Introduction
The protein TNF-alpha-induced protein 3 (TNFAIP3, also known as A20) is a potent suppressor of inflammatory responses [Reference Ma and Malynn1]. Structurally, it contains an amino-terminal ovarian tumor-like unit (OTU) domain capable of deubiquitinating (DUB) activity, while the fourth carboxyl-terminal zinc finger (ZnF) domain contains its E3 ubiquitin ligase activity [Reference Bosanac2]. The seventh ZnF has recently been shown to bind to linear ubiquitin motifs, which is important for its regulatory function downstream of TNF-receptor signaling (Fig. 1) [Reference Tokunaga3,Reference Verhelst4]. These functions allow A20 to abrogate inflammation through modifying the ubiquitination of specific proteins in the signaling pathways of TNFα [Reference Lee5], toll-like receptor (TLR) [Reference Boone6], nucleotide-binding oligomerization domain-containing protein 2 (NOD2) [Reference Hitotsumatsu7], and interleukin 1 receptor (IL1R) [Reference Heyninck and Beyaert8]. A20-deficient mice demonstrate hypersensitivity to TNFα and die perinatally from spontaneous multi-organ inflammation [Reference Lee5], whereas A20ZF4 or A20°TU knock-in mice develop pronounced response to TNFα injection and dextran sodium sulfate (DSS)-induced colitis [Reference Lu9]. Finally, mice harboring tissue-specific A20 deficiencies develop distinct abnormal pathologies including polyarthritis [Reference Matmati10], colitis [Reference Hammer11,Reference Vereecke12], lupus-like systemic condition [Reference Kool13], keratinocyte hyperproliferation [Reference Lippens14], and intestinal polyposis [Reference Shao15].
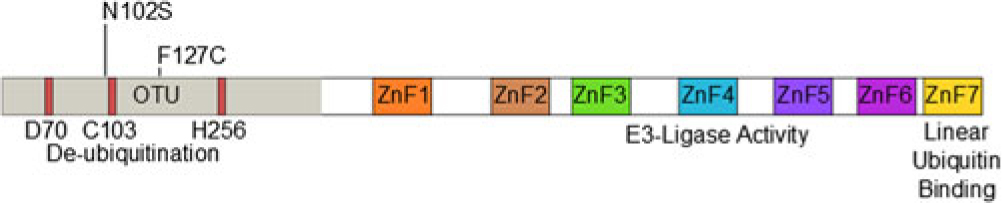
Fig. 1. Domain structure of A20 and SNP locations.
Given A20’s regulatory role on inflammation and the NF-κB pathway, its implications in immune-related disorders are not unexpected. Genetic studies have linked A20 polymorphisms to the development of multiple chronic inflammatory conditions, including rheumatoid arthritis (RA) [Reference Plenge16,Reference Thomson17], celiac disease [Reference Trynka18–Reference Dubois20], inflammatory bowel disease (IBD) [Reference Majumdar, Ahuja and Paul21–23], systemic lupus erythematosus (SLE) [Reference Adrianto24–Reference Kim26], systemic sclerosis [Reference Dieude27], Sjogren’s syndrome [Reference Musone28], psoriasis [Reference Nair29], and Behcet’s disease [Reference Li30]. Many point mutations cause detrimental alterations to protein function or expression, diminishing A20’s capacity to abolish inflammation [Reference Lodolce31–Reference Zhou33].
The Los Angeles County Health System and the Keck Hospital of the University of Southern California (USC) serve the region’s diverse racial and ethnic population – a significant percentage consisting of first- or second-generation immigrants. Over 60% of the patients self-report a Hispanic background, providing the unique opportunity to study genetic polymorphisms in this understudied population. In this study, we sequenced the exons encoding A20 of over 700 patients who received care in our clinics. We also evaluated the minor allele frequency (MAF) of a well-studied single-nucleotide polymorphism (SNP). Among the Hispanic patients, we identified two SNPs with significantly different MAF than reported in the literature among Caucasians. Subsequent in-vitro studies confirm that these SNPs inhibit the ability of A20 to regulate inflammation, suggesting that they may carry critical clinical implications.
