Introduction
There are approximately 75,000 adults and 15,000 children living with epilepsy in Ontario. Reference Bowen, Snead, Chandra, Blackhouse and Goeree1 ,Reference Ng, Maxwell and Yates2 The condition is clinically heterogeneous and has diverse etiologies with age dependent presentation. Reference Thomas and Berkovic3 Twin and family studies show that epilepsy is a highly heritable condition,Reference Myers, Johnstone and Dyment4 and a proportion of affected individuals will develop epilepsy due to pathogenic variant(s) in a single gene (e.g., monogenic). These individually rare, highly penetrant disease-causing variants have previously been difficult to diagnose by molecular means until the advent of next-generation sequencing (NGS). Several studies now show that NGS-based strategies can provide a molecular diagnosis in 15%–30% of individuals with epilepsy and, for specific clinical presentations, the rate can be higher. Reference Myers, Johnstone and Dyment4,Reference Mercimek-Mahmutoglu, Patel and Cordeiro5 As a result, the application of NGS has rapidly transformed the way in which genetic epilepsy is diagnosed. The sequencing of dozens to even hundreds of genes is now possible with a wide variety of “epilepsy panels” available through commercial laboratories. Reference Chambers, Jansen and Dhamija6 There are different panels available to the clinician that include “targeted” panels, “comprehensive” panels and panels generated by a “filter” for select genes. Generally, a targeted panel has fewer genes than a comprehensive panel but higher depth of coverage and complete overall coverage of the target sequence, while the comprehensive panels may comprise several hundred genes – typically with slightly less coverage of the target sequence (97%–99%). A filtered panel is a panel that includes a subset of clinically relevant genes that are selected from a broad gene capture and sequencing experiment (such as an exome). The introduction of NGS testing into clinical care has subsequently changed the knowledge requirements for physicians involved in the care of persons with epilepsy. Physicians must now have a baseline expertise in the field of genetics; they must consider the potential benefit(s), harms, and limitations of pursuing genetic testing, understand what type of genetic testing is most appropriate, may be required to select the most suitable commercially available genetic panel, inform and counsel patients and their families prior to pursuing testing, and disseminate the results (whether positive, negative, or uncertain) to others within the circle of care. Reference Myers, Johnstone and Dyment4,Reference Scheffer7, Reference Poduri8
The benefits of a molecular diagnosis are significant. An accurate genetic diagnosis can direct treatment toward disease-modifying therapies and/or medications known to be effective in certain epilepsy syndromes. The information may also prevent the addition of a potentially harmful medication. Genetic diagnosis can also clarify the long-term prognosis and limit further diagnostic investigations that have associated risks and costs. A genetic diagnosis may spare patients the morbidity, and society the costs, of invasive and expensive evaluations for epilepsy surgery. Further, genetic diagnoses often help to identify potential comorbidities allowing for optimization of surveillance, early treatment, and associated reduction in disease morbidity. A genetic diagnosis will also provide informed risks for genetic counseling purposes and provide prenatal testing options. Finally, and importantly, a genetic diagnosis can have a psychosocial benefit to the individual and their families.
The Epilepsy Panels Implementation Working Group
Within Canada, each province and territory has its own single-payer system of healthcare with differing, but similar, clinical care delivery models. Ontario’s healthcare system is funded by the Ministry of Health and Long-Term Care (MOHLTC) through the Ontario Health Insurance Plan (OHIP). As previously described, genetic testing for epilepsy patients living in Ontario has historical disparities in terms of who is offered testing and what testing is conducted. Reference Jain, Andrade and Donner9 This, in addition to the known advantages of a molecular diagnosis for persons with epilepsy, prompted the former Genetic Testing Advisory Committee (GTAC; 2014–2017), established by the MOHLTC for advice on the provision of genetic tests in Ontario, to develop and disseminate the criteria for panel testing to guide physicians caring for individuals with epilepsy in Ontario Reference Jain, Andrade and Donner9 (http://www.health.gov.on.ca/en/pro/programs/gtac/). While this has aided clinicians in determining who should and should not be tested, genetic testing for epilepsy continues to be sent out of country (OOC) to several commercial laboratories offering different epilepsy panels, creating inequity in patient care at considerable cost to the MOHLTC, who funds testing for eligible patients as an OHIP-funded service through the Ministry’s Out-of-Country/Out-of-Province Prior Approval Program for Laboratory and Diagnostic Services Reference Jain, Andrade and Donner9 administered by the Laboratories and Genetics Branch (LGB). Furthermore, because this testing is conducted by a variety of commercial laboratories with no requirements to report back to the MOHLTC, there is little information as to whether Ontario patients are benefitting from this test in terms of a satisfactory diagnostic yield or any other outcome measure of clinical utility. As such, it was recognized that clinical testing within Ontario could provide benefits to patients, physicians, and the public healthcare system, in terms of consistency, improved access, the development of local expertise, and the generation of relevant outcome data. Furthermore, locally performed genetic testing has been shown to improve diagnostic yield. Reference Baldridge, Heeley and Vineyard10 Given a need for improved care, the Epilepsy Panels Implementation (EPI) Working Group was formed by LGB with the purpose of developing a programmatic approach to implementing epilepsy panel testing in Ontario as a province-wide service.
