INTRODUCTION
Hepatitis E virus (HEV) is a small non-enveloped RNA virus classified in the genus Hepevirus, the only member of the Hepeviridae family [Reference Purcell and Emerson1, Reference Meng2]. There are two species of HEV, mammalian and avian HEV. Antibodies against mammalian HEV have been identified in numerous species worldwide including humans, pigs, wild boars, rodents, dogs, cats, cows, sheep, and goats [Reference Purcell and Emerson1].
HEV is often responsible for waterborne outbreaks of viral hepatitis in humans, and for a large proportion of cases with sporadic acute hepatitis in endemic regions, such as large parts of Asia, Africa and Latin America [Reference Purcell and Emerson1, Reference Meng2]. In epidemics in these regions disease morbidity mainly affects young adults, a high mortality of up to 25% is seen in infected pregnant females during the third trimester [Reference Khuroo3]. Human HEV is mainly transmitted through the faecal/oral route through contaminated water or food, although vertical and bloodborne transmissions have also been reported [Reference Balayan4–Reference Gérolami, Moal and Colson9]. Human-to-human transmission is rare. In developed countries most hepatitis E cases are associated with travel from endemic regions. However, in the last few years an increasing number of sporadic cases have been reported in patients with no known epidemiological risk factor [Reference Dalton10–Reference Lopes Dos Santos16].
The HEV RNA is about 7·2 kb long and contains three open reading frames, ORF1–ORF3, flanked by short untranslated regions. ORF1 encodes for viral non-structural proteins containing several conserved domains, functioning as the putative methyltransferase, protease, helicase, and RNA-dependent RNA polymerase. ORF2 encodes for the viral capsid protein and ORF3 for a small phosphoprotein with uncertain function [Reference Chandra17–Reference Zafrullah19].
Based on divergences of complete genomes, four phylogenetically distinct genotypes of mammalian HEV have been identified, HEV genotypes 1–4. Genotype 1 has so far only been isolated from humans, although one pig in Cambodia has been shown as infected with this genotype [Reference Caron20]. This genotype is the main cause of sporadic and epidemic hepatitis E in Asia, Africa and Latin America. Genotype 2 has so far only been isolated from humans infected in Mexico and sub-Saharan Africa [Reference Buisson21–Reference Nicand24]. Genotype 3 has a worldwide distribution and has been isolated from humans, swine, wild boars, mongooses and rabbits both in endemic and non-endemic countries, while genotype 4 strains have been isolated from humans, wild boars, pigs and deer in Asia, particularly in Japan, China, Vietnam, Taiwan, and India. It is evident that there is a domestic source of genotype 3 infections, but the infection route is generally not known although spread from animals to humans by ingestion of undercooked liver and deer meat has been documented [Reference Khuroo, Kamili and Jameel6, Reference Takahashi8, Reference Gérolami, Moal and Colson9]. Genotype 3 infection leads to hepatitis mainly in persons aged >45 years [Reference Dalton10]. In Sweden several cases of non-travel-related HEV genotype 3 infections in humans have occurred and previous studies demonstrated that genotype 3 is also present in Swedish pigs [Reference Lopes Dos Santos16, Reference Gyarmati25, Reference Xia26]. However, little is known regarding the prevalence of HEV in the Swedish pig population and the prevalence of HEV in the Swedish wild boar population is not known at all. The genetic relationship between strains from humans, pigs and wild boars in Sweden is also not known. In order to gain more information on the prevalence of zoonotic transmission of genotype 3 infections, samples from Swedish domestic pigs and wild boars were collected. Partial ORF1 and ORF2 of the HEV genomes were sequenced, analysed and compared with the corresponding regions in strains from humans.
MATERIALS
Human serum samples
A total of 102 human serum samples were used for typing HEV strains. The samples, collected between 1993 and 2009, were reactive for anti-HEV IgM and/or IgG at the Swedish Institute for Infectious Disease Control. All samples were reactive for anti-HEV IgG and 57 of those were also reactive for anti-HEV IgM. The samples were from Swedish patients with clinical signs of hepatitis not caused by hepatitis A, B, C or D virus. Most patients had a recent travel history. The sera were tested for HEV IgG and IgM by a commercial ELISA using two recombinant HEV antigens corresponding to a structural region of HEV (Diagnostic Biotechnology, Singapore). Between 1993 and 2006, all samples were also tested for HEV IgG by the then commercially available ELISA kits from Abbott Laboratories (USA). During 2009 the samples were also tested for anti-HEV reactivity by another commercially available ELISA, Recomwell (Mikrogen GmbH, Germany). All reactive sera were tested for HEV RNA by polymerase chain reaction (PCR) and the amplified fragments were sequenced.