Methods
Study Population
This study was approved by the institutional review board of the USC (Protocol no. IRB #HS-09-00543). Patients who received care at the Gastroenterology clinics of the Los Angeles County hospital (LAC+USC) were recruited randomly. All patients were recruited when they presented for routine medical care and had a variety of reasons for being in clinic. Buccal swabs containing DNA were obtained from those who agreed to participate and provided written informed consent, along with their self-identified gender (male or female), race/ethnicity (Caucasian, Hispanic, Middle Eastern, African-American, Asian, or other), and whether they were born abroad (yes or no).
Sequencing and Genotyping
Genomic DNA was extracted from buccal swabs using QuickExtract DNA Extraction Solution (Epicentre, Madison, WI, USA). Briefly, swabs were immersed in 500 µL of extraction solution. Tubes were vortexed for 10 s and then incubated at 65°C for 1 min, then mixed again for 15 s, and finally incubated at 98C for 2 min. DNA samples were stored at −70°C. DNA concentrations were measured by Nanodrop (Thermofisher, Waltham, MA, USA). For rs146534657 (NM_001270507.1) and rs2230936 (NM_001270507.1), genomic DNA samples (∼1 ug) were used to amplify exon 3 of TNFAIP3 using the following primers: exon 3 forward 5’-TTGCTGGGTCTTACATGCA G-3’ and exon 3 reverse 5’-CCCACCATGGAGCTCTGTTA -3’. PCR primers used to sequence the remaining exons of A20 were published previously [Reference Zhu34]. PCR products were then enzymatically digested with exonuclease I and shrimp alkaline phosphatase (both from NEB, Ipswich, MA, USA) to prepare samples for direct Sanger sequencing using the exon 3 reverse primer. For rs6920220 (NC_000006.11), genomic DNA samples were subjected to qPCR using specific Taqman primers and probes (Applied Biosystems, Foster City, CA, USA).
In-Vitro Experiments
Human A20 cDNA was obtained from Dharmacon (Lafayette, CO, USA) and cloned into a mammalian expression vector (pCMV, Agilent Santa, Clara, CA, USA). A20 with the rs146534657:A>G (p.N102S) SNP, or A20 with the rs2230926:T>G (p.F127C) SNP were constructed from the wild-type A20 plasmid by QuikChange Lightning site-directed mutagenesis (Agilent, Santa Clara, CA, USA). Wild-type, p.N102S, or p.F127C A20 constructs were transfected into A20 knockout 293 cells, generated previously [Reference Nakamura35] containing the NF-κB Firefly luciferase reporter gene at concentrations of 10, 50, 100, or 500 ng. Total plasmid concentrations were normalized using empty pCMV vector. Cells were subsequently stimulated with TNFα at a concentration of 10 ng/mL for 8 h, and luciferase activity was assayed and normalized to Renilla luciferase. Protein levels of wild-type A20, and the A20 mutant-types p.N102S and p.F127C obtained from the transfected A20 knockout 293 cells, were quantified by Western blot using antibodies toward A20 (Cell Signaling, Danvers, MA, USA) and normalized to GAPDH (Merck Millipore, Darmstadt, Germany).
Statistical Analysis
Comparative statistics was performed with Student’s t-test for continuous variables, and chi-squared test for categorical variables. All analyses were executed on IBM SPSS Statistics for Windows, version 25 (IBM Corp., Armonk, NY, USA).
Results
Participant Demographics
In total, 721 participants were recruited over a 10-year period. Approximately half were male, and 64.9% were born abroad (Table 1). The majority self-identified as Hispanic or non-Hispanic, followed by Caucasian, Asian, African-American, Middle Eastern, and others.
Table 1. Demographics of study participants
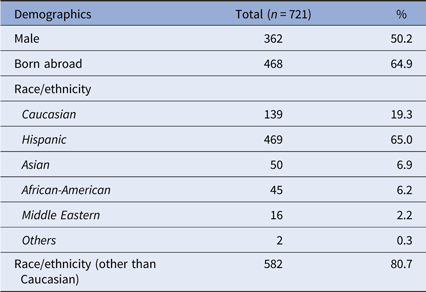
Demographic data of the participants are presented, along with the numbers of participants with each characteristic and the respective percentage (of the total 721 participants).