The EPI Working Group members were identified for their clinical expertise in the field of epilepsy clinical care and their experience in, and understanding of, the use of clinical genetic testing in epilepsy in adults and children in the province of Ontario. As well, the EPI Working Group members were selected to provide widespread regional representation across the province of Ontario. To fulfill the Working Group’s mandate, members were organized into two subgroups – Clinical and Technical. The Clinical Group was tasked with providing a testing algorithm to guide test utilization, develop performance indicators, and provide details for resources and capacity. The Technical Group was focused on the technical performance and service standards, as well as quality assurance, ongoing evaluation processes and resources and capacity as they relate to technical aspects of the test. The Clinical and Technical Groups proceeded simultaneously via teleconference meetings, and each was responsible for documenting their separate discussions, developing the assigned content, and coordinating and supporting each other to produce one cohesive final report with recommendations reflecting the consensus of the Working Group as a whole. This process took place from July 2017 to February 2018.
Guiding Principles
Early in the process, the EPI Working Group highlighted several key elements to include in the overall plan. The first was improved access and education to facilitate gene panel testing throughout the province. At present, OOC testing is requested by geneticists, metabolic specialists, and a few neurologists in tertiary care centers. In order to inform and educate additional care providers, epilepsy genetic testing was to be incorporated into a video-conference-based extension of community healthcare outcomes (“ECHO”) model Reference Komaromy, Ceballos, Zurawski, Bodenheimer, Thom and Arora11 of knowledge translation for epilepsy in Ontario. Secondly, it was recognized that genes present on commercially available panels did not, necessarily, show significant scientific evidence of being clinically relevant to epilepsy. Therefore, an evidence-based approach to the gene composition of a panel, and the coverage of the genetic variation within the gene, was considered a priority. Lastly, the test was to be implemented as part of a sustainable program in order to continually evaluate and improve epilepsy testing going forward.
Here, we present the recommendations generated by the EPI Working Group to implement a comprehensive and unique epilepsy genetic testing strategy for the province of Ontario.
Recommendations for the Implementation of an Ontario-Based Epilepsy Genetic Test
Recommendation 1: An ECHO-Based Program Should Be Incorporated to Inform and Educate Ordering Providers When Requesting and Interpreting a Genetic Panel Test
Project ECHO is a program developed by Dr. Sanjeev Arora in 2003 to help primary care providers in rural New Mexico manage patients with hepatitis C. Reference Arora, Thornton and Murata12 Project ECHO’s virtual hub-and-spoke educational model harnesses case-based learning via video conferencing to facilitate primary care providers’ learning in conjunction with experts at academic health centers, thus increasing the ability of participating primary care providers to manage complex medical cases in their own practice. This cadre of ECHO participants can then serve as an expert resource for other providers in their community.
The ECHO model thus breaks down the walls between specialty and primary care. It is designed to link expert specialist teams at an academic “hub” with primary care providers in local communities – the “spokes,” or community partners, of the model. Together, they participate in weekly Tele-ECHO clinics, which are like virtual grand rounds, combined with mentoring and patient case presentations.
The Ontario Epilepsy Network has developed an ongoing Project ECHO Ontario – Epilepsy Across the Life Span, in which each District Epilepsy Centre and Regional Epilepsy Centre in Ontario, as well as Thunder Bay, will serve as a hub that will recruit primary care provider, pediatricians, and community neurology spokes in their respective regions (https://oen.echoontario.ca/) (Figure 1). The Project ECHO system is administered by The Hospital for Sick Children and a common, accredited continuing medical education (CME) curriculum is available for all hubs, one in Epilepsy in Child and Youth and the other in Epilepsy in Adults. The genetics of epilepsy will be incorporated into both curricula in terms of pre- and postgenetic test counseling, as well as the guidelines and indications for genetic testing in epilepsy. Project ECHO’s relevance to genetic testing is through the insight gained by “spokes,” including significant knowledge that will enable consistent and informed choices for genetic tests to be made throughout the province for patients with epilepsy. These caregivers will serve as a local resource for epilepsy care in their respective communities.
