The selective serotonin reuptake inhibitor (SSRI) antidepressants are used as first-line treatment of depression, but up to 50% of people with depression do not adequately benefit from SSRI treatment. Reference Thase, Entsuah and Rudolph1,Reference Trivedi, Rush, Wisniewski, Nierenberg, Warden and Ritz2 A significant proportion of the individual differences in response to SSRI is familial. Reference Franchini, Serretti, Gasperini and Smeraldi3 When searching for a genetic predictor of SSRI response, the serotonin transporter gene (SLC6A4) is the natural candidate as it encodes the molecular target of SSRI antidepressants. Several functional polymorphisms have been found in the SLC6A4 gene. The common length polymorphism constituted by a deletion of 44 base pairs in the promoter region, referred to as 5-HTTLPR, has been explored in most detail and implicated in depression aetiology. Reference Uher and McGuffin4 The long allele has been associated with more efficient transcription, Reference Lesch, Balling, Gross, Strauss, Wolozin and Murphy5 resilience to depressogenic effects of adversity Reference Uher and McGuffin4 and better response to SSRI antidepressants. Reference Smeraldi, Zanardi, Benedetti, Di, Perez and Catalano6 A meta-analysis of 14 studies with 1435 participants showed that long-allele carriers had higher probability of response and remission than short-allele homozygotes. Reference Serretti, Kato, De and Kinoshita7 Therefore, SLC6A4 was the prime candidate for response to the SSRI escitalopram and the first gene to be genotyped and tested in the Genome Based Therapeutic Drugs for Depression (GENDEP) project. However, significant heterogeneity of 5-HTTLPR effect was reported in non-European populations. Reference Kim, Lim, Kim, Kim, Chang and Carroll8,Reference Serretti, Kato, De and Kinoshita7 Whereas no effect of 5-HTTLPR on efficacy of citalopram was found in a large mixed-ethnicity sample, Reference Kraft, Peters, Slager, Jenkins, Reinalda and McGrath9 an ethnicity-sensitive re-analysis of the same sample found an effect in the expected direction among White non-Hispanic participants. Reference Mrazek, Rush, Biernacka, O'Kane, Cunningham and Wieben10 As the 5-HTTLPR allele frequencies and its linkage disequilibrium with other polymorphisms within the SLC6A4 gene vary markedly across populations, Reference Gelernter, Cubells, Kidd, Pakstis and Kidd11 it is possible that the discrepancies are a result of the influence of another polymorphism that is in population-specific linkage disequilibrium with the 5-HTTLPR.
Three polymorphisms have been proposed to modulate the functionality of 5-HTTLPR: single nucleotide polymorphisms rs25531 and rs2020933, and a 17bp variable tandem repeat in the second intron (STin2). The rs25531 is located within the repeats constituting the 5-HTTLPR. Two initial reports disagreed about the location of this putatively functional A/G single nucleotide polymorphism being either within Reference Hu, Oroszi, Chun, Smith, Goldman and Schuckit12 or immediately upstream Reference Kraft, Slager, McGrath and Hamilton13 of the 5-HTTLPR insertion/deletion segment. However, sequence alignment has demonstrated that these two reports point to the same polymorphism, the rs25531 (A/G), which is located in the sixth repeat of the 5-HTTLPR outside the deleted segment and therefore can co-occur with either long or short variant of the 5-HTTLPR. Reference Wendland, Martin, Kruse, Lesch and Murphy14 The minor G allele of the rs23551 creates a consensus binding sequence for the transcription factor AP-2, which is involved in antidepressant action. Reference Braganca, Eloranta, Bamforth, Ibbitt, Hurst and Bhattacharya15,Reference Damberg16 It has been proposed that the rs25531 influences the functionality of the serotonin transporter and recoding of 5-HTTLPR long alleles based on the rs25531 has been suggested. Reference Hu, Lipsky, Zhu, Akhtar, Taubman and Greenberg17 The functionality of this single nucleotide polymorphism has however not been consistently replicated and it is still unclear whether the recoding of the 5-HTTLPR is warranted. Reference Uher and McGuffin4,Reference Wendland, Martin, Kruse, Lesch and Murphy14,Reference Martin, Cleak, Willis-Owen, Flint and Shifman18 Therefore, in the present study, the rs25531 is evaluated separately from the 5-HTTLPR. The rs2020933 single nucleotide polymorphism is located in the first intron, approximately 2.5kb 3′ to the 5-HTTLPR and contributes to the variation of serotonin transporter expression. Reference Martin, Cleak, Willis-Owen, Flint and Shifman18 The STin2 two major alleles (STin2.10 and STin2.12) correspond to 10 or 12 repeats of a 17bp sequence and the STin2.12 allele has been associated with enhanced transcription. Reference MacKenzie and Quinn19
It has been proposed that the moderation of antidepressant response by the 5-HTTLPR may depend on age. Reference Joyce, Mulder, Luty, McKenzie, Miller and Rogers20 This suggestion is supported by data showing age-related variation in serotonergic function, Reference Tauscher, Verhoeff, Christensen, Hussey, Meyer and Kecojevic21–Reference Yamamoto, Suhara, Okubo, Ichimiya, Sudo and Inoue23 but tests of interaction between age and 5-HTTLPR have not been reported in most pharmacogenetic studies.