Pig samples
Faecal samples (n=240) were collected from 22 pig farms selected at random in south Sweden. The samples were from piglets aged between 2 and 4 months. Fresh faecal samples were collected from ten pigs per farm. For each farm, each pig sampled was kept in a box separate from the others. Faecal samples were stored at -20°C until processed. All pig samples were tested for HEV RNA by real-time PCR as described previously [Reference Gyarmati25].
Wild boar samples
A total of 159 wild boar blood samples were selected from samples sent in by hunters. The wild boars were from nine different counties in southern Sweden. Sixty-four (40%) were piglets and 95 (60%) were yearlings. The age of the sampled wild boars was estimated by the hunter and classified as either piglet or yearling. Samples exhibiting excessive haemolysis or foul smell were not tested. The sera were stored at −20°C until processed. All samples were tested for HEV RNA by real-time PCR as described previously [Reference Gyarmati25].
METHODS
Homogenization of faecal samples
Pig faecal samples were diluted about 1:10 in TE buffer (Tris EDTA; pH 7·6), homogenized in grinding tubes as described previously [Reference Gyarmati25] and stored at −80°C.
RNA extraction
HEV RNA was extracted from 200 μl human sera as previously described [Reference Lopes Dos Santos16]. HEV RNA was extracted from 140-μl faecal sample supernatant or wild boar sera with the QIAamp viral RNA mini kit (Qiagen, Germany) and stored at −80°C.
cDNA synthesis
Five microlitres of RNA were used for cDNA synthesis in a 20-μl mix that included 100 U Superscript II (Invitrogen, USA), which was performed according to the manufacturer's instructions.
Screening for HEV in pig and wild boar samples
TaqMan® assay for faecal samples
Assays were run in 12 μl reaction mix per tube as described previously [Reference Gyarmati25], with 2 μl cDNA added as template.
TaqMan assay for serum samples from wild boars
RNA from wild boar serum samples were analysed by a combined reverse transcription (RT) and PCR program using the Qiagen OneStep RT–PCR kit (Germany). From each RNA sample, 2·5 μl was analysed. Assays were run in 12 μl reaction mix per tube with the following cycling conditions: 50° for 30 min, 95°C for 2 min followed by two-step cycling 45 times at 94°C for 15 s and 56°C for 60 s. Fluorescence was monitored during the annealing step of each cycle.
PCR amplification in ORF1
A nested PCR was carried out in a 50 μl reaction with 5 μl cDNA, 10× Taq buffer, 2 mm MgCl2 (Applied Biosystems, Roche Molecular Systems, USA), 4 U Taq polymerase, 0·2 mm dNTP (Thermo Scientific, Abgene®, UK) or as a combined RT–PCR protocol with 5 μl of the extracted RNA (OneStep master mix, OneStep RT–PCR enzyme mix; Qiagen). The primers ISP-4232 and EAP-4576 [Reference Zhai, Dai and Meng27] (0·06 μl of 0·2 mm of each) were used. RT was performed at 50°C for 30 min. The PCR reaction was performed for 40 cycles, each cycle consisting of denaturation at 94°C for 20 s, annealing at 60°C for 30 s and extension at 72°C for 60 s. Cycling conditions for the second round were identical to the first. Five microlitres of the first-round product was added as template for the second round and 2·5 mm MgCl2 and 0·06 μl of 0·2 mm of each primer ISP-4232 and IAP-4561 [Reference Zhai, Dai and Meng27] were used.
PCR amplification in ORF2
The PCR in ORF2 was performed, as described previously [Reference Mizuo28], in a 50 μl reaction with 10 μl cDNA as template. Primers HE110 and HE041 were used for the first round, and primers HE110 and HE3159 for the second round.
Sequencing
The amplified products were purified using the EZNA Cycle Pure kit (Omega Bio-Tek, USA) or the Qiagen PCR purification kit (Qiagen). The sequencing reaction was made with BigDye Terminator Cycle Sequencing Ready reaction kit version 3.1 (Applied Biosystems, USA) with the primers used in the PCR as sequencing primers. The ABI Prism 3100 genetic analyser (Applied Biosystems) was used for electrophoresis and data collection.