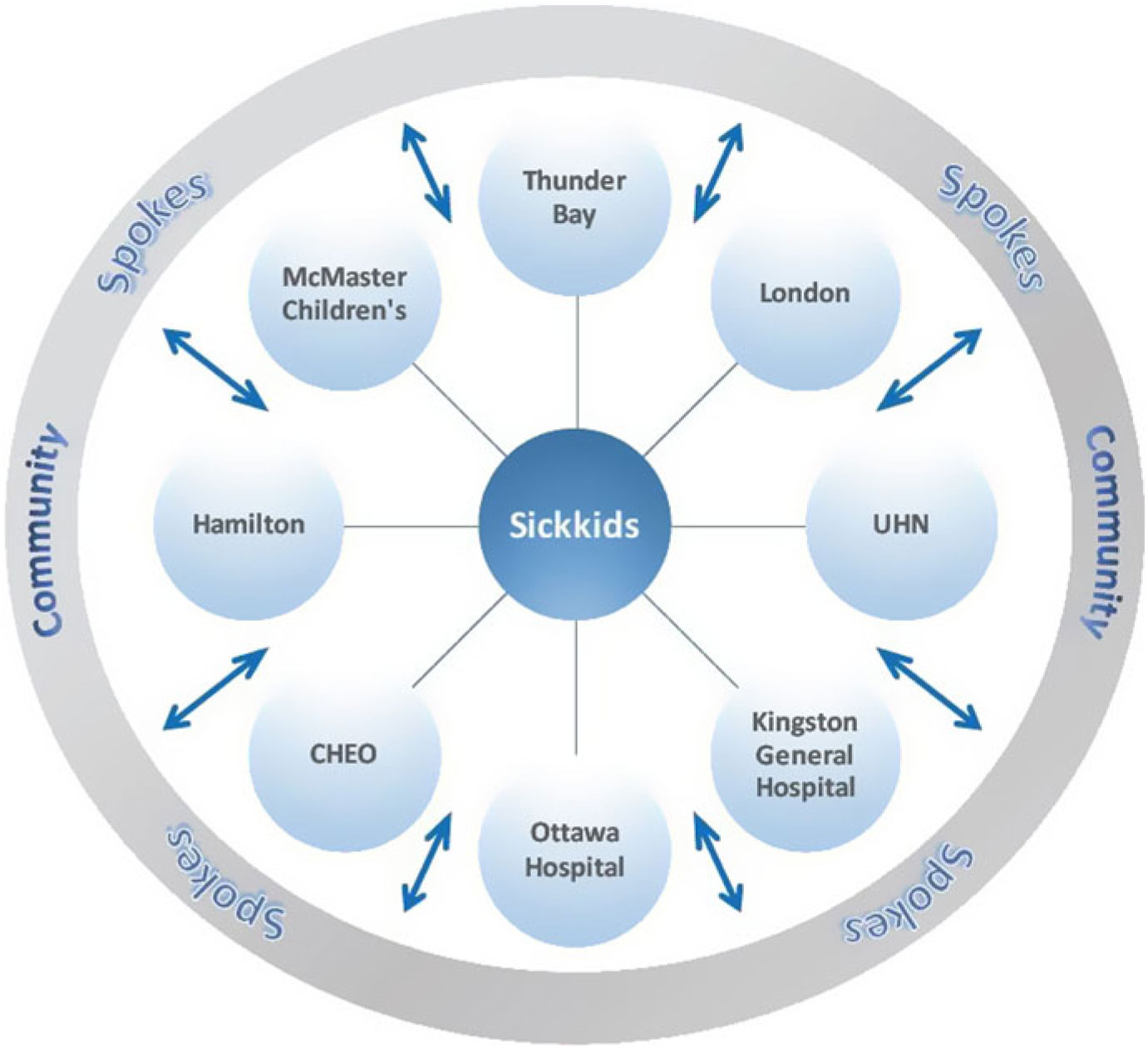
Figure 1: Project ECHO Ontario – Epilepsy across the Hub-and-Spoke Organization. Represents the hub-and-spoke model of ECHO. CHEO, Children’s Hospital of Eastern Ontario.
Recommendation 2: Develop and Implement Multiple Evidence-Based Genetic Panel Tests Based on Specific Clinical Indications for Epilepsy
A key element to the implementation of genetic diagnostic testing for epilepsy in Ontario is that the genes selected have strong supporting evidence for their role in epilepsy. This is in recognition that many currently available epilepsy panels are comprised of hundreds of genes, many of which may lack strong evidence for an epilepsy association. Reference Chambers, Jansen and Dhamija6 In addition, the comprehensive panels offered commercially may be comprised of genes with a heterogeneous mix of clinical indications (e.g., a panel may have genes associated with storage and metabolic diseases in combination with structural brain malformation-related genes). The sequencing of genes with limited evidence for gene–disease association, or a panel comprised of genes with mixed clinical presentations, may result in confusion regarding the medical interpretation of the laboratory report and certainty of potential molecular diagnoses.