Gender may be another important factor influencing the relationship between 5-HTTLPR genotype and antidepressant response. Ovarian steroids have a strong influence on serotonin synthesis, Reference Hiroi, McDevitt and Neumaier24 expression of serotonergic receptors, Reference Biegon and McEwen25–Reference Wissink, van der, Katzenellenbogen and van der Saag27 and the serotonin transporter. Reference Lu, Eshleman, Janowsky and Bethea28,Reference McQueen, Wilson and Fink29 There are gender differences in 5-HTTLPR moderation of response to serotonergic challenges, including tryptophan depletion Reference Walderhaug, Magnusson, Neumeister, Lappalainen, Lunde and Refsum30 and to stressful life events. Reference Uher and McGuffin4,Reference Sjoberg, Nilsson, Nordquist, Ohrvik, Leppert and Lindstrom31 However, previous pharmacogenetic studies have not reported a test of gender×genotype interaction.
The GENDEP project aims to identify differential predictors of response to SSRI and tricyclic antidepressants. The primary aim is to test the hypothesis that the 5-HTTLPR interacts with the drug used (escitalopram v. nortriptyline) in treatment of depression. Specifically, it is expected that short-allele homozygotic status will be associated with poor response to escitalopram but will not influence response to nortriptyline. As the heterogeneity of findings across studies may be a result of the influence of a polymorphism that is in population-specific linkage disequilibrium with the 5-HTTLPR, we systematically explore DNA sequence variation across the SLC6A4 gene, including the three polymorphisms with known effects on gene expression. Additionally, the moderating effects of age and gender are tested.
Method
Study design and sample
The GENDEP project is an open-label part-randomised multi-centre pharmacogenetic study with two active pharmacological treatment arms. Reference Uher, Maier, Hauser, Marušič, Schmael and Mors32 It was designed to establish clinical and genetic determinants of therapeutic response to two antidepressants with contrasting primary modes of action: nortriptyline (a tricyclic inhibitor of monoamine reuptake with strongest affinity for the noradrenaline transporter) and escitalopram (an SSRI). Eight hundred and eleven adults with ICD–10 33 /DSM–IV 34 unipolar major depression of at least moderate severity Reference Wing, Sartorius and Ustin35 were recruited in eight European countries: Belgium, Croatia, Denmark, Germany, Italy, Poland, Slovenia and the UK. To minimise confounding by population stratification, recruitment was restricted to individuals of White European parentage. Personal or family history of bipolar disorder or schizophrenia constituted exclusion criteria. The study was approved by ethics boards in all participating centres. All participants provided a written consent after the procedures were fully explained.
Participants included 296 men and 514 women between 19 and 72 years old (Table 1). The average participant was in her second episode of moderately severe depression and scored 28.7 (s.d. = 6.7) on the Montgomery–Åsberg Depression Rating Scale (MADRS), Reference Montgomery and Åsberg36 21.7 (s.d. = 5.3) on the 17-item Hamilton Rating Scale for Depression (HRSD–17) Reference Hamilton37,Reference Hamilton38 and 28.0 (s.d. = 9.7) on the Beck Depression Inventory (BDI) Reference Beck, Ward, Mendelson, Mock and Erbaugh39 at baseline.
Table 1 Sample characteristics at baseline. The demographic and clinical characteristics are given by drug and 5-HTTLPR genotype

| Escitalopram (n = 450) | Nortriptyline (n = 345) | |||||
|---|---|---|---|---|---|---|
| l/l | l/s | s/s | l/l | l/s | s/s | |
| Total, n (%) | 162 (36) | 211 (47) | 77 (17) | 135 (38) | 157 (46) | 53 (16) |
| Female gender n (%) | 107 (66) | 127 (60) | 44 (57) | 86 (64) | 103 (66) | 36 (69) |
| Age, years: mean (s.d.) | 41.4 (11.5) | 42.9 (11.4) | 44.5 (12.5) | 41.5 (11.4) | 42.8 (11.5) | 41.3 (13.8) |
| Education, years: mean (s.d.) | 12.2 (3.2) | 12.2 (3.3) | 11.8 (2.9) | 12.4 (3.1) | 11.9 (3.1) | 12.1 (3.1) |
| Married/cohabiting, n (%) | 84 (52) | 103 (49) | 38 (49) | 75 (56) | 78 (50) | 25 (47) |
| Employed/student, n (%) | 95 (59) | 120 (57) | 40 (52) | 78 (58) | 71 (45) | 27 (51) |
| Age at depression onset, years: mean (s.d.) | 32.3 (9.5) | 33.5 (9.8) | 34.3 (9.7) | 31.7 (9.8) | 32.3 (10) | 31.4 (10.4) |
| First episode, n (%) | 61 (38) | 76 (36) | 23 (30) | 41 (30) | 42 (27) | 17 (32) |
| Current episode duration, weeks: mean (s.d.) | 18.2 (12.3) | 19.8 (12.1) | 19.7 (10.0) | 17.1 (11.0) | 20.5 (14.6) | 17.7 (13.7) |
| History of taking antidepressants, n (%) | 62 (38) | 96 (45) | 30 (39) | 68 (50) | 84 (54) | 22 (42) |
| Baseline severity (MADRS), mean (s.d.) | 28.5 (6.0) | 28.2 (6.7) | 28.4 (7.6) | 28.7 (6.7) | 29.3 (6.9) | 29.9 (6.5) |
Participants with no contraindications were randomly allocated to receive flexible dosage of nortriptyline (50–150 mg daily) or escitalopram (10–30 mg daily) for 12 weeks. Individuals with relative or absolute contraindications (medical conditions incompatible with the use of the antidepressant and history of intolerance or non-response to one study medication) for one of the drugs were allocated non-randomly to the other antidepressant. Other psychotropic medication was not allowed with the exception of occasional use of hypnotics. Adherence was monitored by weekly self-report and plasma levels of antidepressants were measured at week 8. Adverse effects of medication were measured using the UKU Side-Effect Rating Scale Reference Lingjaerde, Ahlfors, Bech, Dencker and Elgen40 and Self-Report Antidepressant Side-Effect Checklist. Reference Uher, Farmer, Henigsberg, Rietschel, Mors and Maier41
Outcome measurement
The response to antidepressant medication is a complex phenotype that includes changes in a number of symptoms occurring over a period of up to 12 weeks following the initiation of an antidepressant. In GENDEP, the responses to escitalopram and nortriptyline were assessed by weekly administration of three established measures of depression severity: the clinician-rated 10-item MADRS, the HRSD–17 and the self-report 21-item BDI were administered at week 0 (baseline) and then weekly for 12 weeks. All raters were trained and achieved high interrater reliability, with intraclass correlation of at least 0.9. Reference Uher, Farmer, Maier, Rietschel, Hauser and Marusic42 A psychometric analysis found that the MADRS was the most internally consistent and informative of the three scales and that depressive symptoms could be described in more detail by three symptom dimensions derived by categorical item factor analysis: observed mood, cognitive symptoms and neurovegetative symptoms. Reference Uher, Farmer, Maier, Rietschel, Hauser and Marusic42 In the pharmacogenetic analyses we used the MADRS score as a primary continuous outcome variable. Findings of interest were further explored using the three symptom dimensions.