Phylogenetic analysis
The sequences obtained were analysed in the programs DNAstar SeqMan and Sequencing Analysis. The sequences were aligned with the corresponding region of 554 sequences obtained from GeneBank. Phylogenetic analysis was carried out with the PHYLIP package version 3.65 [Reference Felsenstein29]. Evolutionary distances were calculated using the F84 algorithm in the dnadist program with a transition/transversion ratio of 4·29. Phylogenetic trees were constructed using the unweighted pair-group method using arithmetic averages (UPGMA) and the neighbour-joining method in the neighbor program of the PHYLIP package. The trees were visualized using the program Tree View, version 1.6.6. Bootstrap analysis of 1000 replicas was performed with the programs seqboot and consense in the PHYLIP package.
RESULTS
HEV RNA could be amplified in 58/102 human anti-HEV-positive samples (Table 1). Most of the samples with detectable HEV RNA were from patients with anti-HEV IgM (48/58), while 10 (22%) were from patients with detectable anti-HEV IgG only without anti-HEV IgM. Sixty-seven (64%) of the 102 patients had been infected while in Asia or the Middle East. However, 17 had been infected in Europe (Table 1). The HEV strains could be sequenced in 56 samples, 46 of which were from patients infected outside Europe. All these patients were infected with genotype 1 (Table 1). One genotype 3-infected patient was a newly arrived immigrant from Mongolia. It was not known if he was infected en route to Sweden. The HEV strains in sera from the 17 patients infected in Europe were all but one of genotype 3 (Table 1). The patient infected with genotype 1 in Sweden, was infected while taking care of his brother, who became infected during a visit in Pakistan [Reference Lopes Dos Santos16]. The patients infected with genotype 3 were all aged >46 years, and the majority (7/9), were male (Table 2).
Table 1. Country of infection of humans with IgG or IgM reactivity reactive to IgM and IgG and to IgG alone against hepatitis E virus (HEV) in relation to HEV genotype obtained by sequencing HEV strains in the serum samples

Table 2. Strains of HEV genotype 3, sex, age, and country of infection of patients positive for the virus
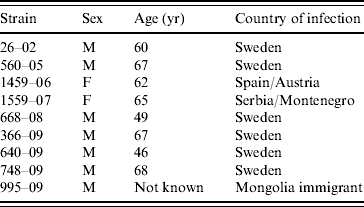
The age of the Mongolian immigrant was not known.
HEV RNA could be amplified by real-time PCR in 71 (29·6%) of 240 faecal samples collected from pigs aged 2–4 months from 22 pig farms (Fig. 1). HEV RNA was detected in samples from piglets in 16 (72·7%) of the farms. Thirty of the HEV RNA samples were sequenced in ORF1 and 26 samples were sequenced in ORF2. All were genotype 3 strains.

Fig. 1. Map of Sweden showing the counties labelled with the official county letter. Counties where samples were collected are indicated with arrows. The arrows are labelled with the official county letter followed by number of pigs sampled/number of PCR-positive pigs/number of wild boar sampled /number of PCR-positive wild boar in the county indicated. AB, Stockholm county; C, Uppsala county; D, Sörmland; E, Östergötland; F, Jönköping county; G, Kronoberg county; H, Kalmar county; K, Blekinge county; M, Skåne region; N, Halland county; O, Västra Götaland region; S, Värmland county; T, Örebro county; U, Västmanland county; W, Dalarna county; X, Gävleborg county; Y, Västernorrland; Z, Jämtland county; AC, Västerbotten county; BD, Norrbotten county; I, Gotland municipality.
Thirteen (8·2%) of 159 samples from wild boars had detectable HEV RNA by real-time PCR. The infected wild boars were from five of the nine investigated counties (Fig. 1). Eight (14·8%) of the wild boar piglets, and five (4·8%) of the older wild boars were infected. ORF1 was partially sequenced in seven and ORF2 in six wild boar samples, all were of genotype 3. In five of these samples both ORF1 and ORF2 were partially sequenced.
Phylogenetic comparison of the sequences demonstrated that the genotype 3 strains subdivided into two groups, designated 3-I and 3-II both in ORF1 and ORF2 as previously shown for ORF2 (Figs 2 (a), 3, 4; Supplementary Table 1 (available online) [Reference Lopes Dos Santos16]). Both groups contained strains from pigs and humans intermixed in the tree. Based on analysis of the sequences, the 3-I group contained strains of subtypes 3a, b, c, h, and j as well as three additional groups of strains with undefined subtypes (Fig. 2). The subtype 3a strains were from the USA, South Korea, and Japan. Those of subtype 3b were from Japan and China, the 3h strain was from Mongolia, and the 3j strain from Canada (Fig. 2). Ten Swedish strains were found in this group. One strain was from a woman infected in Serbia/Montenegro. The HEV strain infecting her was similar to strains from South Korea. One separate branch was formed by strains from a Swedish man and a wild boar. One additional branch was formed by two strains from German wild boars and a strain from a Swedish man. A third branch was formed by six strains from Swedish piglets at farm 11 (Fig. 2, b). These subdivisions of the strains were confirmed in ORF2 (Supplementary Fig. 1, available online).