Methods to assess the pathogenicity of a specific variant within an established disease gene have been well described. Reference Richards, Aziz and Bale13 However, standardized guidelines or consistent methodologies for the assessment of the gene, itself, have been less well studied and reported in the literature. This pertains directly to clinical relevance. Currently, there is a recognized need in the medical and genetics community for improved resources to assess these disease–gene associations in epilepsy and other diseases.
ClinGen (Clinical Genome Resource) is a National Institutes of Health-funded resource that sets out to define, annotate, and curate the clinical significance of reported disease genes. Reference Rehm, Berg and Brooks14 Annotation for the genes involved in epilepsy has been initiated by a ClinGen-based working group Reference Helbig, Riggs and Barry15 and is available online (http://www.clinicalgenome.org). Other elements of the curated resource will include the “actionability” of any given disease gene, as well as the aforementioned variant calls of the known disease genes in association with ClinVar. The ClinGen database is dynamic, and it will broaden as additional knowledge is acquired.
The use of a standardized method, such as ClinGen, is tailor-made for the Ontario EPI program. Any clinical laboratory offering genetic testing for epilepsy would be expected to use the methods outlined by ClinGen to assess the clinical significance of genes offered on a panel (or to use the available information at the ClinGen website). In addition, the “actionability” of a gene will be assessed in a consistent manner, and this will highlight the genes that have a direct impact on management (e.g., ALDH7A1 and treatment with pyridoxine). This would then be used to inform best turn-around-times (TATs) for a genetic test result.
Recommendation 3: Implement a Robust Evaluative Programmatic Framework
The broad and uniform implementation of an evidence-based gene testing program requires a robust framework for ongoing evaluation. It is expected that the clinical laboratories responsible for epilepsy panel testing in Ontario will involve an ongoing consultation process with local experts (clinical and laboratory), ad hoc working groups, and the end users of the panel test, in order to facilitate this process and ensure that performance outcomes of the epilepsy panels are captured and reviewed every 1–2 years. Such measures will include a review of available, epilepsy-related databases and literature to include any genes reported to be associated with epilepsy and meeting the ClinGen requirements for “Strong” or “Definitive” evidence or to exclude any genes based on limited (or lack of) evidence. Annual reports will include the number of tests conducted by the clinical laboratory, positive results (diagnostic yield), negative results, and uncertain results, including the average number of variants of uncertain significance per patient. Other laboratory indicators, such as TATs, must also be included. These measures will be reviewed by a Program Advisory Group, which will be appointed by the MOHLTC for the purpose of evaluating and monitoring the proposed services, as well as make recommendations for continuous improvement, including any changes to the genes included on each panel. This group is expected to be comprised of two to three independent experts in the field of clinical testing.
Lastly, the Ordering Physician will be required to provide the clinical laboratory with information pertaining to (1) the clinical interpretation of molecular results for their patient, (2) if there was any change in management as a result of a positive diagnosis, or lack of a diagnosis, and (3) if additional tests/consultations were ordered. This information will be expected to be collected within 3 months of sending the final results report to the Ordering Physician. This information will be collected and maintained by the clinical laboratory and be summarized as part of the ongoing evaluation process. The summary will therefore provide information regarding, among others outcomes, the diagnostic yield and positive predictive values of the test by panel, gene, and indication. This will require significant resources (e.g., a genetic counselor and the management of a database).
Clinical Requirements for the Ontario-Based Tests
Pretest Evaluation and Counseling
Individuals must undergo pretest evaluation and counseling for genetic testing. This may be performed by a genetic counselor, geneticist, or other clinician with suitable training and knowledge base (e.g., metabolics specialist, neurologist, pediatrician, or family doctor) ordering the test. This is an expectation that must be met for all genetic tests and will be a focus of the genetics portion of the Project ECHO CME accredited curriculum. The counseling will include a discussion of the benefits and risks as well as limitations of genetic testing (false negative) and the high likelihood of detecting variants of unclear significance (VUS) that may require testing of additional family members, as outlined below, in an attempt to resolve the variant classification.
Clinical algorithms have been summarized in Figures 2 and 3 and Supplementary Appendix 1. These are meant as a guide for the clinician considering genetic testing.
