaction. Tukey’s multiple range tests were conducted in order to detect treatment differences among the interactions. In cases in which data were nonparametric or not homoscedastic, data transformations (such as logarithms, square roots and reciprocals) were used to meet ANOVA criteria. Normality was assessed using the Shapiro–Wilks’ test, whereas homoscedasticity was determined using Levene’s test. For all statistical analyses, the level of significance was set at P<0·05.
Results
Phenotypic characterisation of dietary performance
After the 1-month feeding trial, no significant differences were found for feed intake, whole-body protein content or percentage survival for all treatments (Table 3). Compared with the FM diet, two SMI diets (SMI and SMI+AA) resulted in decreased specific growth rate (P<0·001), feed efficiency (P=0·01), protein (P=0·004), fat (P=0·001) and energy (P=0·004) retention, and whole-body fat content (P=0·001) (Table 3). No significant differences were found between the SMI and SMI+AA diets for any of these measures.
Table 3 Growth performance and nutrient utilisation of turbot after 30-d diet feeding trial (Mean values with their standard errors; n 3)

FM, fishmeal diet; SMI, soyabean meal-incorporated diet; SMI+AA, SMI diet with dietary essential amino acids supplementation.
a,b Mean values within a row with unlike superscript letters were significantly different (P<0·05).
* Specific growth rate (%/d)=100×(ln final body weight−ln initial body weight)/d.
† Feed intake (% body weight/d)=100×feed consumption/(30 d×(initial body weight+final body weight)/2).
‡ Feed efficiency=wet weight gain (g)/total feed consumed (g).
§ Nutrient retention=(100×(final body weight×final carcass nutrient content)−(initial body weight×initial carcass nutrient content)/nutrient intake, where nutrient refers to protein, lipid and energy.
|| Percentage survival=100×(final fish number/initial fish number).
Postprandial modulations of amino acid transporters by dietary proteins
Postprandial gene expressions of major amino acid transporters in the intestine were measured (Fig. 1). After refeeding, the expression levels of intestinal PepT1 and y + LAT1 were significantly increased and peaked at 2 and 8 h, respectively, whereas the mRNA levels of B 0 AT1 and SNAT2 were markedly decreased (Fig. 1). Compared with the FM diet, SMI and SMI+AA diets did not significantly influence the postprandial gene expressions of PepT1 or SNAT2. However, the SMI diet significantly decreased the peak level of y + LAT1 and reduction of B 0 AT1 after refeeding in the intestine (Fig. 1). Supplementation of EAA to the SMI diet significantly shifted the postprandial gene expression pattern of B 0 AT1 to that of the FM diet, but it showed no significant impact on postprandial peak values of y + LAT1 (Fig. 1).

Fig. 1 Postprandial expressions of peptide and amino acid transporters in intestine of juvenile turbot. Values are means (n 6), with their standard errors and were analysed by two-way ANOVA followed by Tukey’s multiple range test. a,b,c,d Mean values among all treatments with unlike letters are significantly different when the interaction was significant (P<0·05). A,B,C,D Mean values among four time points with unlike letters were significantly different (P<0·05). T, time points; D, diets; T×D, interaction between T and D; fishmeal diet (![]() ); SMI, soyabean meal-incorporated diet (
); SMI, soyabean meal-incorporated diet (![]() ); soyabean meal-incorporated diet with dietary essential amino acids supplementation (
); soyabean meal-incorporated diet with dietary essential amino acids supplementation (![]() ); SQRT indicates that data were transformed and statistically analysed with square roots; B
0
AT1, B0-type amino acid transporter 1; PepT1, peptide transporter 1; SNAT2, sodium-coupled neutral amino acid transporter 2; y
+
LAT1, y+L-type amino acid transporter 1; RPSD, RNA polymerase II subunit D.
); SQRT indicates that data were transformed and statistically analysed with square roots; B
0
AT1, B0-type amino acid transporter 1; PepT1, peptide transporter 1; SNAT2, sodium-coupled neutral amino acid transporter 2; y
+
LAT1, y+L-type amino acid transporter 1; RPSD, RNA polymerase II subunit D.
