In Japan, since 2012, diarrheagenic Escherichia coli have been classified into six categories: enterohemorrhagic/Shiga toxin-producing E. coli (EHEC/STEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAggEC) and ‘other diarrheagenic E. coli’. ‘Other diarrheagenic E. coli’ is defined as follows: not categorized by the above five definitions but is considered a pathogen of gastroenteritis, such as E. coli strains isolated from many patients involved in an outbreak and characterized as the same by biochemical analysis [1, Reference Buchan and Carroll2].
EAggEC heat-stable enterotoxin 1 (EAST1) was originally found as an enterotoxin of EAggEC [Reference Savarino3]. Although its virulence is not clear, food poisoning cases have occurred where E. coli carrying the astA for EAST1 was commonly detected in patients. In such cases, it is classified as ‘other diarrheagenic E. coli’. In Osaka, Japan, in 1996, the first case of an outbreak caused by E. coli that did not have well-characterized virulence genes other than astA was reported [Reference Zhou4]. Since then, outbreaks caused by E. coli carrying astA have been reported in various places, with many due to the O166:H15 serotype, and others have been attributed to O169:HNM, OUT, etc. [Reference Zhou4–Reference Ishimura6]. The number of patients per outbreak was less than 200.
From 27 to 28 June 2020, many elementary and junior high school students in Yashio, Saitama, Japan presented with gastroenteritis symptoms such as diarrhoea and abdominal pain. Patients with symptoms were distributed in all 15 public elementary and junior high schools in Yashio and the Yashio Board of Education. A school lunch supplied by a private school meal supply facility was the common food stuff ingested by all the patients.
As many as 6732 students, teachers and other staff had consumed the school lunch, and 2958 of them developed symptoms. The major symptoms were diarrhoea (2716, 91.8%), abdominal pain (2581, 87.3%), fever (671, 22.7%) and other symptoms. The subsequent investigation involved the collection of faecal specimens from 19 patients from 8 of 15 schools and the Yashio Board of Education who were not receiving antibiotics. In addition, 27 food samples (lunch served on 24, 25 and 26 June) stored by meal type at the Yashio Board of Education and the private school meal supply facility, and nine faecal specimens from the food preparation staff were collected. Also, four swab samples of the kitchen facilities were collected.
Faecal specimens were examined for the presence of salmonella, shigella, vibrios, campylobacter, Escherichia coli, Staphylococcus aureus, Bacillus cereus, Clostridium perfringens and Norovirus.
Direct examination of culture plates from 18 of 19 patients' samples and five of nine food workers' samples showed growth of lactose-fermenting colonies that appeared to be E. coli on modified Drigalski agar (Eiken Chemical, Tokyo, Japan). No food poisoning-associated bacteria or virus were detected other than E. coli. The colonies were confirmed for the presence of virulence-associated genes for diarrheagenic E. coli and tested for biochemical characteristics. Individual colonies were suspended in distilled water and heated at 100 °C for 10 min, then centrifuged at 20 000 × g for 5 min. The supernatant was used as a template DNA solution for multiplex ExEC (elt, estA1, estA2, stx1, stx2, stx2f, invE) and EpAll (eae, aggR, astA, afaD) PCR [Reference Kudoh, Iyoda, Kawanishi and Watanabe7, Reference Ito8]. The thermocycling conditions were 94 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 50 °C (for EpAll; 55 °C) for 30 s and 72 °C for 1 min, followed by 72 °C for 10 min. The PCR products were mixed with MIDORI GREEN DIRECT (Nippon Genetics, Tokyo, Japan) and subjected to 2% agarose gel electrophoresis. The PCR products were detected under blue/green LED light. The astA amplicon was detected in 14 isolates from 14 patients' faecal specimens. No pathogenic genes were detected in the faecal specimens of four patients and five food workers. The 14 isolates in which astA was detected showed the same characteristics as typical E. coli: glucose and lactose fermenting, indole reaction positive, Voges-Proskauer negative, motility positive, lysine decarboxylase positive and citrate utilization negative. The serotype of the 14 isolates was evaluated with E. coli antisera (Denka, Tokyo, Japan and SSI Diagnostica, Hillerød, Denmark) and E. coli O-genotyping PCR [Reference Iguchi9]. All isolates were confirmed as E. coli O7:H4. Based on the above results, the 14 isolates from 14 patients were identified as E. coli serotype O7:H4, which lacks well-characterized virulence genes other than astA.