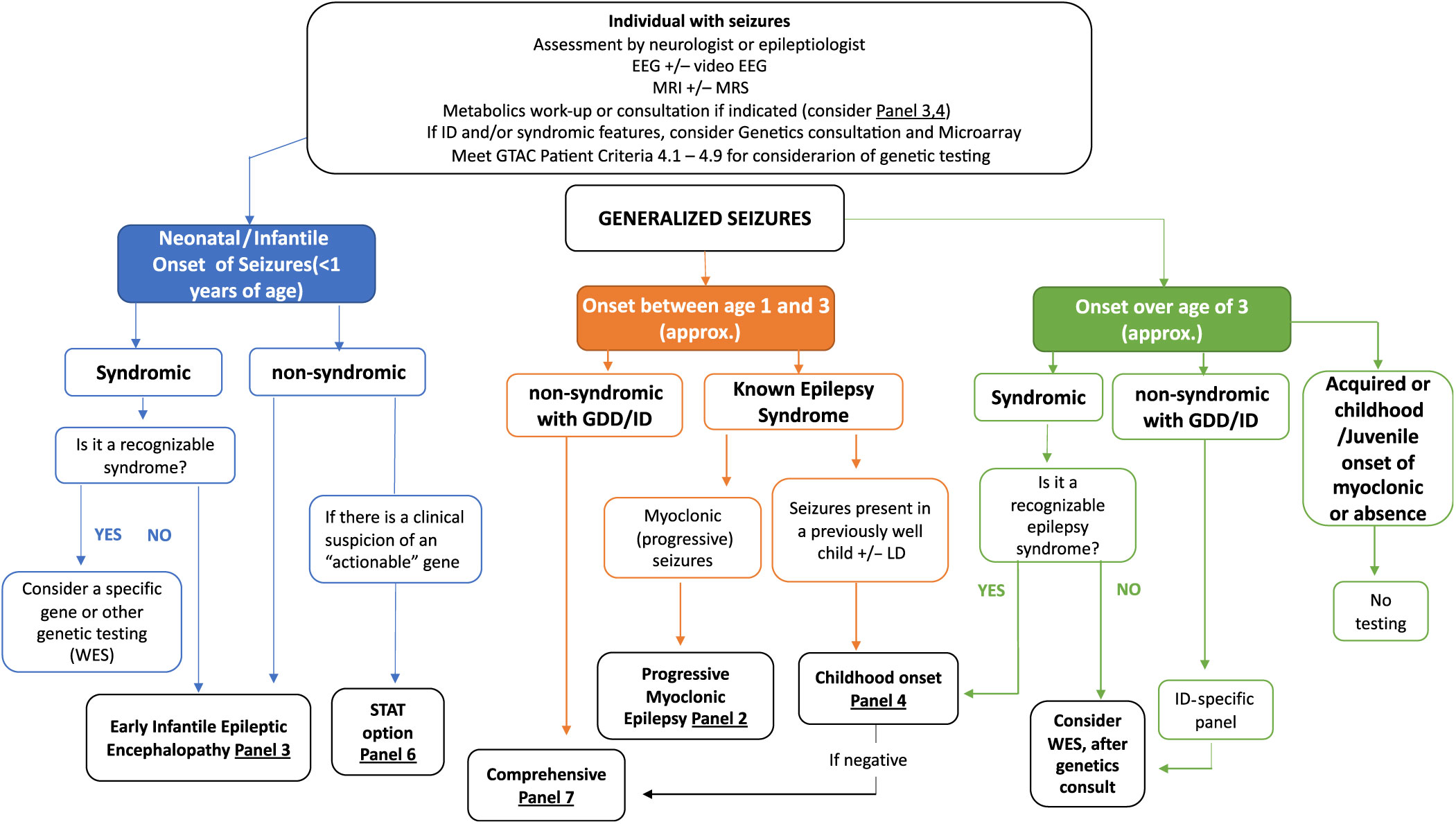
Figure 2: Suggested Ontario clinical algorithm for generalized and early-onset epilepsy panel testing. This is a guide to aide clinicians for panel selection. In Ontario, individuals with epilepsy are required to have the appropriate work-up as defined in criteria for panel testing Reference Jain, Andrade and Donner9 (http://www.health.gov.on.ca/en/pro/programs/gtac/). This includes an assessment by a neurologist or epileptologist with appropriate laboratory, imaging and electroencephalogram (EEG) studies. If the individual is young (<1 year) and does not present with an obvious recognizable syndrome, consider Panel 3 (Early Infantile Epileptic Encephalopathy Panel). If the patient has multisystem disease, which includes epilepsy, consider whole exome sequencing (WES). If there is clinical suspicion of an “actionable” gene consider Panel 6 (STAT Option). If the individual is 1–3 years (approximately), nonsyndromic, with global developmental delay (GDD) or intellectual disability (ID), consider Panel 7 (Comprehensive Panel). If the individual presents with myoclonic epilepsy, consider Panel 2 and if the seizures present in a previously well child, consider Panel 4 (Childhood-Onset Epilepsy Panel). If older than 3 years of age and in context of ID/GDD, consider an ID-specific panel or WES if considered syndromic.
