Clostridium difficile is the leading cause of healthcare-associated diarrhoea in adults and the aetiological agent of antibiotic-associated colitis [Reference Rupnik, Wilcox and Gerding1]. During the last decade, C. difficile infection (CDI) epidemiology has experimented significant changes (i.e. emergence of hypervirulent strains) that have led to increased severity and mortality rates, reaching as high as 15% in some outbreaks [Reference Kuijper, Coignard and Tull2].
In Chile, two early studies in 2000 demonstrated that CDI occurred with similar frequencies as in developed countries of North America and the European Union [Reference Alvarez3, Reference Herrera4], yet the toxinotype and ribotype of the clinical isolates was not analysed. Recently, we have reported the presence of the epidemic ribotype 027 in Chile causing two severe clinical cases of CDI, one of which ended in colectomy [Reference Hernandez-Rocha5]. Simultaneously, the largest outbreak of CDI in Chile occurred in 2011 at the Dr. Alejandro del Río emergency hospital in Santiago, infecting 186 patients over an 8-month period (June 2011 to April 2012) [Reference Sociedad Chilena de6]. This outbreak was caused by hypervirulent ribotype 027 stains and presented a mortality rate that reached a disturbing 20% [Reference Sociedad Chilena de6]. Here, we report the most common PCR ribotypes of C. difficile over an 11-month period (July 2011 to June 2012) from a university hospital in Santiago, Chile.
Specimens were obtained from the Hospital Clínico de la Pontificia Universidad Católica de Chile. This is a teaching tertiary-level healthcare centre with 450 beds, of which 60 correspond to critical care services. During an 11-month period (July 2011–June 2012), liquid or soft stool specimens from 392 hospitalized patients suspected of having CDI were analysed by multiplex real-time PCR GeneXpert (Cepheid, USA). Eighty-one patients were positive for C. difficile and all positive samples were also positive for toxigenic culture of C. difficile, causing cytophatic effects in monolayers of Vero cells, as described previously [Reference Hernandez-Rocha7]. An average of 7·4 cases of CDI per month was observed, with the lowest and highest incidence observed in July 2011 with zero cases and October of 2011 with 16 cases, respectively. The incidence rate of CDI during this period was 2·6 CDI cases/1000 discharges, which is fivefold lower than national rate of CDI cases reported in USA [Reference Evans and Safdar8] but sixfold higher than those results reported in a multicenter European study [Reference Bauer9]. The mean age of the study population was 58·2 ± 2·7 years, and 66% of the patients received antibiotics (i.e. cephalosporins, β-lactams, carbapenems, quinolones, macrolides, clindamycin, metronidazole, vancomycin), and no mortality associated with CDI was reported.
PCR ribotyping was performed according to a previously described method [Reference Bidet10] with minor modifications. In brief, DNA of three colonies from each stool sample was isolated [Reference Hernandez-Rocha5] and subjected to standard ribotyping using C. difficile 16S rRNA and 23S rRNA primers [Reference Bidet10]. PCR reaction was done by adding 1 µl crude template DNA diluted 1:10 to a 25 µl PCR mixture containing 10 mm Tris-HCl (pH 8·3) and 50 mm KCl (GeneAmp 1 × PCR Buffer II; Applied Biosystems, USA), 4·0 mm MgCl2, 0·4 mm dNTP, 1·5 U Taq DNA polymerase (Fermentas, USA). The cycling conditions were 3 min at 94 °C, followed by 35 cycles of denaturation at 94 °C for 60 s, annealing at 57 °C for 60 s, and elongation at 72 °C for 1 min. A final elongation step of 5 min at 72 °C was performed. PCR products were analysed on 5% non-denaturing polyacrylamide gels for 45 min at 150 V and stained with ethidium bromide. Images were analysed using Gel Compare II Software (Applied Maths, Belgium). Band patterns were compared with those of the C. difficile Ribotype library at TechLab (USA) and at Leeds Institute of Molecular Medicine, University of Leeds (UK). A total of 27 PCR ribotypes were identified, the most abundant being 012 (14·8%), 027 (12·3%), 046 (12·3%), 014/020 (9·9%), 001 (4·9%), and 023 (4·9%) (Fig. 1a, b ). Three unidentified ribotypes infecting a total of four patients were sent to the Anaerobe Reference Laboratory for confirmation. Ribotypes 027, 082, 097 and 251 caused relapsing CDI in four patients (4·9% of CDI patients).
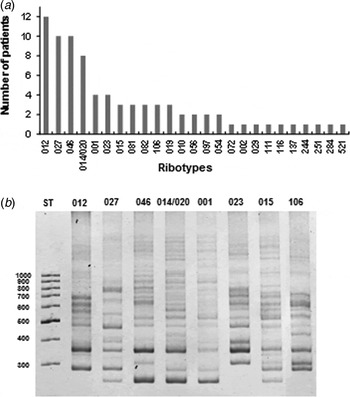
Fig. 1. (a) Frequency of C. difficile PCR ribotypes of 81 clinical isolates isolated between July 2011 and June of 2012 from a university hospital. (b) PCR ribotyping results show the band pattern of the ribosomal intergenic space regions of clinical isolates of the most frequent C. difficile clinical isolates. ST is 1000-bp standard.
