Introduction
The PUF proteins are a family of RNA-binding proteins with a highly conserved PUF domain, including the classical PUF proteins (PUM1/2 and FBF1/2) and two evolutionary distant homologues (PUM3 and NOP9; Wang et al., Reference Wang, Ogé, Perez-Garcia, Hamama and Sakr2018; Najdrová et al., Reference Najdrová, Stairs, Vinopalová, Voleman and Doležal2020). The PUF family members are widely distributed in eukaryotes and specifically recognize and bind the motif sequence through PUF repeats. This post-transcriptional regulation is mediated using RNA-binding proteins, which can control RNA splicing, polyadenylation, capping, modification, localization, stability, and spatiotemporally specific translation, and is known as RNA metabolism (Blackinton and Keene, Reference Blackinton and Keene2014). Generally, PUM proteins regulate mRNA stability and translation by directly binding to the target mRNAs through the PUM homology domain (PUM-HD; Edwards et al., Reference Edwards, Pyle, Wharton and Aggarwal2001; Wang et al., Reference Wang, Zamore and Hall2001). Pum3 (Pumilio RNA binding family member 3, also known as KIAA0020 or PUF-A in humans) is conserved in a wide range of eukaryotes, including fungi, plants, protozoans and metazoans (Liang et al., Reference Liang, Hart, Dong, Siddiqui, Sebastian, Li, Albert, Miao, Lindner and Cui2018; Joshna et al., Reference Joshna, Saha, Atugala, Chua and Muench2020; Najdrová et al., Reference Najdrová, Stairs, Vinopalová, Voleman and Doležal2020; Son et al., Reference Son, Jang and Park2021). The PUM-HD in classical PUMs (PUM1 and PUM2) consists of eight tandem PUF repeats, 36 amino acids contain the motif, forming a crescent-shaped structure (Wang et al., Reference Wang, Zamore and Hall2001); however, PUM3 consists of 11 PUF repeats in PUM domain, which is arranged in an L-shape and exhibits the ability to bind to DNA and single-stranded and double-stranded RNA without sequence specificity (Najdrová et al., Reference Najdrová, Stairs, Vinopalová, Voleman and Doležal2020). Therefore, the non-classical PUM homologue PUM3 may also play a significant role at the transcriptional and post-transcriptional levels.
Extensive studies in mammals have revealed that classic PUMs (PUM1 and PUM2) proteins play multiple roles in various biological processes (Goldstrohm et al., Reference Goldstrohm, Hall and McKenney2018). Previous studies in invertebrates have suggested that PUF family proteins are essential for germline stem cell maintenance and embryonic development in Drosophila and C. elegans (Lin and Spradling, Reference Lin and Spradling1997; Zhang et al., Reference Zhang, Gallegos, Puoti, Durkin, Fields, Kimble and Wickens1997; Kraemer et al., Reference Kraemer, Crittenden, Gallegos, Moulder, Barstead, Kimble and Wickens1999; Parisi and Lin, Reference Parisi and Lin1999). Recent studies in mammals have also shown that PUMs have key roles in germ-cell development, neurodegeneration, early embryonic development and postnatal growth. In particular, PUMs have a strong correlation with a variety of human cancers, and also have a strong promoting role in the occurrence and development of cancer using post-transcriptional regulation (Miles et al., Reference Miles, Tschöp, Herr, Ji and Dyson2012; Naudin et al., Reference Naudin, Hattabi, Michelet, Miri-Nezhad, Benyoucef, Pflumio, Guillonneau, Fichelson, Vigon, Dusanter-Fourt and Lauret2017; Munschauer et al., Reference Munschauer, Nguyen, Sirokman, Hartigan, Hogstrom, Engreitz, Ulirsch, Fulco, Subramanian, Chen, Schenone, Guttman, Carr and Lander2018; Elguindy and Mendell, Reference Elguindy and Mendell2021; Silva et al., Reference Silva, Kohata and Shigunov2022; Yoon et al., Reference Yoon, Lee, Choi, Ko, Lee, Cho, Park, Lee, Kim and Lee2022). In conclusion, classical PUM proteins control various cellular functions by post-transcriptionally regulating the fate of target RNAs in both invertebrates and mammals.