Single-Nucleotide Polymorphisms
Twenty-eight samples from Hispanic individuals were used in a pilot study in which all exons within the A20 gene were sequenced in their entirety. This subset analysis revealed two SNPs, both within exon 3 which contains the OTU enzymatic domain responsible for DUB activity (Fig. 1). The rs146534657 locus is located directly upstream to the catalytic cysteine of the OTU, where the missense mutation 305A>G converts the amino-acid N102S (NP_001257436.1:p. Asn102Ser). This SNP has a reported MAF of 0.11–1.3% [Reference Sherry36], but is significantly overrepresented within this cohort at an MAF of 3.12% (45/1442) (Table 2). The significant majority of individuals with this SNP (40/44) self-reported as Hispanic (χ 2 = 14.229, P = 0.014), including one with alleles GG. As there are 40 Hispanic participants with this SNP with 41 alleles present, it can be determined that there are 39 with the heterozygote genotype (AG) and one homozygote (GG). The MAF of this polymorphism among the Caucasian participants was 1.08%, conforming to literature and the MAF seen in the 1000 Genomes database [Reference Auton37], whereas it was 4.37% among Hispanics. Compared to Caucasians, the odds of acquiring this SNP as a Hispanic is 4.05 (95% CI 1.24–13.18).
Table 2. Distribution of the SNP rs146534657:A>G
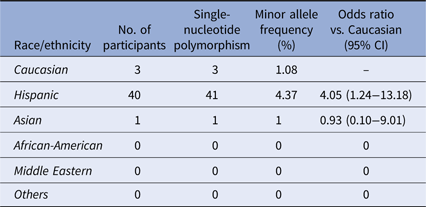
Distribution of rs146534657:A>G among the ethnic/racial groups. The MAF of this SNP (0.11–1.3%) per NCBI dbSNP is similar to the Caucasian participants of our cohort. The MAF of this SNP among the Hispanic participants of our cohort is significantly different.
The second SNP rs2230926 identifies a missense mutation 380T>G which converts the amino-acid F127C (NP_001257436.1:p. Phe127Cys). The MAF of this SNP among our cohort is 4.30% (62/1442) (Table 3), slightly lower than the 6.13–17.85% reported in the literature [Reference Sherry36]. A significant difference in the distribution of this SNP among the racial/ethnic groups was found (χ 2 = 24.886, P < 0.001), with the highest MAF of 14.44% among African-Americans. Notably, the 1000 Genomes database demonstrates a total MAF of 14% with only 2.3% of Mexicans living in Los Angeles carrying the SNP [Reference Auton37]. However, differences in the racial and ethnic characterization between our cohort and the 1000 Genomes database preclude a direct statistical comparison between the two. Twenty-eight Hispanic participants with this SNP carried the heterozygote genotype and four carried the homozygote genotype. Eleven African-American participants carried the heterozygote genotype and one had the homozygote genotype of this SNP.
Table 3. Distribution of the SNP rs2230926:T>G
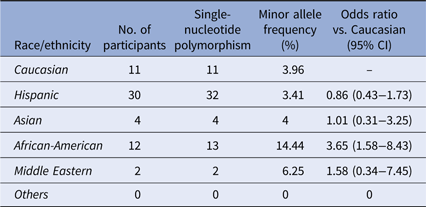
Distribution of rs2230926:T>G among the ethnic/racial groups. The MAF of this SNP (6.13–17.85%) per NCBI dbSNP is lower among the Caucasian participants of our cohort. The MAF of this SNP among African-American participants is significantly higher.
Many genome-wide association studies utilize the Illumina array chips to locate SNPs associated with autoimmune diseases [Reference Zhu34,Reference Liu38,Reference Zhang39]. We explored the MAF of the point mutation at rs6920220, one of the sites targeted by the Illumina chipset located within the intergenic gene region of A20, among our participants. 714 participants were screened for this SNP with using specific Taqman primers and probes. We again discovered heterogeneous distribution among the races/ethnicity within our cohort (χ 2 = 24.8235, P < 0.001) (Table 4). Compared to Caucasian participants, this SNP is less prevalent among Hispanics and Asians. According to the 1000 Genomes database, this SNP has an MAF of 9% [Reference Auton37]; our population had an MAF of 12.5%.