Genotyping
Adequate DNA samples were available for 795 of the 811 participants. A sample of 5–8 ml of blood was collected in ethylenediamine tetra-acetic acid and frozen. DNA was extracted using a standard procedure. Reference Freeman, Smith, Curtis, Huckett, Mill and Craig43
A two-stage method was used to type the 5HTTLPR and rs25531. In the first stage, short and long alleles were determined by polymerase chain reaction (PCR). In the second stage, the A/G alleles of rs25531 single nucleotide polymorphism within the 5HTTLPR repeats were identified using restriction fragment length polymorphism. The PCR product was digested with the Msp1 enzyme, which cuts the amplified PCR product at two or three places, depending on the rs25531 genotype. The combinations of products that determine the genotypes are shown in Appendix 1.
Five of the microsatellite markers listed on the UCSC bioinformatics website (http://genome.ucsc.edu/ accessed in September 2004) were selected for investigation and genotyped on 20 individuals, using PCR and electrophoresis, to check whether common polymorphisms at these loci are present. Only STin2 (SLC6A4, intron2) and STin4 (SLC6A4, intron4) proved to be polymorphic, and thus were carried forward for full analysis on the GENDEP sample. Fluorescent forward primers were used for amplification of these microsatellites. Genotyping of the PCR products was performed using the ABI3130 sequencer.
Single nucleotide polymorphisms were selected that tag DNA sequence variation within the SLC6A4 gene and its 1000 bp margins using the SNPTagger program (www.broad.mit.edu/mpg/tagger) run in Windows XP and HAPMAP data on the CEPH CEU population with European ancestry (CEPH NCBI Build 35/UCSC hg17/May 2004 coordinates). Reference De Bakker, Yelensky, Pe'er, Gabriel, Daly and Altshuler44 Criteria for single nucleotide polymorphism selection included a minimal allele frequency of 5% in the White population and a pairwise r 2 = 0.8. Ten single nucleotide polymorphisms met these criteria (Table 2). According to SNPTagger, these 10 single nucleotide polymorphisms provided 92% coverage of the DNA sequence variation in the SLC6A4 gene. The single nucleotide polymorphisms were genotyped using the SNPlexTM method. This included oligonucleotide ligation assay/PCR technology for allelic discrimination and ligation product amplification. The assay products were run on the ABI3130 sequencer and data were exported and analysed using GENEMAPPER software version 3.7 for Windows XP (Applied Biosystems). Eleven samples were genotyped in blind duplicates and the agreement was 100% for all markers.
Table 2 Linkage disequilibrium, allele frequency and Hardy–Weinberg equilibriuma

| HTTLPR | rs2020933 | rs2066713 | rs2020939 | STin2 | rs8076005 | rs2020942 | STin4 | rs140700 | rs4583306 | rs140701 | rs4325622 | rs3813034 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HTTLPR | 0.04 | 0.09 | 0.05 | 0.07 | 0.00 | 0.09 | 0.07 | 0.01 | 0.07 | 0.08 | 0.05 | 0.05 | |
| rs2020933 | 0.90 | 0.01 | 0.03 | 0.00 | 0.17 | 0.01 | 0.04 | 0.04 | 0.03 | 0.04 | 0.04 | 0.04 | |
| rs2066713 | 0.47 | 0.39 | 0.42 | 0.89 | 0.14 | 0.97 | 0.47 | 0.06 | 0.46 | 0.47 | 0.31 | 0.31 | |
| rs2020939 | 0.25 | 0.72 | 0.94 | 0.38 | 0.14 | 0.42 | 0.88 | 0.06 | 0.86 | 0.86 | 0.71 | 0.71 | |
| STin2 | 0.40 | 0.27 | 0.99 | 0.84 | 0.11 | 0.92 | 0.43 | 0.06 | 0.43 | 0.43 | 0.27 | 0.27 | |
| rs8076005 | 0.07 | 0.74 | 1.00 | 0.90 | 0.86 | 0.14 | 0.16 | 0.42 | 0.15 | 0.15 | 0.17 | 0.17 | |
| rs2020942 | 0.46 | 0.38 | 0.99 | 0.93 | 1.00 | 1.00 | 0.46 | 0.06 | 0.46 | 0.47 | 0.31 | 0.31 | |
| STin4 | 0.29 | 0.81 | 0.99 | 0.94 | 0.91 | 0.95 | 0.99 | 0.08 | 0.97 | 0.98 | 0.81 | 0.81 | |
| rs140700 | 0.40 | 0.24 | 1.00 | 0.89 | 0.89 | 0.98 | 1.00 | 1.00 | 0.08 | 0.07 | 0.09 | 0.09 | |
| rs4583306 | 0.29 | 0.81 | 0.99 | 0.93 | 0.90 | 0.95 | 0.99 | 0.99 | 1.00 | 0.97 | 0.79 | 0.79 | |
| rs140701 | 0.29 | 0.81 | 0.99 | 0.93 | 0.90 | 0.95 | 0.99 | 0.99 | 0.96 | 1.00 | 0.80 | 0.80 | |
| rs4325622 | 0.24 | 0.79 | 0.76 | 0.89 | 0.67 | 0.96 | 0.75 | 0.95 | 1.00 | 0.95 | 0.94 | 1.00 | |
| rs3813034 | 0.24 | 0.79 | 0.76 | 0.89 | 0.67 | 0.96 | 0.75 | 0.95 | 1.00 | 0.95 | 0.94 | 1.00 | |
| MAF | 0.40 | 0.06 | 0.39 | 0.43 | 0.42 | 0.18 | 0.39 | 0.43 | 0.09 | 0.43 | 0.43 | 0.46 | 0.46 |