However, the physiological function of PUM3 in mammals remains unclear. A previous study has shown that PUM3 (Puf-A) is essential for eye development and primordial germ-cell (PGC) migration in zebrafish (Kuo et al., Reference Kuo, Wang, Chang, Chang, Huang, Lin, Yu, Li and Yu2009). In mouse embryonic stem cells, the loss of PUM3 can increase the Zscan4-positive 2-cell embryo-like state cells in a p53-dependent manner (Takahiro et al., Reference Takahiro, Kimura, Nakano and Yamaguchi2021). Recently, studies have shown that PUM3 is located in the nucleolus and regulates ribosome biogenesis during the promotion of cancer progression (Fan et al., Reference Fan, Lee, Yu, Tzen, Chou and Chang2013; Lin et al., Reference Lin, Lee, Chou, Shih and Chang2021; Cho et al., Reference Cho, Huang, Hung, Hung, Cheng, Liu, Kuo, Wang, Yu and Yu2022). To explore the role of Pum3 in mouse development, we characterized the expression levels of Pum3 among different tissues. The result showed that Pum3 mRNA was widely expressed in multiple organs, but predominantly in the ovary. Furthermore, using immunohistochemical staining, we found that the PUM3 protein was abundantly expressed in follicles at different stages. MII oocytes exhibited a higher level of PUM3 in the cytoplasm. To reveal the function of Pum3 during oocyte in vitro maturation (IVM), we found that knockdown of Pum3 in GV oocytes did not affect GVBD and polar body extrusion (PBE). In addition, we detected preimplantation embryo development for PUM3-deficient oocytes and the control group. However, the zygotes in the knockdown group developed normally, as in the control group. Taken together, our data suggested that PUM3 is dispensable for mouse oocyte IVM and early embryo development after fertilization.
Materials and methods
Animals
Animals were housed under controlled environmental conditions in the animal facility at Nanjing Medical University, Nanjing, China, with free access to water and food. Illumination was provided between 8 a.m. and 8 p.m. daily.
Oocyte collection, siRNA microinjection and oocyte IVM
Female ICR mice were primed intraperitoneally with 5 IU PMSG to stimulate the development of multiple follicles. After 48 h, mice were sacrificed for GV oocyte collection or injected with 5 IU of hCG to induce ovulation. The ovaries were washed three times with 0.9% (w/v) NaCl. Cumulus–oocyte complexes (COCs) were aspirated from follicles (2–8 mm) in mHTF medium (Irvine Scientific, USA) containing 1.0 mM IBMX (Sigma, USA) using an 18-gauge needle attached to a 10-ml syringe.
Three Pum3 (NM_177474.5) siRNAs and nonsense siRNA were designed and chemically synthesized by GenePharma (Shanghai, China) and mixed for injection. The siRNA target sequences are shown in Table 1. The COCs were digested in 0.2% hyaluronidase for 3 min at 37°C. When all cumulus cells were separated from oocytes, 10 pl of the siRNA mixture (300 nmol/l) and nonsense RNA were microinjected into the germinal vesicle (GV)-stage oocytes using the Eppendorf Cell Tram Vario system (Eppendorf, Hamburg, Germany). After microinjection, oocytes were cultured in KSOM medium (Millipore, Germany) containing 1.0 mM IBMX (Sigma, USA) under mineral oil at 37°C and 5% CO2 in the incubator for 16 h.
Table 1. Sequence of primers usedfor RT-qPCR and siRNA

After siRNA injection, GV oocytes were washed and cultured under normal KSOM medium without IBMX addition for 30, 45, 60, 75, or 90 min. Then, oocytes were scored as either germinal vesicle intact or having undergone GVBD. The percentage of either GV or GVBD was calculated for each time point. The polar body was counted and calculated.
Embryo culture in vitro
Matured oocytes were used for in vitro fertilization as previously described (Yan et al., Reference Yan, Liang, Deng, Long, Chen, Chai, Suo, Xu, Kuang, Wu, Lu and Lyu2015). Approximately 6 h later, these two-pronuclear zygotes were then washed and cultured in mHTF medium drops. The siRNA mixture or nonsense siRNA was microinjected into zygotes immediately. Then, all embryos were transferred into KSOM medium and cultured under normal conditions. Embryos (from the 2-cell stage to blastocysts) were observed and recorded at 1.5, 2.5, 3.5, and 4.5 days post fertilization.
RNA isolation and real-time polymerase chain reaction (PCR)
RNA was extracted from ∼50 oocytes or embryos with the RNAprep Pure Micro Kit (Tiangen, China) according to the manufacturer’s instructions. Reverse transcription was performed using a PrimeScript RT Master Mix (Vazyme, China). Real-time PCR was performed using Taq polymerase (Vazyme, China) according to the product manual and gene-specific primers. The number of PCR cycles ranged from 22 to 35, depending on the linearity of the reaction. All gene expression analyses were performed with samples from at least three independent experiments. All primers for real-time RT-qPCR are listed in Table 1.