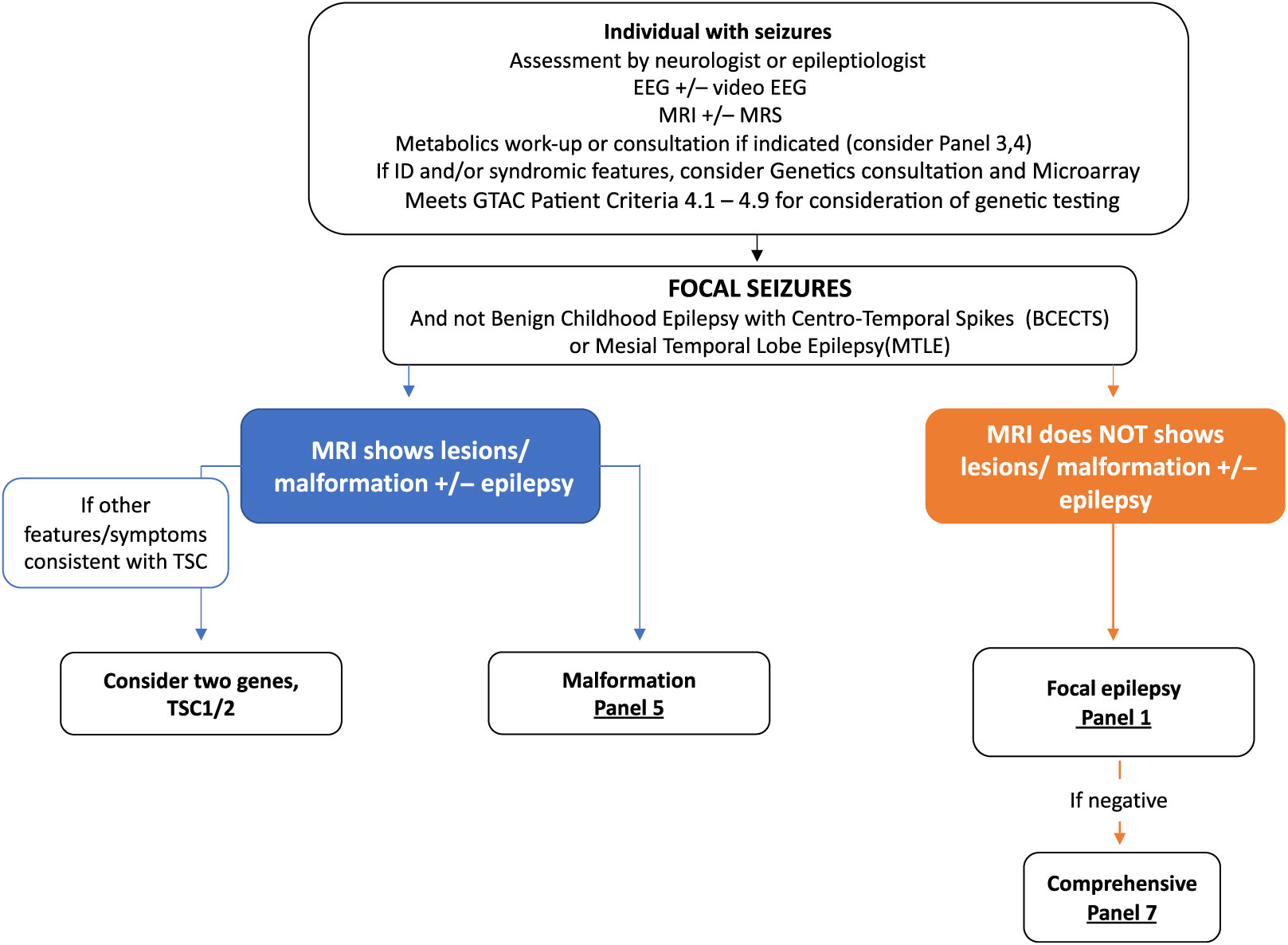
Figure 3: Suggested Ontario clinical algorithm for focal seizures panel testing. This is a guide to aide clinicians for panel selection. In Ontario, individuals with epilepsy are required to have the appropriate work-up as defined in criteria for panel testing Reference Jain, Andrade and Donner9 (http://www.health.gov.on.ca/en/pro/programs/gtac/). This includes an assessment by a neurologist or epileptologist with appropriate laboratory, imaging, and EEG studies. If clinical assessment and imaging studies show evidence of tuberous sclerosis, consider testing TSC1 and TSC2. If imaging consistent with a brain malformation consider Panel 5 (Brain Malformation Panel associated with epilepsy) and Panel 1 if EEG studies shows evidence of focal epileptiform activity but no malformation on imaging studies.
Post-Test Management Considerations
Post-test management considerations are varied and will depend on the specific results. These considerations should be discussed in the pretest counseling phase of testing.
Providers requesting testing are expected to communicate the results of the genetic test to the patient and family and take appropriate action depending on the outcome of the results. A positive result may require referral to another service, further (invasive and noninvasive) investigations, and for other individuals in the healthcare team to be made aware. A positive result may also require appropriate medical management and/or surveillance measures to be initiated. Post-test genetic counseling also needs to be offered, and at-risk family members should be offered the option of a referral for genetic counseling (+/− carrier testing) as appropriate.