Table 4. Distribution of the SNP rs6920220:G>A
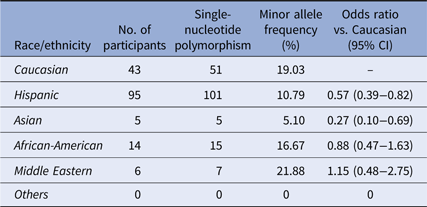
Distribution of rs6920220:G>A among the ethnic/racial groups. Compared to the Caucasian participants of our cohort, the MAF of this SNP is less prevalent among the Hispanic and Asian participants.
Functional Significance of SNPs
Since A20 is a well-known negative regulator of TNFα-induced NF-κB activity, we evaluated the functional significance of rs146534657:A>G and rs2230926:T>G with an NF-κB luciferase reporter assay. After stimulating with TNFα, cells pre-transfected with 10 ng of plasmid containing rs146534657:A>G (N102S) demonstrated a 2-fold increase in NF-κB luciferase activity (P < 0.01) compared to cells transfected with 10 ng of wild-type A20 plasmid. Similarly, cells transfected with 10 ng of plasmid containing rs2230926:T>G (F127C) A20 showed a 1.7-fold increase in NF-κB luciferase activity (P < 0.01) compared to cells transfected with the same concentration of wild-type A20 plasmid after TNFα stimulation (Fig. 2A). These deficits introduced by the SNPs gradually diminished with a higher quantity of transfected plasmids (i.e., 50, 100, and 500 ng) correlating to increasing protein concentrations (Fig. 2B), suggesting a baseline activation of NF-κB after TNFα stimulation regardless of A20 level or activity.
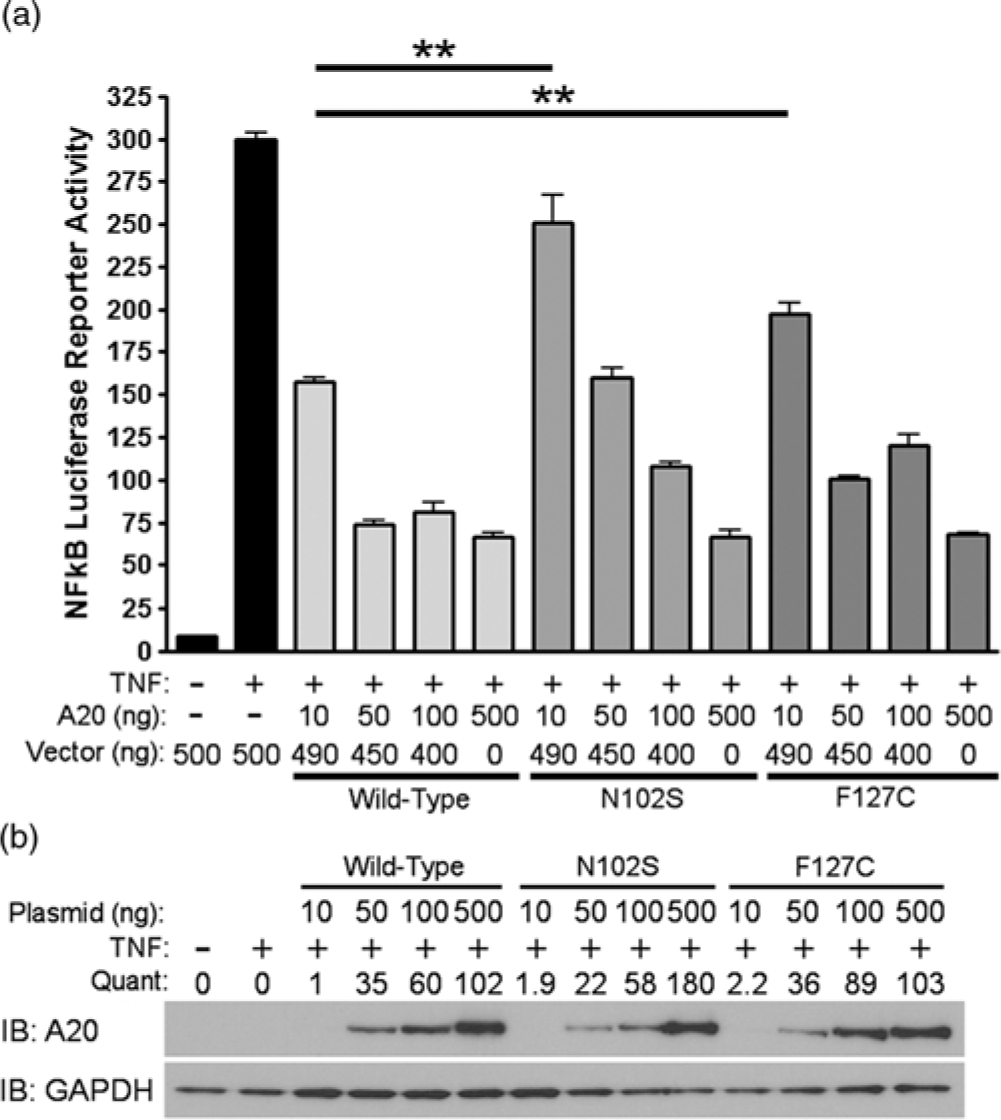
Fig. 2. Functional consequences of rs146534657:A>G and rs2230926:T>G. (A) Wild-type A20 plasmid or plasmids containing the rs146534657:A>G (N102S) and rs2230926:T>G (F127C) were transfected into A20 knockout 293 cells at the indicated concentrations along with an NF-kB firefly luciferase reporter. Cells were stimulated with 10 ng/ml of TNFα (TNF) for 8 h then harvested for luciferase assay. Firefly luciferase activity was normalized to a co-transfected Renilla luciferase reporter. Total plasmid concentrations were normalized using empty pCMV vector. (B) Protein from these cells was also used in Western blot. Quantifications of transfected A20 band intensities shown as immuno-blots (IB) are normalized to the housekeeping protein GAPDH.
Discussion