Histology, immunohistochemistry and immunofluorescence
Tissues were fixed overnight in Hartman’s fixative (Sigma). Fixed ovaries were embedded in paraffin and cut into 5-μm-thick serial sections. Sections were processed for haematoxylin and eosin (H&E) staining and immunohistochemistry according to standard protocols. Immunostaining for PUM3 (abcam, 1:50) was performed, following citrate buffer antigen retrieval, by incubation with primary antibodies and detected using biotin–streptavidin horseradish peroxidase (HRP) detection systems (ZSGB-BIO). A minimum of three randomly chosen discontinuous sections were used to determine positive cells in tubules.
Quantification and statistical analysis
All data in bar and line graphs are expressed as means ± standard deviation (SD) of the mean. All experiments were repeated at least three times unless noted. Statistical significance between two groups of data was evaluated using Student’s t-test (two-tailed) comparison using GraphPad Prism software 5 or Microsoft Excel t-test function. Statistical significance is indicated by ns for no significant difference, *P-value <0.05, **P-value <0.01, ***P-value <0.001.
Results
High expression of PUM3 in oocytes and granulosa cells
To identify the expression patterns of Pum3 RNA in different mouse tissues, we extracted total RNA from various tissues of adult mice. Pum3 RNA levels were detected using real-time quantitative reverse transcription PCR (RT-qPCR). Our results showed that Pum3 RNA was widely expressed in multiple tissues, including the ovary, epididymis, testis, kidney, liver, thymus, lung, spleen, brain, uterus and intestine (Figure 1A), especially with the highest expression in the ovary tissue. Therefore, we speculated that Pum3 may play an important role in the female reproductive system. Then we detected the expression of PUM3 protein in mouse ovary using immunohistochemistry (IHC) staining. Compared with the negative control (5% BSA), adult ovary sections stained with PUM3 antibody displayed special and strong positive signals in oocytes, granulosa cells and theca cells. However, a false-positive signal also appeared in the stroma cells of the negative control section, suggesting a non-specific signal (Figure 1B). At the same time, we also paid attention to the oocyte PUM3 expression in follicles at different stages. There were obvious signals in the oocyte cytoplasm from primordial follicles to mature antral follicles (Figure 1C). In addition, we also obtained GV-stage and MII-stage oocytes by ovulation induction. Immunofluorescence staining showed that the MII oocyte, GV oocyte and its cumulus granulosa cells produced strong positive signals (Figure 1D). We also noticed that the PUM3 protein formed abundant granules in both GV and MII oocyte cortex. The nucleus had a weak signal and no fluorescent signal was detected in the nucleolus of GV oocytes. The nuclei of granulosa cells had a positive signal (Figure 1C, D). In MII oocytes, PUM3 does not appear to overlap the spindle. The fluorescence intensity of oocyte PUM3 at the MII stage was significantly higher (29%) than in the GV stage (Figure 1E). In summary, Pum3 is highly expressed in the mouse ovary, especially in oocytes and granulosa cells, which is similar to the PUM1 expression pattern (Li et al., Reference Li, Zhu, Zang, Cao, Xie, Liang, Bian, Zhao, Hu and Xu2022).