As mentioned above, classification of VUS may require further evaluation in the context of the patient and family structure. This may require the testing of relatives, in particular the parents, to assess de novo status or determine if two variants are in cis (coupled) or trans (repulsion). A negative result may require additional testing be performed including a reflex to a broader epilepsy gene panel available by the clinical laboratory or, if appropriate testing criteria are met, genome-wide sequencing.
Technical Considerations
Required Panel Core Genes
The goal for implementation of Ontario-based epilepsy testing is to provide panel test(s) that includes genes that meet evidence-based criteria as proposed by ClinGen, or other initiatives, as well as provide a comprehensive and beneficial test for individuals living with epilepsy and the care physicians. An initial “core” complement of epilepsy gene panels is listed in Supplementary Appendix 1. The genes listed would be expected to be present on any proposed panel for testing in Ontario.
The recommended clinical indications for each panel (e.g., Focal Epilepsy Panel) are based on commercially available panels, the literature, and the experience of the EPI Working Group Technical and Clinical Subgroups. We recognize that panel composition will change as novel epilepsy genes are identified. As such, a testing program that includes an ongoing reevaluation of gene content and the performance of the respective panels will be an absolute requirement for implementation. These panels were generated in January of 2018, and the gene composition of the panels was updated in August of 2019.
The Recommended Gene Panels include: (1) Focal Epilepsy; (2) Progressive Myoclonic Epilepsy; (3) Early Infantile Epileptic Encephalopathy; (4) Childhood-Onset Epilepsy; (5) Epilepsy associated with brain malformations; (6) a “STAT”or “Urgent” Epilepsy panel; and (7) a Comprehensive Epilepsy Panel. The specific genes on these panels are outlined in Supplementary Appendix 1. In addition, Figures 2 and 3 suggest clinical algorithms to aide clinicians when deciding on the most appropriate panel. These algorithms are not meant to represent a required approach but are meant to serve as a guide for panel selection.
Specific Technical Considerations
The platform and technical considerations for epilepsy gene testing in Ontario will include the selection of the NGS instrument, and the DNA library preparation technical procedures. The laboratory must ensure that the performance of these two components provides gene testing data quality that is equivalent to Sanger sequence analysis for single-nucleotide variants (SNVs), in/dels, and multiplex ligation PCR amplification (MLPA) for detection of copy number alterations. The sequencing instrumentation used should enable quality and performance standard assessment. A number of specific requirements were also deemed to be important by the Technical Group. These include:
1. Coverage for every nucleotide in every coding exon ± 10 bp of adjacent intronic sequence.
In order to ensure adequate sensitivity and specificity, the minimum recommended depth of coverage should be set at a level that ensures detection of heterozygous SNVs and small insertions/deletions (in/dels) for all nucleotides in the target region. A minimum sequencing depth of 20–30× for any single nucleotide in the target region is recommended for NGS technologies currently used for diagnostic applications. If this criterion is not met, the laboratory should also perform Sanger sequencing of the region. There may also be regions that are routinely refractory to NGS (e.g., GC rich regions or long stretches of short repeats). The regions that have previously been shown to have a significant proportion of pathogenic variants, and do not meet minimum coverage requirements should undergo Sanger sequencing. Sanger sequencing may not be possible for certain regions (i.e., pseudogenic regions), and these need to be defined as part of the validation and clearly described in the clinical report. The authors recognize that a minimum 20–30-fold depth of coverage, per nucleotide is not sufficient to detect copy number variants. Reference Schenkel, Kerkhof and Stuart16 , Reference Kerkhof, Schenkel and Reilly17 Should a laboratory opt for, and propose, an NGS-based assessment of copy number variants (CNVs), the depth of coverage, at the “exon” level should be a minimum of 200-fold, and a minimum of 50-fold at the “nucleotide” level. Reference Schenkel, Kerkhof and Stuart16 , Reference Kerkhof, Schenkel and Reilly17
2. Testing for Copy Number Variants (CNVs)
CNVs detectible at least at exon level resolution (the standard for MLPA) must be performed for the select core genes with CNV assessment requirement routinely as part of the panel/subpanel screen (see Core Genes and Panels section in Supplementary Appendix 1). In addition to the required CNV assessment of these selected genes, given emerging evidence of pathogenic CNVs across the genome, routine CNV assessment for all remaining genes is suggested, but not mandatory. Since targeted MLPA approaches for routine CNV screening of multiple genes is not a feasible and cost effective option, comprehensive approaches are recommended. These can include, and are not limited to, NGS-based CNV analysis or microarray CNV analysis.