Postprandial free amino acid influx was modulated by dietary proteins
The postprandial kinetics of the free amino acid concentrations in plasma and muscle are shown in Tables 4 and 5, and in online Supplementary Tables S2 and S3. Plasma free amino acid concentrations were markedly increased and peaked 2–8 h after refeeding, before returning to basal levels. Compared with the FM diet, the SMI diet provided significantly lower free amino acids entering into plasma, whereas dietary EAA supplementation fully compensated for these deficits (Table 4 and online Supplementary Table S2). Postprandial free amino acid concentrations in muscle were markedly increased and peaked 8 h after refeeding. The SMI diet led to significantly reduced postprandial peak values of free amino acids and shorter duration of total EAA concentrations in muscle. EAA supplementation in the SMI diet did not significantly affect postprandial peak values of the majority of free amino acids in muscle (Table 5 and online Supplementary Table S3).
Table 4 Changes of plasma individual free essential amino acid (EAA) concentrations in turbot after refeeding (µg/µl) (Pooled standard errors)

T, time points; TAA, total free amino acids; FM, fishmeal diet; SMI, soyabean meal-incorporated diet; SMI+AA, SMI diet with dietary EAA supplementation; D, diet; LG10 indicates that data were transformed and statistically analysed with log transforms; SQRT indicates that data were transformed and statistically analysed with square roots.
a,b,c,d,e,f,g,h Mean values among all treatments within a row with unlike superscript letters were significantly different (P<0·05).
* Treatment means represent the average values for three tanks per treatment and were analysed by two-way ANOVA (n 3). Tukey’s test was conducted for individual means only if there was a significant interaction (P<0·05).
Table 5 Changes of muscle individual free essential amino acid (EAA) concentrations in turbot after refeeding (µg/g) (Pooled standard errors)

T, time points; TAA, total free amino acids; FM, fishmeal diet; SMI, soyabean meal-incorporated diet; SMI+AA, SMI diet with dietary EAA supplementation; LG10 indicates that data were transformed and statistically analysed with log transforms; SQRT indicates that data were transformed and statistically analysed with square roots; D, diet.
a,b,c,d,e,f,g,h Mean values among all treatments within a row with unlike superscript letters were significantly different (P<0·05).
* Treatment means represent the average values for three tanks per treatment and were analysed by two-way ANOVA (n 3). Tukey’s test was conducted for individual means only if there was a significant interaction (P<0·05).
Postprandial modulations of amino acid-sensing pathways by dietary proteins
Postprandial activation of amino acid-sensing pathways was examined in muscle (Fig. 2(A)) and liver (Fig. 2(B)). Two-way ANOVA statistical analysis showed that the phosphorylation of TOR and AAR pathways were both significantly affected by refeeding time courses and different diets (P<0·01). The TOR pathway was markedly activated by refeeding both in muscle and liver, characterised by the phosphorylation of protein kinase B (Akt) on Ser473, TOR on Ser2448, ribosomal protein S6 (S6) on Ser235/236 and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) on Thr37/46 (Fig. 2). Contrarily, the AAR-related molecule eukaryotic initiation factor 2 α (eIF2α) on Ser51 was markedly activated during starvation (time 0 h) and inhibited after refeeding in all three diets. Compared with the FM diet, the SMI diet led to significantly lower levels and shorter duration of phosphorylation of Akt, TOR, S6 and 4E-BP1 in both muscle and liver tissues (Fig. 2). However, the SMI diet significantly induced stress-responsive eIF2α phosphorylation and activating transcription factor 4 (ATF4) (Fig. 2). Supplementation of EAA to the soyabean diet did not have a significant effect on the activation of these nutrient-sensing molecules except for phosphor-eIF2α in liver and muscle, and ATF4 levels in liver (Fig. 2).

Fig. 2 Dietary modulations of nutrient-sensing responses involving the total and phosphorylation levels of proteins related to target of rapamycin (TOR) and amino acid response signalling pathways in muscle (A) and liver (B). A representative blot is shown from replicated examinations (n 6). Values are means with their standard errors and were analysed by two-way ANOVA followed by Tukey’s multiple range test. a,b,c,d,e,f,g,h; A,B,C; x,y,z Values with unlike letters are significantly different (P<0·05). FM, fishmeal diet (![]() ); SMI, soyabean meal-incorporated diet (
); SMI, soyabean meal-incorporated diet (![]() ); SMI+AA, SMI diet with dietary essential amino acids supplementation (
); SMI+AA, SMI diet with dietary essential amino acids supplementation (![]() ); T, time points; D, diets; T×D, interaction between T and D; LG10 indicates that data were transformed and statistically analysed with log transforms; Akt, protein kinase B; S6, ribosomal protein S6; 4E-BP1, eukaryotic initiation factor 4E-binding protein 1; eIF2α, eukaryotic initiation factor 2α; ATF4, activating transcription factor 4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase (see Fig. 1 legend for details).