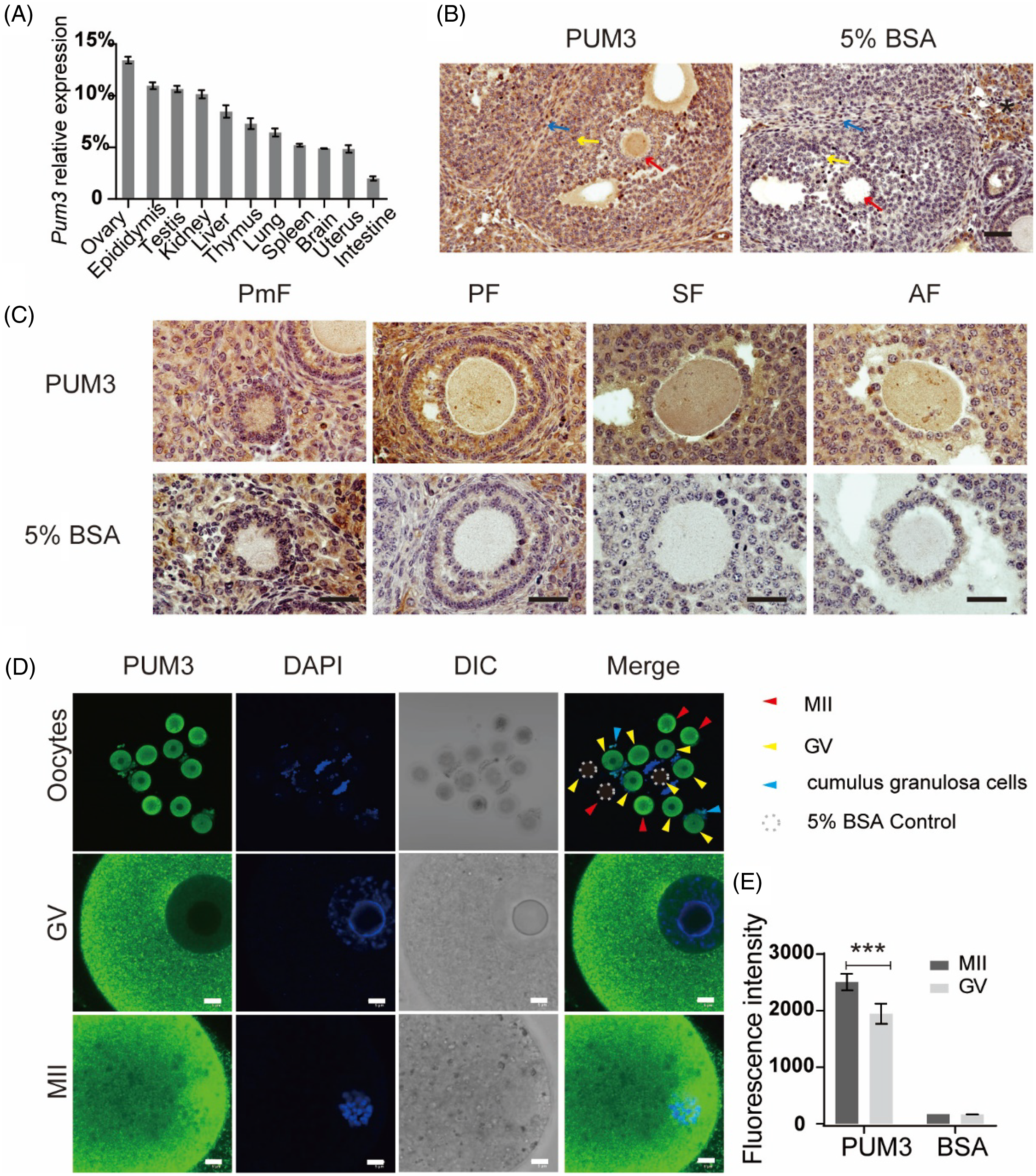
Figure 1. Characterization of Pum3 gene expression in the ovary. (A) RT-qPCR detection of Pum3 mRNA levels in various tissues. (B) Expression of PUM3 protein was detected using immunohistochemistry staining in the adult wild-type ovary. Scale bar: 200 μm. Red arrows: oocyte; yellow arrows: granulosa cells; blue arrows: theca cells; asterisk: no specific signal in stroma cells. (C) PUM3 expression in different stages of follicles: primordial follicle (PmF), primary follicle (PF), secondary follicle (SF) and antral follicle (AF). Scale bars: 20 μm. (D) Immunofluorescence images of PUM3 in GV and MII oocytes. Dotted circle: antibody negative control group incubated with 5% BSA. Red arrowheads: MII oocytes; yellow arrowheads: GV oocytes; blue arrowheads: granulosa cells. Scale bars 5 μm. (E) Fluorescence intensity of PUM3 in MII and GV oocytes; ***P < 0.001.