3. Ability to resolve pseudogenic regions
For pseudogenic regions, it can be difficult to determine whether an identified variant resides in the pseudogene or the gene targeted on the panel. For example, SYNGAP1 has a partial pseudogene, and a long-range PCR approach is required to map CNVs identified by NGS. Some such regions may not be resolvable for CNV analysis without special considerations in the NGS design and custom analytical procedures. Any such regions must clearly be described, in detail, or identified as limitations of the assay in clinical reports and testing validation documents. This applies to CNVs, SNVs and in/dels.
4. Technical confirmation of NGS findings
At the discretion of the laboratory, variants classified as American College of Medical Genetics (ACMG) category “Pathogenic,” “Likely Pathogenic” and “Variants of Uncertain Significance (VUS)” should be confirmed by Sanger sequencing or other appropriate methodology, when there is significant risk the variant is a false-positive. While the Sanger sequencing of SNVs is recommended, it would not be considered a mandatory prerequisite. However, for SNVs that are not confirmed by Sanger sequencing, we would require the laboratory to provide evidence for, and a description of, the procedures used to minimize the laboratories false-positive or false-negative test rate and procedures to ensure sample identity and integrity (e.g., the tracking of samples using molecular barcoding). We would also highly recommend Sanger sequencing of small insertions and deletions (in/dels) or other complex variants from the source DNA specimen.
5. Validation should include a range of specimens
The laboratory must demonstrate that variants such as SNVs and in/dels can be detected with the equivalent accuracy of previously established methodology (e.g., Sanger sequencing). While size of the detectable insertions, deletions and in/dels may vary depending on the sequence context and NGS technologies, size range validations should aim to include single to >50 nucleotides variants, and exon/gene deletions and duplications for CNV analysis.
Since mosaic pathogenic variants have been reported in epilepsy genes, Reference Stosser, Lindy and Butler18 validation of analytical procedures (e.g., a dilution series) should be designed to ensure detection of a predefined degree of mosaicism (e.g., 20%) for a limited subset of genes (see Supplementary Appendix 1).
Discussion
The comprehensive testing strategy summarized in this report is based on the consensus opinion of a working group of neurologists, medical geneticists, and laboratory scientists with expertise in genetic testing and epilepsy genetics. The incorporation of ECHO is unique for a genetic testing program as it facilitates the education of additional providers throughout Ontario in order to provide increased access to genetic testing for persons with epilepsy. Furthermore, the recommended testable epilepsy-related genes are based on an evidence-based framework (ClinGen), providing meaningful results that are interpretable by the ordering care providers with reduced risk of confusion. Lastly, the intent is for a program based on continuous improvement, current literature, laboratory performance metrics, and clinical outcome data collected from the ordering physician. This comprehensive programmatic approach to genetic testing is unique and will provide a robust mechanism providing high-quality testing.
The proposed epilepsy testing herein is based on a panel approach. This includes the option of a targeted or comprehensive panel approach or alternatively a filtered “exome” panel. Broader whole “exome” approaches to test for rare diseases, which include those with epilepsy or encephalopathies, are also advocated. Eligible Ontario patients can access exome sequencing if they meet the ministry’s eligibility criteria for testing, which are based on the GTAC report, “The Use of Genome-Wide Sequencing for Undiagnosed Rare Genetic Diseases,” to be considered for testing as an OHIP-funded service (http://www.health.gov.on.ca/en/pro/programs/gtac/reports.aspx). For those with epilepsy plus additional comorbidities such as dysmorphic features, multisystem involvement, or progressive clinical course, an exome sequencing strategy may be the better initial test. However, for those with isolated epilepsy (± refractory, ± early onset) without additional comorbidities, the criteria for exome sequencing would not be met and a panel-based approach is an option that offers a comparable diagnostic rate. Reference Myers, Johnstone and Dyment4
This implementation plan should serve as a helpful guide for other broad clinical indications being considered for provincially based patriation of testing, such as ataxia or neuromuscular disorders. It may also serve as a guide for other provinces in Canada or countries with a similar healthcare delivery model, wishing to implement an evidence-based genetic testing program. An evidence-based approach to diagnostic testing is key to responsible test implementation and utilization, particularly in a single-payer healthcare system such as Ontario.
Acknowledgements
We would like to acknowledge Cynthia Ho and P. Gordon for helpful discussion.
Statement of Authorship
DAD and ANP drafted the manuscript. DAD, ANP, DJS, BS, KMB, AH, MP, LBL, OJ, and OCS generated the data to be used for the manuscript. OCS was the Lead for both Technical and Clinical committees. KEM, GUE, and TH critically reviewed the manuscript.
Disclosures
The authors have no conflicts interest to declare.
Supplementary Material
To view supplementary material for this article, please visit https://doi.org/10.1017/cjn.2019.304