| HWE χ2 | 0.87 | 0.05 | 2.00 | 1.75 | 12.33 | 0.13 | 2.40 | 1.46 | 1.22 | 0.53 | 1.56 | 4.82 | 5.18 |
| HWE P | 0.3500 | 0.8292 | 0.1574 | 0.1853 | 0.0004 | 0.7201 | 0.1210 | 0.2264 | 0.2695 | 0.4659 | 0.2121 | 0.0282 | 0.0228 |
In addition to the SLC6A4 markers, a panel of 35 single nucleotide polymorphism markers selected as being informative for population stratification in a European population Reference Seldin, Shigeta, Villoslada, Selmi, Tuomilehto and Silva45 were genotyped in all individuals to establish the presence of and allow effective control for any population stratification.
The linkage disequilibrium measures (D′ and R 2) were calculated using the Haploview program version 4 for Windows XP. Reference Barrett, Fry, Maller and Daly46 Markers were included irrespective of genotype distribution as deviations from the Hardy–Weinberg equilibrium are expected in a cases-only sample. Reference Wittke-Thompson, Pluzhnikov and Cox47 However, Hardy–Weinberg equilibrium was tested and results are given in Table 2.
Statistical analysis
To use all available weekly data on response to antidepressants and provide unbiased estimates in the presence of missing values, effects of genotype markers were tested using linear mixed models with individual random intercept and slope and fitted with maximum likelihood. All models included the fixed effects of time (linear and quadratic), baseline depression severity, history of using antidepressants, drug, age and gender, and random effects of the individual and the recruitment centre. To account for correlations between repeated observations, the intercept and slope of time were allowed to vary randomly between participants. The results of regression are presented as standardised regression coefficients (β) with 95% confidence intervals. Standardised regression coefficients denote the number of standard deviations on the outcome measure per unit of the predictor variable. As the standard deviation of the MADRS was 7, a β of 0.14 corresponds to a difference of one point on the MADRS.
As there was strong prior evidence for the influence of 5-HTTLPR on response to serotonergic antidepressants, the 5-HTTLPR was first tested as a single marker without correcting for multiple testing and the model was further developed using a stepwise approach. The effect of genotype and its interactions with drug, gender and age were tested as fixed explanatory variables in the mixed linear models. Competing nested models were compared using likelihood ratio tests. The previously reported recessive short allele model Reference Serretti, Kato, De and Kinoshita7,Reference Smeraldi, Zanardi, Benedetti, Di, Perez and Catalano6 wascomparedwithafull genotype model. To test the hypothesis that a polymorphism in linkage disequilibrium with the 5-HTLPR modulates its effect, other markers within the SLC6A4 gene were then added in order of linkage disequilibrium with the 5-HTTLPR and retained if model fit improved significantly according to likelihood ratio tests (P<0.05).
Haplotypes for markers within the SLC6A4 gene that were in linkage disequilibrium (D′>0.7) were estimated 12 times using a Bayesian procedure in Phase, version 2.1 for Windows XP Reference Stephens and Donnelly48,Reference Stephens, Smith and Donnelly49 and used as multiply imputed data-sets in the mixed linear models. Reference Cordell50 This method has been demonstrated to provide unbiased estimates and to have an adequate confidence interval coverage even when haplotypes are estimated based on cases only. Reference Cordell50
The ancestry-informative markers were analysed using 10 000 burn-ins and 10 000 repetitions under an admixture model with correlated allele frequencies in STRUCTURE version 2.2 for Windows XP (http://pritch.bsd.uchicago.edu/software.html). Reference Falush, Stephens and Pritchard51 This analysis found no clustering within the sample. Stratification was further estimated using the genomic control approach Reference Devlin and Roeder52 implemented in PLINK for Windows XP (http://pngu.mgh.harvard.edu/~purcell/plink). Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira and Bender53 Based on the same set of ancestry-informative single nucleotide polymorphisms and all candidate gene markers, PLINK estimated a genomic inflation factor of 1, confirming absence of significant stratification in the GENDEP sample.