Presence of tcdB and binary toxin was determined using GeneXpert PCR (Cepheid). C. difficile tcdA was detected by PCR amplification of a 3087 bp fragment between 5090 bp and 17 bp downstream of the start and stop codon of tcdA, respectively, as described previously [Reference Rupnik11]. All 81 isolates were positive for tcdB, while 10 tcdA−tcdB+ variants were detected in ribotypes 012 (1/12), 027 (1/10), 046 (1/10), 014/020 (2/8), 001 (1/4), 023 (1/4), 106 (1/3), 019 (1/3) and 137 (1/1). Twenty-two clinical isolates were positive for binary toxin, i.e. 012 (1/12), 27 (6/10), 046 (2/10), 014/020 (2/8), 082 (1/3), 019 (2/3), 106 (1/3), 081 (1/3), 010 (1/2), 054 (2/2), 111 (1/1), 116 (1/1), 521 (1/1). Interestingly, tcdA−tcdB+cdtAB+ variants occurred in ribotypes 012 (1/12), 014/020 (2/8), 027 (1/10), 046 (1/10); the majority of these ribotypes are described as tcdA+tcdB+cdtA− , while ribotype 027 is designated tcdA+tcdB+cdtAB+ .
Antimicrobial susceptibility testing to calculate the minimum inhibitory concentration of all clinical isolates was performed with Etest strips (AB Biodisk, USA) as described previously [Reference Oka12]. Antibiotic resistance to amoxicillin (Mox), rifaximin (Rif), erythromycin (Ery) and clindamycin (Clin), but not to vancomycin and metronidazole was observed (Table 1). Only eight isolates of ribotype 012 were resistant to Mox, Rif and Clin, of which only four were resistant to Ery (Table 1). Only four isolates of ribotype 027 were resistant to Mox, Ery and Clin (Table 1). Three strains of ribotype 046 were resistant to Mox and Rif, while only one exhibited resistance to Mox and Ery (Table 1). Ribotype 072 strain showed resistance to Mox, Ery and Clin, while ribotype 010 isolates had resistance only to Ery and Clin (Table 1). No strains belonging to ribotype 014/020 exhibited resistance to the tested antibiotics (Table 1). None of the tcdA−tcdB+cdtAB+ variants that occurred in ribotypes 012 (1/12), 014/020 (2/8), 027 (1/10), 046 (1/10) exhibited antibiotic resistance.
Table 1. Ribotypes with antibiotic resistance
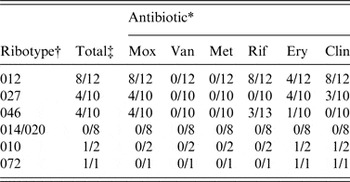
* Resistance to antibiotics was considered at a minimum inhibitory concentration of: Mox (amoxicillin) ⩾8 µg/ml; Van (vancomycin) ⩾32 µg/ml; Met (metronidazole) ⩾32 µg/ml; Rif (rifaximin) ⩾32 µg/ml; Ery (erythromycin) ⩾8 µg/ml; Clin (clindamycin) ⩾8 µg/ml.
† Ribotypes the four most common ribotypes as well as those with presence of antibiotic-resistant isolates are shown.
‡ Number of resistant and total isolates per ribotype are given.
We showed that the most common ribotypes between 2011 and 2012 at this particular hospital in Santiago, Chile, were 012 followed by 027, 046 and 014/020. This situation differs from that reported at the Dr. Alejandro del Río Hospital, where an outbreak that affected 186 patients was caused by hypervirulent ribotype 027. The low incidence of CDI in the study hospital could be due to the implementation of the measures suggested by the Society for Healthcare Epidemiology of America (SHEA) [Reference Cohen13]. The prevalence of ribotype 012 is as intriguing as in Costa Rica, where an outbreak associated with a C. difficile ribotype 012 variant, with increased virulence, was recently reported [Reference Quesada-Gomez14]. Ribotypes 012, 014/020 and 027 are also prevalent in European countries [Reference Bauer9, Reference Argamany15, Reference Freeman16]. The prevalent ribotype 046 found in this study has also been shown to be prevalent in neonatal pigs [Reference Noren, Johansson and Unemo17] and in hospitals in Poland [Reference Pituch18]. Chile, does not currently have a national surveillance policy for C. difficile; therefore, this report complements a recent report published by the Institute of Public Health of Chile concerning the rapid spread of NAP1/027/ST1 in Chilean public hospitals [Reference Aguayo19].
ACKNOWLEDGEMENTS
This work was supported by a grant from the Fondo Nacional de Ciencia y Tecnología de Chile FONDECYT Regular 1151025 to D.P-S.; Fondo Nacional de Innovación en Salud CONICYT-FONIS SA12|2197, FONDECYT Regular 1131012 to M.A-L; FONDECYT Inicio 1130502 to C.H-R.
Angela Plaza-Garrido is a PhD student in the Molecular Biosciences programme at the Gut Microbiota and Clostridia Research Group at Universidad Andrés Bello.
DECLARATION OF INTEREST
None.