); T, time points; D, diets; T×D, interaction between T and D; LG10 indicates that data were transformed and statistically analysed with log transforms; Akt, protein kinase B; S6, ribosomal protein S6; 4E-BP1, eukaryotic initiation factor 4E-binding protein 1; eIF2α, eukaryotic initiation factor 2α; ATF4, activating transcription factor 4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase (see Fig. 1 legend for details).
Postprandial modulations of the metabolism by dietary proteins
Postprandial glucose and TAG reached peak concentration at 2 and 8 h, respectively, after refeeding. However, the peak values of both GLU and TAG were lower in SMI diets (SMI and SMI+AA), compared with those in the FM group (Fig. 3(A)).

Fig. 3 The dietary modulations of postprandial metabolism. (A) Plasma glucose (GLU) and TAG levels. Expression of selected enzymes involved in (B) glycolysis, (C) gluconeogenesis, (D) lipogenesis, (E) TAG synthesis and (F) fatty acid oxidation were analysed (n 6). Values are means with standard errors and were analysed by two-way ANOVA followed by Tukey’s multiple range test. a,b,c,d,e,f,g,h; A,B,C; x,y,z Values with unlike letters are significantly different (P<0·05). Fishmeal diet (![]() ); soyabean meal-incorporated diet (
); soyabean meal-incorporated diet (![]() ); soyabean meal-incorporated diet with dietary essential amino acid supplementation (
); soyabean meal-incorporated diet with dietary essential amino acid supplementation (![]() ); GK, glucokinase; EF1α, elongation factor 1α; PK, pyruvate kinase; FBPase, fructose 1,6-bisphosphatase; G6Pase, glucose 6 phosphatase; FAS, fatty acid synthase; SREBP1, sterol regulatory element-binding protein 1; DGAT, diacylglycerol O-acyltransferase homolog; ACOX1, acyl-CoA oxidase 1; CPT1A, carnitine palmitoyltransferase 1 isoforms A (see Fig. 1 legend for details).
); GK, glucokinase; EF1α, elongation factor 1α; PK, pyruvate kinase; FBPase, fructose 1,6-bisphosphatase; G6Pase, glucose 6 phosphatase; FAS, fatty acid synthase; SREBP1, sterol regulatory element-binding protein 1; DGAT, diacylglycerol O-acyltransferase homolog; ACOX1, acyl-CoA oxidase 1; CPT1A, carnitine palmitoyltransferase 1 isoforms A (see Fig. 1 legend for details).
The gene expressions of key metabolic enzymes in the liver were determined. In the FM group, the expressions of enzymes for glycolysis (GK and PK) (Fig. 3(B)), fatty acid synthesis (SREBP1 and FAS) (Fig. 3(D)) and TAG synthesis (DGAT1 and DGAT2) (Fig. 3(E)) were significantly up-regulated and peaked at 2–8 h after refeeding. The gene expressions of gluconeogenic enzymes (FBPase and G6Pase) were markedly down-regulated (Fig. 3(C)). In contrast, these postprandial modulations were not obvious in the two SMI diets with lower transcription levels than those of the FM diet. No significant difference was observed for the gene expression involved in fatty acid oxidation (CPT1A and ACOX1) among different treatments (Fig. 3(F)).
Discussion
Turbot is an aquaculture species that is highly sensitive to the replacement of dietary FM by plant sources of protein. In previous reports, FM replacements over 20 % using maize gluten meal( Reference Regost, Arzel and Kaushik 27 ) and over 25 % using soyabean meal concentrate( Reference Day and GonzÁlez 28 ) were found to reduce growth rates in turbot. In accordance with these studies, we observed that 45 % dietary FM replacement by soyabean meal reduced growth, feed utilisation and nutrient retention, irrespective of EAA supplementation.