Fig. 2. UPGMA dendrogram based on 346 nucleotides of ORF1 in 309 HEV strains. The genotypes and subdivision of genotype 3 into two groups are shown on the branches. The branch of 3-I strains is shown. The subtypes are indicated on the branches. Origin of strains from Swedish pigs is indicated on the branch, by the designation of the pig farm. Strains sequenced and described in this study are shown in bold. Strains from animals are indicated in italic. Bootstrap values of 1000 replications are indicated below the branches. Accession numbers of sequences obtained from GenBank are given at the nodes or for sequences within groups 1–4 in Supplementary Table 1 (available online).

Fig. 3. UPGMA dendrogram based on 346 nucleotides of ORF1 in 309 HEV strains. The genotypes and subdivision of genotype 3 into two groups are shown on the branches. The branch formed by 3-II strains is shown. Origin of strains from Swedish pigs is indicated on the branch, by the designation of the pig farm. Strains sequenced and described in this study are shown in bold. Strains from animals are indicated in italic. Bootstrap values of 1000 replications are indicated below the branches. Accession numbers of sequences obtained from GenBank are given at the nodes or for sequences within groups 1–4 in Supplementary Table 1 (available online).

Fig. 4. UPGMA dendrogram based on 276 nucleotides of ORF2 in 640 HEV strains. The genotypes and subdivision of genotype 3 into two groups are shown on the branches. The details of the branch formed by 3-II strains are shown. The subtypes are indicated on the branches. Origin of strains from Swedish pigs is indicated on the branch, by the designation of the pig farm. Strains sequenced and described in this study are shown in bold. Strains from animals are indicated in italic. Bootstrap values of 1000 replications are indicated below the branches. Accession numbers of sequences obtained from GenBank are given at the nodes or for sequences within groups 1-3 in Supplementary Table 1 (available online). The details of branch 3-I strains are not shown here but are available in Supplementary Fig. 1 (available online).
Most of the Swedish strains were found in group 3-II. This group contained strains of subtypes 3e, 3f, and 3g (Fig. 3). The 3f strains originated from Europe, Thailand, and Mongolia, while the 3e strains originated from Germany and Japan. The 3g strain was from a pig from Kyrgyzstan. Moreover, in the 3-II group, the strains from the Swedish piglets formed separate branches according to origin. The strains from farms A, 1, 3, 4, 12, and 46, were of subtype 3f while the strains from farms 10 and BT were of subtype 3e (Fig. 3). One strain from farm A was similar to seven strains from farm 1, and clustered with those on the same branch. Another three strains from farm 1 were divergent and formed a separate branch (Fig. 3). Six strains from Swedish wild boars belonged to subtype 3f. All strains from Halland county were grouped as a separate branch. One of the other strains was distantly similar to a strain from a man infected in Sweden (Fig. 3). The humans infected in Spain all were infected by strains similar to strains from Spanish pigs. The similarities and sub-clustering of the strains were confirmed on analysis of ORF2 (Fig. 4).
DISCUSSION
This study confirmed the prevalence of hepatitis E in humans, domestic pigs and wild boars in Sweden. Most identified human cases were travel related, mainly from Asia, and were infected with the HEV genotype 1. However, those infected in Sweden or in other European countries were all infected with genotype 3. This was the only genotype found in Swedish domestic pigs and wild boars. Based on the geographical distribution of infected animals and human cases, this study demonstrates a probable endemic circulation of HEV genotype 3 in Sweden.