A majority of genetic studies to date have focused on populations of European descent [Reference Rosenberg40,Reference Need and Goldstein41], while few feature the comparison of genetic and clinical differences among Caucasians and other races [Reference Liu38,Reference Afzali and Cross42]. Risk alleles may differ significantly across races and ethnicity, as evidenced by the unique genetic signature of African-American patients with RA and the near absence of mutations in the bacterial sensor NOD2 in Asian populations, which was a major advancement in the genetics of IBD among Europeans and Americans [Reference Liu38,Reference Hugot43–Reference Leong47]. Taken together, the inclusion of participants representing diverse racial and ethnic background in genetic studies to identify population-specific mutations is crucial.
We selected A20 as a candidate gene to investigate because of its regulatory role on NF-κB activation and multiple other inflammatory signaling pathways. We identified two potential SNPs in the A20 gene which may have critical implications on chronic inflammatory conditions. Based on findings from our in-vitro assays, the locations of these SNPs within the exon 3 and their proximities to the OTU where deubiquitination occurs, these mutations may severely compromise the ability of A20 to control inflammation. These SNPs within exon 3 were identified after screening 28 Hispanic patients for all A20 exons; therefore, we focused specifically on these exon 3 SNPs within the entire cohort. Furthermore, we evaluated the SNP included in the Illumina array located at the intergenic region close to A20. We found this mutation to be significantly underrepresented among the Hispanics and Asians in our cohort, possibly limiting its potential as a susceptibility variant among these racial and ethnic groups.
The rs6920220 SNP has been associated with autoimmune and inflammatory conditions such as IBD and RA in the literature [Reference Stahl48]. This SNP was included in the study to determine if the frequency of rs6920220 in Hispanics is as common as the other two SNPs being studied. The MAF of this SNP was found to be less prevalent among Hispanic and Asian patients in our cohort compared to Caucasian patients.