Cellular uptake and transport of peptide and amino acids in the intestine represent a critical step for protein absorption. PepT1 is the major peptide transporter across the intestine brush border membrane in fish( Reference Verri, Romano and Barca 29 ). We observed that PepT1 expression peaked at 2 h after refeeding, which is consistent with previous reports stating that the expression of PepT1 was up-regulated by refeeding and down-regulated by fasting in sea bass( Reference Terova, Corà and Verri 30 ). B 0 AT1 and y + LAT1 were also identified in fish( Reference Margheritis, Terova and Cinquetti 31 , Reference Yang, Tan and Zhu 32 ) and had a major role in mediating EAA transport across the apical and basolateral sides of the brush border membrane, respectively( Reference Bröer 23 , Reference Bröer, Juelich and Vanslambrouck 33 ). Intestinal B 0 AT1 was down-regulated, whereas y + LAT1 peaked at 8 h after refeeding, highlighting their different mechanisms in response to feed ingestion. SNAT2, a proven amino acid availability sensor in mammals( Reference Hyde, Cwiklinski and MacAulay 34 ), was down-regulated after refeeding, which is similar to observations from mammalian studies( Reference Hyde, Christie and Litherland 35 ). Among the transporters characterised, gene expression of PepT1 and SNAT2 levels was not influenced by different diets. However, B 0 AT1 and y + LAT1 were less responsive to the SMI diet than the FM diet, suggesting that different protein sources resulted in differential responses of amino acid transporters, which in turn would possibly have influenced amino acid absorption and transport efficiency, accompanied by the change of plasma and muscle free amino acid pools in the present study.
Our results demonstrated that postprandial peak values of plasma free amino acids in circulation occurred 2 h after refeeding and were transported to muscle in <8 h. This was similar to observations of rainbow trout( Reference Karlsson, Eliason and Mydland 36 ), but slower than those of rats( Reference Vary and Lynch 37 ). Postprandial peak values of free amino acids in the SMI group were much lower than in the FM group. Supplementation of EAA to match the amino acid profile of the FM diet in the SMI diets increased postprandial free amino acid levels in plasma, but not in muscle. A similar result was found in turbot after partial FM replacement by maize gluten meal( Reference Regost, Arzel and Kaushik 27 ). Crystalline amino acids were reported to have a lower retention in muscle compared with protein-bound amino acids( Reference Berge, Lied and Espe 38 ). This asynchronous amino acid retention may explain the inefficiency of free amino acid supplementation in the SMI diet.
Intracellular sensing of amino acid availability is mediated mainly by two distinct, yet complimentary, pathways – the AAR and the TOR pathways – both of which regulate protein synthesis and metabolism( Reference Laplante and Sabatini 8 , Reference Kilberg, Shan and Su 39 ). TOR activation is the primary driving force for postprandial anabolism( Reference Kimball and Jefferson 6 , Reference Ma and Blenis 7 ) and is required for postprandial protein synthesis in response to amino acid availability( Reference Dickinson and Rasmussen 40 ). Consistent with findings in mammals( Reference Laplante and Sabatini 8 ) and rainbow trout( Reference Seiliez, Gabillard and Skiba-Cassy 41 ), we observed that refeeding activated TOR signalling. A previous study reported that increased amino acid levels enhanced S6 kinase 1 and S6 phosphorylation in rainbow trout primary hepatocytes( Reference Dai, Panserat and Plagnes-Juan 13 ). In the present study, SMI diets reduced the postprandial peak values of free amino acids compared with the FM diet. Accordingly, it reduced the levels and duration of postprandial phosphorylation of TOR, Akt, S6 and 4E-BP1, suggesting a hypoactivated TOR signalling status. However, the SMI diet induced higher eIF2α phosphorylation and ATF4 levels, indicating hyperactivated AAR signalling and inhibited cellular protein synthesis( Reference Kilberg, Shan and Su 39 ). In particular, AAR enhanced total 4E-BP1 in muscle during fasting and decreased its phosphorylation in FM replacement diets, further inhibiting protein translation in cells( Reference Sayano, Kawakami and Kusada 42 ). The combination of hypoactivated TOR signalling and hyperactivated AAR signalling would reduce postprandial protein synthesis( Reference Laplante and Sabatini 8 , Reference Kilberg, Shan and Su 39 ). Chronically, it would lead to lower protein accretion, evidenced by decreased protein retention after 1 month of SMI diet feeding.