To investigate the cause of this outbreak, 27 stored food samples and four swab samples were tested for E. coli serotype O7:H4 carrying astA. Twenty-five grams of individual food samples were homogenized in 225 ml m-EC Broth (Oxoid, Basingstoke, Hampshire, UK), and each swab was placed in 10 ml m-EC Broth, incubated statically at 42 °C for 18 h, then plated onto modified Drigalski agar, sorbitol MacConkey agar (Oxoid) and CHROMagar STEC (CHROMagar, Paris, France). Five colonies similar to the isolates from patients were tested using PCR assay, biochemical characteristics and serotype. Escherichia coli serotype O7:H4 carrying astA was detected in two seaweed salad samples served on 26 June of the 27 food samples collected from the Yashio Board of Education and the private school meal supply facility. At the same time, a quantitative test for E. coli in the seaweed salad was performed; however, E. coli could not be quantified. No E. coli was detected in food samples other than the seaweed salad samples, or in the swab samples.
The seaweed salad was made from six kinds of seaweed rehydrated with water and boiled vegetables with dressing. Coliforms were detected in the red seaweed (Gigartina tenella) by the manufacturer during self-inspection. Thus, we investigated whether E. coli carrying astA was detected in the red seaweed, and a positive result was obtained. When we traced back the red seaweed, it turned out to be the product imported from overseas in 2017. Coliforms were not detected by inspection at the time of import nor at the time of sale to the manufacturer. In addition, no complaints about the same lot of seaweed, besides this case, have been identified. Therefore, the source of the E. coli contamination of red seaweed could not be identified.
Pulsed-field gel electrophoresis (PFGE) was performed to compare the 17 isolates obtained from 14 patients, two seaweed salads and one red seaweed [Reference Pei10]. All isolates showed the same DNA banding pattern in PFGE after digestion with the restriction enzyme XbaI (Roche Diagnostics, Mannheim, Germany) (Fig. 1). The sensitivity of the 17 isolates to the following antibiotics was examined using Sensi Discs (Becton Dickinson, Franklin Lakes, NJ, USA) according to the performance standards for antimicrobial disk susceptibility tests of the Clinical and Laboratory Standards Institute: amikacin, ampicillin, ceftazidime, cefotaxime, cefoxitin, chloramphenicol, ciprofloxacin, colistin, fosfomycin, gentamicin, imipenem, kanamycin, meropenem, nalidixic acid, norfloxacin, streptomycin, sulfamethoxazole/trimethoprim and tetracycline. All isolates were resistant to ampicillin and cefotaxime.