Power analysis
The sample size required for detecting an interaction between antidepressant and the 5-HTTLPR genotype was estimated based on data from published European studies that provided continuous outcome measures by genotype group. Reference Smeraldi, Zanardi, Benedetti, Di, Perez and Catalano6,Reference Zanardi, Serretti, Rossini, Franchini, Cusin and Lattuada54 These studies showed that 5-HTTLPR influenced outcome with an effect size of 0.65–1.00 s.d. on a continuous outcome scale. Assuming a recessive model with minor allele frequency of 0.4, an effect size of 0.65 in participants treated with escitalopram and no effect of genotype on outcome in participants treated with nortriptyline, 270 participants per treatment arm were needed to detect a drug×genotype interaction at α = 0.05 with a power of 0.8. The GENDEP sample provided a power of more than 0.9.
Results
Genotyping
The 5-HTTLPR, intron 2 and intron 4 tandem repeats and 11 single nucleotide polymorphisms were typed in 795 (98%) of the 811 participants. Sample demographic and clinical characteristics at baseline were unrelated to the 5-HTTLPR genotype (all P>0.05, Table 1). There was also no significant relationship between the 5-HTTLPR genotype and the mode of allocation (random v. non-random, χ2 = 1.34, P = 0.246) or the drug allocated among non-randomised participants (χ2 = 0.0001, P = 0.994). The 16 participants with missing or inadequate DNA samples did not differ from the genotyped participants on depression severity, age or gender (all P>0.05). Table 2 shows the linkage disequilibrium among the 14 genotyped markers and minor allele frequencies. Frequencies of the 5-HTTLPR, s25531, rs2020933 and STin2 genotypes are given in Table 3.
Table 3 Distribution of genotypes at the 5-HTTLPR, rs25531, rs2020933 and STin2 locia
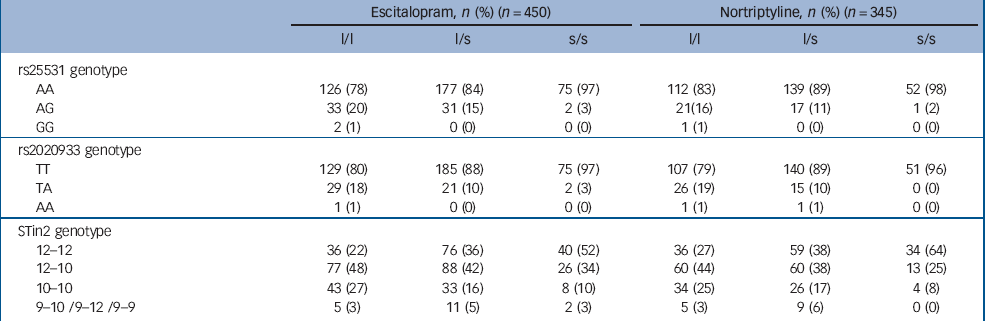
| Escitalopram, n (%) (n = 450) | Nortriptyline, n (%) (n = 345) | |||||
|---|---|---|---|---|---|---|
| l/l | l/s | s/s | l/l | l/s | s/s | |
| rs25531 genotype | ||||||
| AA | 126 (78) | 177 (84) | 75 (97) | 112 (83) | 139 (89) | 52 (98) |
| AG | 33 (20) | 31 (15) | 2 (3) | 21 (16) | 17 (11) | 1 (2) |
| GG | 2 (1) | 0 (0) | 0 (0) | 1 (1) | 0 (0) | 0 (0) |
| rs2020933 genotype | ||||||
| TT | 129 (80) | 185 (88) | 75 (97) | 107 (79) | 140 (89) | 51 (96) |
| TA | 29 (18) | 21 (10) | 2 (3) | 26 (19) | 15 (10) | 0 (0) |
| AA | 1 (1) | 0 (0) | 0 (0) | 1 (1) | 1 (1) | 0 (0) |
| STin2 genotype | ||||||
| 12-12 | 36 (22) | 76 (36) | 40 (52) | 36 (27) | 59 (38) | 34 (64) |
| 12-10 | 77 (48) | 88 (42) | 26 (34) | 60 (44) | 60 (38) | 13 (25) |
| 10-10 | 43 (27) | 33 (16) | 8 (10) | 34 (25) | 26 (17) | 4 (8) |
| 9-10/9-12/9-9 | 5 (3) | 11 (5) | 2 (3) | 5 (3) | 9 (6) | 0 (0) |
The 5-HTTLPR
The 5-HTTLPR genotype moderated response to the serotonergic antidepressant in the expected direction. A significant three-way interaction between the 5-HTTLPR, drug and gender (P = 0.026; Table 4) indicated that this moderating effect is conditional on the type of antidepressant drug and on gender. Specifically, male carriers of the 5-HTTLPR long allele had a better response to escitalopram than male short-allele homozygotes (Fig. 1). The best-fitting model included a recessive effect of the 5-HTTLPR short allele, its two-way interactions with gender and drug, as well as the three-way interaction. Age did not significantly interact with genotype.

Fig. 1 Moderation of antidepressant response by 5-HTTLPR genotype and gender: (a) men treated with escitalopram, (b) women treated with escitalopram, (c) men treated with nortriptyline, (d) women treated with nortriptyline. Depression severity measured by the Montgomery–Åsberg Depression Rating Scale (MADRS) in observed cases over the 12 weeks of treatment. Details of genotype are given in key; l, long allele; s, short allele. Note that values for weeks 9–12 are less reliable because of missing values resulting from drop-out and switching.