TOR and AAR signalling not only control protein synthesis but also regulate metabolic gene expression, both in mammals and in fish( Reference Guo and Cavener 11 , Reference Dai, Panserat and Mennigen 26 , Reference Yecies and Manning 43 , Reference Dibble and Manning 44 ). In the present study, postprandial increased expressions of key enzymes involved in glycolysis and lipogenesis were associated with the activation of TOR signalling after refeeding, which is consistent with previous reports demonstrating that postprandial activation of hepatic GK and lipogenesis require TOR activation in rainbow trout( Reference Dai, Panserat and Mennigen 26 ). Furthermore, we observed that SMI diets reduced tissue amino acid concentrations and TOR signalling activities, and suppressed the expression of genes involved in glycolysis and lipogenesis. As previous findings in trout hepatocytes showed that increased amino acid availability effectively up-regulated fatty acid synthetic and glycolytic genes expression in a TOR-dependent manner( Reference Dai, Panserat and Plagnes-Juan 13 , Reference Lansard, Panserat and Plagnes-Juan 45 ), we presume that SMI diets reduced postprandial tissue amino acid concentration, which in turn suppressed TOR activation and subsequently down-regulated fatty acid synthetic and glycolytic gene expression. However, the soyabean meal-induced increase of eIF2α phosphorylation and ATF4 levels was associated with down-regulation of hepatic lipogenic gene expression. These results were compatible with previous reports, concerning mice, that activation of the AAR signalling pathway reduced gene expression involved in fatty acid and TAG synthesis, alongside liver TAG and adipose tissue weight( Reference Guo and Cavener 11 ). Thus, the reduced postprandial lipogenesis gene expression in SMI groups may be related to the combination effects of hypoactivated TOR signalling and hyperactivated AAR signalling. Chronically, it would lead to a lower body fat content of turbot. Therefore, our study provides a reasonable mechanistic explanation for reduced lipogenesis and fat deposition after FM replacement in many studies( Reference Dias, Alvarez and Arzel 46 , Reference Deng, Mai and Ai 47 ).
It has been widely accepted that FM replacement in aquafeeds by plant proteins is limited by factors including low digestibility, imbalanced amino acid profile, anti-nutritional factors, presence or absence of other factors and so on, in plant proteins( Reference Naylor, Hardy and Bureau 1 , Reference Hardy 2 , Reference Gatlin, Barrows and Brown 17 ). However, in fish fewer studies have compared the responses towards different protein sources at the molecular level, nor provided mechanistic explanations of the dietary effects. Our results, together with previous reports( Reference Seiliez, Panserat and Lansard 18 , Reference Dai, Panserat and Mennigen 26 , Reference Seiliez, Gabillard and Skiba-Cassy 41 ), demonstrated that nutrient-sensing networks were conserved in fish, although varied in some aspects, such as the metabolism of glutamine in muscle( Reference Ballantyne 48 ). The present study demonstrated a cascade of sophisticated, but mechanistically connected, responses towards partial FM replacement by soyabean meal in turbot: reduced postprandial influx of free amino acids led to hypoactivated TOR signalling, which in turn reduced protein synthesis and lipogenesis. In contrast, induced AAR responses further aggravated the postprandial anabolism. Such postprandial changes would lead to differences of growth and feed efficiency after long-term feeding. Further understanding the molecular responses of animals to different diets should pave the way for better feed utilisation.
Acknowledgements
The authors thank the Haiyang fish farm for providing the aquaculture system and animal care direction, and technicians at the South China Agricultural Science and Technology Innovation Center for amino acid analysis guidance.
This work was supported by the National Natural Science Foundation of China (grant number 31222055) and the National Key Basic Research Program of China (grant number 2014CB138602). These funders had no role in the design and analysis of the study, nor in the writing of this article.
G. H. and K. M. designed the study. D. X., F. S., H. Z. and W. X. performed data acquisition and data analysis. G. H. and D. X. wrote the manuscript. All authors read and approved the final manuscript.
The authors declare no conflicts of interest.
Supplementary material
For supplementary material/s referred to in this article, please visit http://dx.doi.org/doi:10.1017/S0007114515004535