Phylogenetic analysis of the sequences showed that the previously described subdivision of genotype 3 strains into two groups, 3-I and 3-II, when comparing ORF2 sequences [Reference Chandra17], was confirmed when more samples were analysed both in ORF1 and ORF2. Most of the Swedish strains were found in group 3-II and belonged to subtype 3f, although piglets from two farms were infected by 3e strains. Subtype 3f is the most common subtype found in pigs in Europe, e.g. France, The Netherlands, and Spain [Reference Kaba30–Reference Rutjes34] although subtype 3e has also been found in European pigs [Reference Kaba30, Reference Rutjes34]. This study showed that HEV prevalence in the Swedish pig population is high. The high prevalence in Swedish piglets aged 2–4 months, which is in accord with a study from France [Reference Kaba30], indicates that this is the age when HEV is spreading most rapidly. In the French study piglets aged 2 months had been moved to a farm, where they became infected by the same strains [Reference Kaba30]. This French farm had two strains circulating – subtypes 3e and 3f. In our study each Swedish farm also had one, or sometimes two specific HEV strains, which could be distinguished from all other strains by phylogenetic analysis. The high prevalence of HEV in the majority of the Swedish farms may be due to a mixing of young piglets from different litters leading to an efficient spread of HEV.
The prevalence of HEV in the wild boars in our study is lower than in domestic pigs. However, the age distribution and sample type were not the same in the two groups and it is anticipated that the viral load is lower in sera than in liver, bile or faeces. The lower prevalence in the wild boars may also reflect a slower spread of the virus due to a lower population density than in a pig farm. Most of the HEV strains from wild boars belonged to subtype 3f but one was found in group 3-I and may represent a new subtype. The strains clustered mainly according to origin with all strains from Halland county forming one branch in the phylogenetic tree. This geographical clustering was also found in German wild boars, although the strains were more diverse with representatives from several subtypes [Reference Schielke35, Reference Baechlein36]. Furthermore, in the German study a difference between urban and rural regions could be seen. This could not be studied in Sweden since there are no urban wild boar populations in Sweden. The difference in prevalence of HEV between the two studies may also be attributed to difference in sample material.
Most HEV strains from humans infected in Europe were of subtype 3f. Those infected in Spain had strains related to strains from Spanish pigs, while most of the strains from those infected in Sweden were related to strains from Swedish domestic pigs or wild boars. One Swedish man was infected with a strain related to a German wild boar. The similarity between strains from humans and pigs or deer from the same country has previously been demonstrated [Reference Lopes Dos Santos16, Reference Meng37–Reference Reuter41]. However, the analysis of ORF1 and ORF2 sequences in this study shows that the country or even county of origin of the infecting HEV strain can be determined. Further studies are needed to investigate if there is a permanent spread of specific strains in each pig farm and if there are specific strains spread in the wild boars in the different Swedish counties.
Since HEV infections are frequent in Swedish domestic pigs and wild boars the risk of human infection from them is possible. This can occur for example, when wild boars are handled after being shot by hunters or if undercooked pig and wild boar products are consumed, as has been shown in Japan [Reference Takahashi8]. This study demonstrates that phylogeny can be a useful tool for tracing the source of non-travel-related HEV infections and in determining the geographical origin of HEV genotype 3 strains infecting humans. The usefulness of this tool for tracing human infections is dependent on regular screenings of pig and the wild boar population. It is reasonable to assume that if more information regarding HEV strains circulating in regional pig and wild boar populations was available, the precision and reliability of this model for tracing infections would increase significantly.
CONCLUSION
This study demonstrates that HEV is common in Swedish pigs and wild boars. It also demonstrates that phylogeny based on short sequences from ORF1 and ORF2 is a useful tool for tracing the origin of human HEV infections. By using this tool it was concluded that there is a strong similarity between HEV in humans on the one hand and in pigs and wild boars on the other. This study indicates that in Sweden human HEV infection may be zoonotic. It is evident that pigs and wild boars may be the reservoirs.
NOTE
Supplementary material accompanies this paper on the Journal's website (http://journals.cambridge.org/hyg).
ACKNOWLEDGEMENTS
Thanks are due to Maria Lindberg and Marie Sjölund at Swedish Animal Heath for collecting samples from pigs, and Per Wallgren for establishing the contact with Swedish Animal Health. Thanks are also due to the Swedish hunters that collected and sent in wild boar blood samples and to Maj Hjort for preparing the samples. This study was supported by Pathogen Combat, contract no. FOOD-CT-2005-007081, European Union Network of Excellence ‘Network for the prevention and control of Zoonoses (Med-Vet-Net)’, contract no. 506122. Support was also received from the ‘Ivar and Elsa Sandberg Foundation’, Sweden. Part of this work was also supported by the European Commission DG Research Quality of Life Program, 6th Framework (EVENT, SP22-CT-2004-502571).
DECLARATION OF INTEREST
None.