Table 4 Results of the final best-fitting models for overall depression symptoms (Montgomery–Åsberg Depression Rating Scale (MADRS))a

| MADRS | β | s.e. | P | 95% CI |
|---|---|---|---|---|
| 5-HTTLPR (recessive s) | 0.194 | 0.076 | 0.010 | 0.046 to 0.343 |
| Drug | 0.065 | 0.018 | <0.001 | 0.029 to 0.100 |
| Gender | 0.320 | 0.117 | 0.006 | 0.091 to 0.549 |
| Interaction 5-HTTLPR × gender | -0.272 | 0.150 | 0.070 | -0.567 to 0.022 |
| Interaction 5-HTTLPR × drug | -0.537 | 0.194 | 0.006 | -0.917 to -0.157 |
| Interaction gender × drug | -0.219 | 0.095 | 0.021 | -0.405 to -0.034 |
| Interaction 5-HTTLPR × drug × gender | 0.532 | 0.240 | 0.027 | 0.061 to 1.004 |
| rs2020933 (additive) | 0.120 | 0.061 | 0.049 | 0.001 to 0.240 |
The interaction effects were qualified by significant main effects in participants treated with escitalopram and especially men treated with escitalopram. Among participants treated with escitalopram those homozygous for the short allele had significantly worse outcome than long-allele carriers (β = 0.154, 95% CI 0.003–0.305, P = 0.045). The effect of the 5-HTTLPR genotype was strong among males treated with escitalopram (β = 0.333, 95% CI 0.085–0.580, P = 0.008, Fig. 1a) but absent among females treated with escitalopram (β = 0.036, 95% CI −0.155 to 0.228, P = 0.709, Fig. 1b). The 5-HTTLPR genotype did not affect outcome among the participants treated with nortriptyline (β = −0.063, 95% CI −0.231 to 0.106, P = 0.466, Fig. 1c and 1d). The 5-HTTLPR genotype explained 1.1% of variance in the repeated measurement of depression severity among men treated with escitalopram, but only 0.3% of variance in antidepressant response across the whole sample.
The additive or full genotype model provided no advantage over the recessive short-allele model (all likelihood ratio tests had P>0.1). The 5-HTTLPR genotype was unrelated to baseline severity, antidepressant dosage, self-reported adherence, plasma levels of either nortriptyline or escitalopram and adverse effects.
rs25531
The single nucleotide polymorphism rs25531 (A/G; minor allele frequency 0.075) was in linkage disequilibrium with the 5-HTTLPR (D′ = 0.82, R 2 = 0.18).
Given the controversies about the moderation of 5-HTTLPR functionality by the rs25531, Reference Hu, Lipsky, Zhu, Akhtar, Taubman and Greenberg17,Reference Martin, Cleak, Willis-Owen, Flint and Shifman18 we tested whether the recoding of the 5-HTTLPR based on the rs25531 presumed functionality, Reference Hu, Lipsky, Zhu, Akhtar, Taubman and Greenberg17 or its inclusion as a separate marker improved the model fit. This was not the case: the recoding impaired model fit and its inclusion as a separate marker added no information to the prediction of antidepressant response (likelihood ratio test P>0.1). Therefore, the rs25531 was dropped from further analyses. The rs25531-recoded 5-HTTLPR genotype was also not related to baseline severity, antidepressant dosage, self-reported adherence, plasma levels of either nortriptyline or escitalopram and adverse effects.
rs2020933
The single nucleotide polymorphism rs2020933 (A/T; minor allele frequency 0.06) was in linkage disequilibrium with the 5-HTTLPR (D′ = 0.90, R 2 = 0.04). The addition of rs2020933 (additive or dominant coding) to the model as a fixed first-order effect provided a significant improvement in the prediction of antidepressant response. Interaction between rs2020933 and 5-HTTLPR, drug, gender and age did not significantly improve the model. The final best-fitting model included both 5-HTTLPR with its interacting effects and the rs2020933 and led to a significant improvement in the prediction of antidepressant response compared with the model with no genotype data (likelihood ratio test χ2(5) = 13.57, P = 0.0219, Table 4).
The intron 2 simple tandem repeat
The STin2 was in weak linkage disequilibrium with the 5-HTTLPR (D′ = 0.40, R 2 = 0.07). The STin2 did not contribute to the prediction of treatment response (likelihood ratio test P>0.1).
Haplotype analysis
As there was significant linkage disequilibrium between the 5-HTTLPR and the rs2020933, the influence of estimated haplotypes comprising these two markers on antidepressant response was investigated. Although the haplotypes containing the 5-HTTLPR long allele were associated with better response to escitalopram, the inclusion of haplotypes provided no advantage over single marker analysis in predicting drug response.
Other markers within the SLC6A4 gene
Other markers within the SLC6A4 gene had low linkage disequilibrium with 5-HTTLPR and their inclusion as single markers or haplotypes conferred no improvement in the prediction model.
Effects on symptom dimensions
Our previous analyses have indicated that separable dimensions of depressive symptoms may differ in their response to antidepressants. Reference Uher, Maier, Hauser, Marušič, Schmael and Mors32 We therefore investigated the genotype effect on change on the three symptoms dimensions. Reference Uher, Farmer, Maier, Rietschel, Hauser and Marusic42 The 5-HTTLPR and rs2020933 significantly moderated response on the observed mood and cognitive symptom dimensions. The change on the neurovegetative symptom dimension was relatively independent of genotype (Table 5). The effect on the observed mood dimension was stronger than on the MADRS.