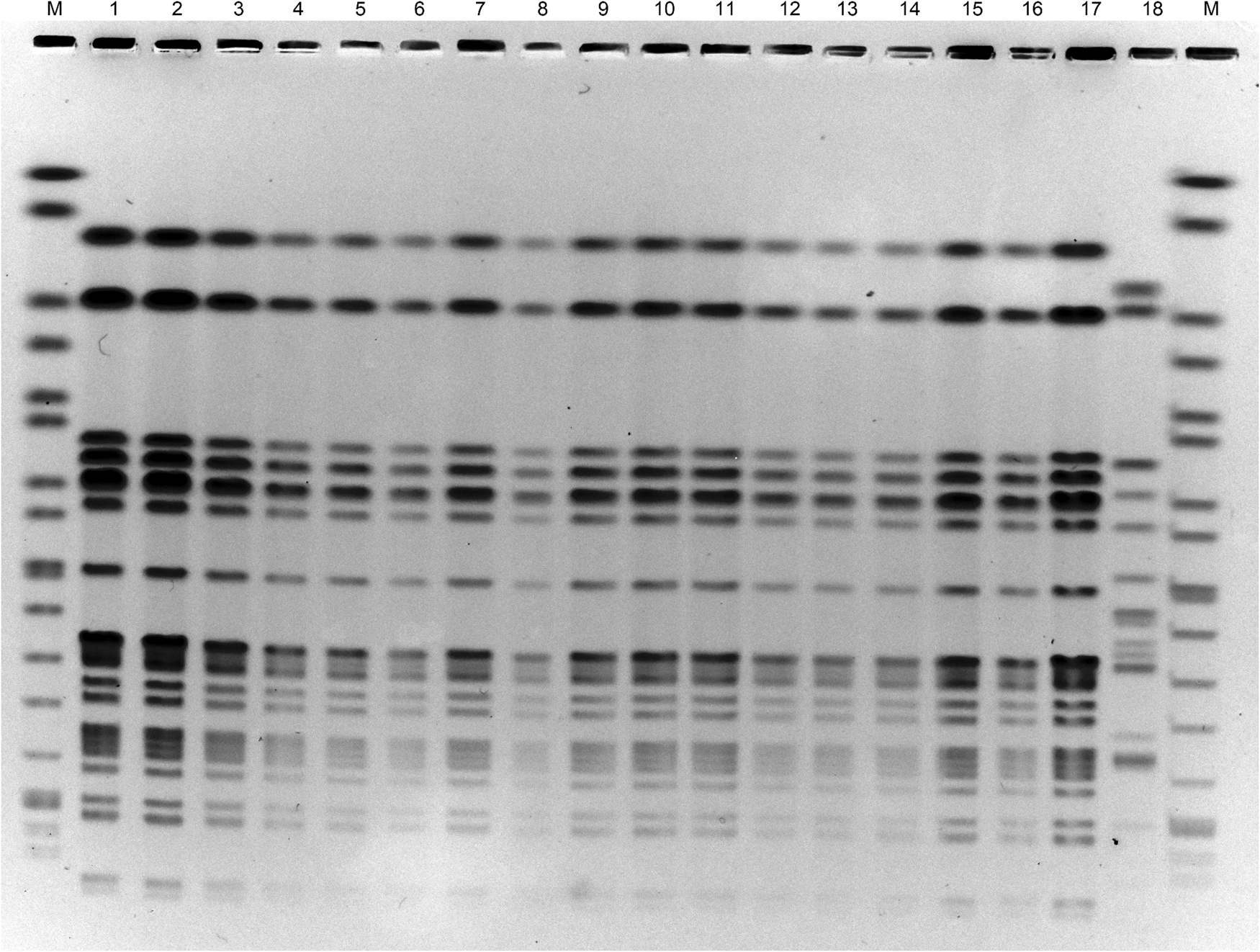
Fig. 1. Pulsed-field gel electrophoresis of DNA of E. coli O7:H4 strains after treating with the restriction enzyme XbaI. Lanes: M, size marker; 1–14, strains from patient's faecal specimens; 15–16, strains from seaweed salad; 17, strain from red seaweed; 18, E. coli O7:H15 strain having no astA.
Furthermore, the whole genomes of E. coli O7:H4 isolates carrying astA from two patients (Fig. 1, lanes 2 and 9), seaweed salad (lane 15) and red seaweed (lane 17) were sequenced using MiSeq (Illumina, San Diego, CA, USA). Core genome single nucleotide polymorphisms (cgSNPs) were analysed as previously described [Reference Kimata11]. Only one locus of cgSNP was detected between the isolates of patients and food samples. Therefore, it was concluded that all isolates were the same clone. Based on the above results, this outbreak of food poisoning was concluded to be due to E. coli serotype O7:H4 carrying astA, and originated from the red seaweed in the seaweed salad.
Food poisoning cases due to E. coli do not necessarily result in the detection of pathogens in food during the follow-up investigation. This outbreak was rare and important, as the same pathogen obtained from patients was detected in food samples and the raw material. While the virulence of EAST1 is not clear, food poisoning cases due to E. coli carrying astA for EAST1 show relatively mild symptoms. Therefore, it is often difficult to identify E. coli carrying astA as a cause of food poisoning in instances with small numbers of patients. However, this outbreak of gastroenteritis occurred in about 3000 people. In order to elucidate the pathogenicity of EAST1, it is desirable to document outbreaks and perform pathogenic studies of E. coli strains carrying astA/EAST1. Since this particular outbreak was attributed to seaweed, it is recommended that marine products be carefully monitored for contamination.
Conflict of interest
None.
Data availability
Short reads and draft genomes of E. coli O7:H4 isolates were deposited in DDBJ/ENA/GenBank under the accession number (PRJDB11800).