This study expands upon current genetics literature in multiple aspects. Access to a diverse population enabled us to validate the existence of racial differences in the distribution and frequencies of SNPs. Sequencing the exon regions of TNFAIP3 revealed two SNPs in close proximity to the OTU catalytic site. While rs2230926:T>G has been well-studied in the context of autoimmune diseases [Reference Zhang39,Reference Zhang49,Reference Damas50], rs146534657:A>G is poorly-characterized with only one report linking it to poor outcome among Chinese RA patients [Reference Zhu34]. The function of this SNP in altering the amino acid directly adjacent to the catalytic cysteine within the OTU and its prevalence among Hispanics highlight clinical implications among this population. Consistent with a prior report on rs2230926:T>G in African-Americans with IBD, we found that this missense mutation significantly impaired the ability of A20 to inhibit inflammatory signaling [Reference Lodolce31]. We also demonstrated for the first time that the polymorphism associated with rs146534657:A>G present at high frequency in the Hispanic population is similarly defective.
The SNP allele frequencies in our cohort were unable to be directly compared statistically to the 1000 Genomes database due to differences in the categorization of cohorts; however, the general frequencies appeared similar. Specific challenges also merit consideration, the most pronounced being the lack of clinical information to correlate the three SNPs with development or progression of autoimmune conditions. Although our study demonstrates the prevalence of these SNPs among different ethnicities, it is possible that these differences in prevalence have no relation to chronic inflammatory conditions such as IBD. Participants’ races and ethnicity were based on self-report, which may lack specificity compared to the genetic determination [Reference Damas50]. Self-identification of ethnicity poses a weakness in our study as some participants may misreport their ethnicity, knowingly or unknowingly. Participants identified as Hispanic originate from multiple diverse ethnic groups which may carry significant genetic differences. Future studies should use genetic sequencing to definitively determine the ethnicity of the study participants. The cohort evaluated in this study was patients presenting to the LAC+USC Gastroenterology Clinic; therefore, a weakness is the suspected higher prevalence of GI pathology compared to a healthy control population. Furthermore, the intronic regions of A20 were omitted from sequencing, potentially excluding some relevant SNPs.
Study Highlights
Genetic studies have identified associations between certain SNPs, such as rs2230926 and s6920220, with inflammatory conditions; however, most studies are focused on populations of European descent. Our study aimed to sequence the A20 gene in a predominantly Hispanic population to identify new SNPs associated with inflammatory conditions, and to compare the frequency of SNPs associated with such conditions in Caucasian patients, such as rs2230926 and rs6920220, with the prevalence in Hispanic patients. We demonstrated for the first time that the polymorphism associated with rs146534657 is present at higher frequency in the Hispanic population and impairs the ability of A20 to inhibit TNFα activity and inflammatory signaling. As previously demonstrated, we showed that the polymorphism at rs2230926, present in higher frequency in African-Americans is similarly defective. Further studies should determine the clinical significance of the rs146534657 mutation and its clinical role in assessing susceptibility to inflammatory conditions such as IBD.
Conclusions
In conclusion, we identified two SNPs within the third exon of A20 affecting protein function with significantly higher MAF among self-reported Hispanics and African-Americans as compared to Caucasians. The SNP rs146534657:A>G is not well described in literature and is significantly overrepresented in Hispanics compared to Caucasians. Further research, with genetically confirmed analysis of ethnicity, is warranted to validate the clinical significance of this mutation to establish it as a susceptibility locus for autoimmune diseases among this population.
Acknowledgments
We thank the USC Libraries Bioinformatics Service for assisting with data analysis. The bioinformatics software and computing resources used in the analysis are funded by the USC Office of Research and Norris Medical Library. AP would like to thank the IBD Support Foundation.
Financial Support
This research was supported by a K-08 award from the NIDDK (DK100462), the Wright Foundation (LS), and the Margaret E. Early Foundation (LS). Additional core facility support was provided by USC Research Center for Liver Diseases, NIH grants Nos. P30 DK048522 and S10 RR022508. This research was also supported by the National Center for Advancing Translational Sciences (NCATS) of the U.S. National Institutes of Health [grant Nos. UL1TR001855 and UL1TR000130].
Disclosures
All authors have no financial and non-financial competing interests to declare.
Ethical Approval
This study was approved by the institutional review board (IRB) of the University of Southern California (Protocol no. IRB #HS-09-00543). Verbal and written consents were obtained from all participants of the study prior to their enrollment.
Availability of Data and Material
Raw trace files are available through the NCBI Trace Archive (link to be supplied upon manuscript acceptance). Cell lines and constructs are available upon request.