Table 5 Results of the final model for specific dimensions of observed mood, cognitive and neurovegetative symptomsa

| β | s.e. | P | 95% CI | |
|---|---|---|---|---|
| Observed mood | ||||
| 5-HTTLPR (recessive s) | 0.231 | 0.075 | 0.002 | 0.083 to 0.378 |
| Drug | 0.056 | 0.018 | 0.002 | 0.020 to 0.091 |
| Gender | 0.344 | 0.116 | 0.003 | 0.117 to 0.572 |
| Interaction 5-HTTLPR × gender | -0.329 | 0.149 | 0.027 | -0.621 to -0.037 |
| Interaction 5-HTTLPR × drug | -0.504 | 0.192 | 0.009 | -0.881 to -0.127 |
| Interaction gender × drug | -0.163 | 0.094 | 0.083 | -0.347 to 0.021 |
| Interaction 5-HTTLPR × drug × gender | 0.554 | 0.238 | 0.020 | 0.086 to 1.022 |
| rs2020933 (additive) | 0.134 | 0.061 | 0.027 | 0.015 to 0.253 |
| Cognitive | ||||
| 5-HTTLPR (recessive s) | 0.252 | 0.080 | 0.002 | 0.095 to 0.409 |
| Drug | 0.051 | 0.019 | 0.007 | 0.014 to 0.089 |
| Gender | 0.308 | 0.123 | 0.013 | 0.066 to 0.551 |
| Interaction 5-HTTLPR × gender | -0.233 | 0.159 | 0.143 | -0.545 to 0.079 |
| Interaction 5-HTTLPR × drug | -0.518 | 0.204 | 0.011 | -0.919 to -0.117 |
| Interaction gender × drug | -0.234 | 0.100 | 0.020 | -0.431 to -0.037 |
| Interaction 5-HTTLPR × drug × gender | 0.464 | 0.254 | 0.068 | -0.034 to 0.962 |
| rs2020933 (additive) | 0.118 | 0.064 | 0.067 | -0.009 to 0.244 |
| Neurovegetative | ||||
| 5-HTTLPR (recessive s) | 0.076 | 0.081 | 0.344 | -0.082 to 0.235 |
| Drug | 0.079 | 0.019 | <0.001 | 0.040 to 0.117 |
| Gender | 0.235 | 0.125 | 0.060 | -0.010 to 0.480 |
| Interaction 5-HTTLPR × gender | -0.202 | 0.160 | 0.207 | -0.517 to 0.112 |
| Interaction 5-HTTLPR × drug | -0.459 | 0.207 | 0.027 | -0.865 to -0.053 |
| Interaction gender × drug | -0.270 | 0.101 | 0.008 | -0.469 to -0.072 |
| Interaction 5-HTTLPR × drug × gender | 0.349 | 0.257 | 0.174 | -0.154 to 0.853 |
| rs2020933 (additive) | 0.073 | 0.065 | 0.263 | -0.055 to 0.200 |
Discussion
The GENDEP data support the previously reported moderation of antidepressant response by the length polymorphism in the promoter region of the serotonin transporter gene. This effect is specific to the serotonergic mode of antidepressant action, appears to be concentrated in men and may be moderated by another polymorphism at the 5′ end of the serotonin transporter gene.
Gender effects
Within the GENDEP sample, the effect of the 5-HTTLPR on response to escitalopram was marked in men but absent in women. Among men treated with escitalopram, the 5-HTTLPR polymorphism is associated with a five-point difference on the MADRS between short-allele homozygotes and long-allele carriers. This finding is consistent with previous reports of gender-specific genetic influences on serotonergic function. Reference Weiss, Abney, Cook and Ober55 It may reflect a biological interaction between the serotonergic system and ovarian hormones. Through the oestrogen alpha receptor, oestrogens stimulate the production of the 5-HT1A receptor that is involved in the regulation of serotonin release and is down-regulated in response to serotonin reuptake inhibitors. Reference Wissink, van der, Katzenellenbogen and van der Saag27 Oestrogens also increase the expression of the serotonin transporter. Reference McQueen, Wilson and Fink29 The impact of experimental manipulations of serotonergic function also depends on gender and hormonal status. Decrease in serotonergic function during tryptophan depletion has different effects on men and women Reference Walderhaug, Magnusson, Neumeister, Lappalainen, Lunde and Refsum30 and oestrogen supplementation may enhance the action of antidepressants in perimenopause. Reference Soares, Almeida, Joffe and Cohen57,Reference Katz, Koslow and Frazer58 It is possible that the impact of the less functional short 5-HTTLPR allele is moderated by the oestrogen-induced stimulatory effect on serotonin transporter expression in hormonally active women. Although this accumulated evidence adds biological plausibility to the observed 5-HTTLPR×gender interaction, this finding requires replication in an independent sample.
Age effects
Although gender appears to be an important factor, age did not significantly interact with genotype in its effect on outcome. Joyce et al Reference Joyce, Mulder, Luty, McKenzie, Miller and Rogers20 reported interaction between age and 5-HTTLPR with age dichotomised at 25. As the GENDEP sample is older, separate analysis of individuals below 25 years of age was not feasible. Therefore, the current negative results should not be interpreted as a refutation of the findings by Joyce and colleagues. Reference Joyce, Mulder, Luty, McKenzie, Miller and Rogers20
The effect of rs2020933
The response to antidepressants in the GENDEP study was also associated with the single nucleotide polymorphism rs2020933, which is located in the first intron of the SLC6A4 gene. The major T allele was associated with a better response to both nortriptyline and escitalopram and there was no significant interaction with gender. The rs2020933 single nucleotide polymorphism has been reported to be functional in an allelic expression imbalance study. Reference Martin, Cleak, Willis-Owen, Flint and Shifman18 It is in significant linkage disequilibrium with the 5-HTTLPR, but because of a large difference in minor allele frequency, only a relatively small proportion of variance in the 5-HTTLPR is shared with the rs2020933. As this genomic region varies significantly between populations, Reference Gelernter, Cubells, Kidd, Pakstis and Kidd11 the interplay between these two functional loci has the potential to explain discordant findings in samples of different ethnic origins. Reference Serretti, Kato, De and Kinoshita7,Reference Kim, Lim, Kim, Kim, Chang and Carroll8 This remains to be tested in non-European samples. However, as the effect of rs2020933 was relatively weak and was not specific to the serotonergic mode of action, it is unlikely to fully explain the influence of the 5-HTTLPR on response to serotonergic antidepressants.
Drug effects on symptom dimensions
Depression is a heterogeneous group of disorders and the genetic mediation of response to antidepressants may differ between separable dimensions of depressive symptoms. For example, it has been demonstrated that change in sleep and appetite-related symptoms is relatively independent of change in core depressive symptoms such as mood and anhedonia. Reference Serretti, Mandelli, Lorenzi, Pirovano, Olgiati and Colombo59 In a psychometric analysis of the GENDEP data, we have identified partially separable dimensions of observed mood, cognitive symptoms and neurovegetative symptoms that differ in their response to the two antidepressants. Reference Uher, Maier, Hauser, Marušič, Schmael and Mors32,Reference Hamilton38 It has been previously reported that the moderation by the 5-HTTLPR of response to antidepressants is relatively specific to core mood symptoms and does not affect changes in sleep-related symptoms. Reference Hu, Rush, Charney, Wilson, Sorant and Papanicolaou60 The present analysis supports this distinction: the 5-HTTLPR and rs2020933 genotypes had the strongest influence on the observed mood dimension, comprising the core depressive symptoms and anxiety. These genotypes also moderated response on the cognitive symptom dimension. However, changes on the neurovegetative symptom dimension which comprises sleep, appetite and libido, were relatively independent of the 5-HTTLPR and rs2020933 genotypes. These findings add to the accumulating evidence indicating that mood and neurovegetative symptoms have distinct pathogenesis.
Effects of genotype on tolerability
It has been reported that tolerability rather than efficacy of antidepressants may be influenced by the 5-HTTLPR genotypes. Reference Murphy, Hollander, Rodrigues, Kremer and Schatzberg61,62 We have therefore explored the relationship between the 5-HTTLPR genotype and adverse effects and related variables. We found that the 5-HTTLPR genotype was not related to escitalopram or nortriptyline dosage, plasma levels, adverse effects, drop-out or self-reported adherence in the GENDEP sample. These results remained negative after recoding according to the rs25531 marker. Thus we can conclude that, in the European population, the 5-HTTLPR genotype is directly related to antidepressant efficacy and there is no significant effect on tolerability.
Strengths and limitations
The GENDEP project is the largest study to date to compare a tricyclic antidepressant with an SSRI and is the second largest sample with pharmacogenetic data on antidepressant treatment outcome. It is however only powered to detect interacting effects of moderate size. The associations between polymorphisms in the SLC6A4 gene and response to antidepressants are of small to moderate size. Subsequently, the reported findings are detected with a modest level of certainty that satisfies the nominal statistical significance threshold but would not survive corrections for the number of polymorphisms that have been reported or suggested to moderate antidepressant response. Therefore, the results of the present study must be interpreted against the background of other published findings. The association of 5-HTTLPR with response to serotonergic antidepressants appears to be a robust and replicable finding in populations of European origin. However, the concentration of effect among males and the additional moderation by the rs2020933 polymorphism are novel findings that have to be tested in other large samples.
In conclusion, the available data suggest that variations in the 5′ end of the serotonin transporter gene have an effect on antidepressant efficacy, which is of modest effect size and depends on type of antidepressant and gender. Future studies should evaluate the population-specificity of this effect and the role of other polymorphisms including the rs2020933.
Appendix
5-HTTLPR and rs25531 genotyping
Restriction fragment length polymorphism analysis of PCR products carrying either the long (L, 16 repeat) or short (S, 14 repeat) alleles of the 5HTTLPR (L/S) and rs25531 (a/g).
Eight combinations were identified within the GENDEP sample: LaLa (324bp, 62bp and 33bp fragment), LaLg (324bp, 174bp, 150bp), LaSa (324bp, 281bp), SaLg (281bp, 174bp, 150bp), SaSa (281bp), LgLg (174bp, 150bp) and SaSg (281bp, 150bp, 131bp).
Acknowledgements
We would like to thank the following collaborators for their contribution: Helen Dean, Joanna Gray, Cerisse Gunasinghe, Amanda Elkin, Desmond Campbell, Richard J. Williamson, Julien Mendlewicz, Mara Barreto, Thomas Schulze, Christine Schmael, Susanne Höfels, Anna Schuhmacher, Ute Pfeiffer, Sandra Weber, Lisbeth Jorgensen, Anne Schinkel Stamp, Caterina Giovannini, Dejan Kozel, Moica Z. Dernovsek, Alenka Tancic, Jerneja Sveticic, Zrnka Kovacic, Pawe Kapelski, Maria Skibiñska, Aleksandra Rajewska, Monika Dmitrzak-Weglarz, Aleksandra Szczepankiewicz, and Elzbieta Cegielska. We would like to specially acknowledge the contribution of Jorge Perez, who was the principal investigator at Brescia, Italy, and who passed away in October 2007, and the important contribution made by Dr Andrej Marušič, the principal investigator at Ljubljana, Slovenia, who passed away in June 2008.









eLetters
No eLetters have been published for this article.