Pum3 knockdown oocytes display normal GVBD and PBE during in vitro maturation (IVM)
As previously reported, PUM1, a member of the classic PUM family, is a maternal factor regulating oocyte development in humans and mice (Mak et al., Reference Mak, Fang, Holden, Dratver and Lin2016, Reference Mak, Xia, Cheng, Lowther and Lin2018; Luo et al., Reference Luo, Ke, Liu, Qin, Mak, Ma, Zhao and Chen2018). As PUM3 is highly expressed in oocytes at different stages, we suspected that PUM3 may play an important role during mouse oocyte development. To identify this we knocked down Pum3 in GV oocytes using microinjection siRNA. The Pum3 mRNA was successfully knocked down in GV oocytes using siRNA (siPum3), and the knockdown efficiency of siPum3 was as high as 96% (Figure 2A). By removing the nuclear maturation inhibitor IBMX from the culture medium, GV oocytes were released and then entered germinal vesicle breakdown (GVBD). Surprisingly, oocytes from the siPum3 group displayed a similar speed and ratio of GVBD compared with the control group (Figure 2B). The proportion of oocytes entering GVBD was slightly higher in the knockdown group compared with the control group, but this was not significant at each test point and, finally, two groups reached a comparable level (96.37%) in the knockdown group and 93.37% in the control group, therefore Pum3 deletion did not affect the development of GVBD in mouse oocytes. In addition, we also assessed the rate of PBE and found that knockdown of Pum3 did not affect the PBE process (Figure 2C). The PBE rates of the control and siPum3 groups reached 68.46% and 77.74%, respectively. Taken together, these results suggested that PUM3 is not essential for in vitro maturation of mouse oocytes.
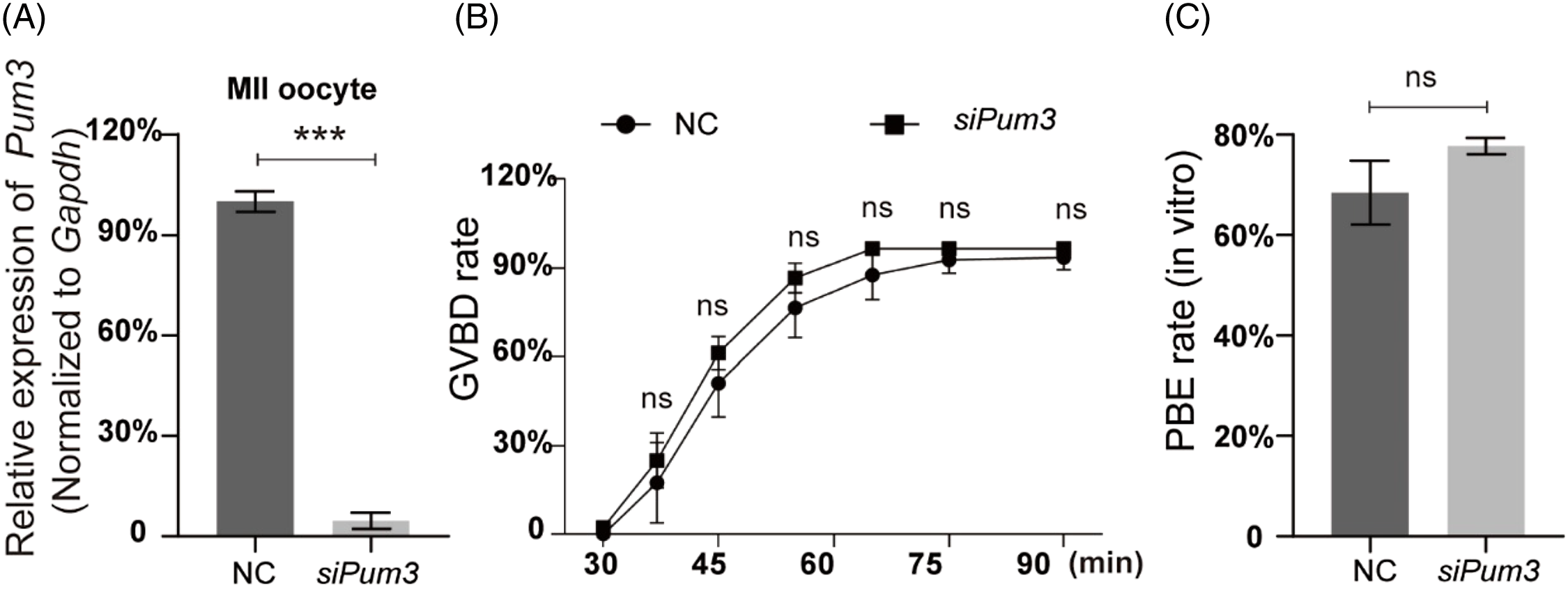
Figure 2. IVM results of Pum3 knockdown oocytes. (A) RT-qPCR of Pum3 knockdown in oocyte cells at 48 h after siRNA infection. (B, C) Percentages of GVBD (B) and PBE (C) in the Pum3 knockdown and control groups. ***P < 0.001; ns, not statistically significant, P > 0.05.
Pum3 knockdown does not affect early embryo development in vitro
Although Pum3 knockdown did not affect the IVM process of mouse oocytes, its high expression in the oocyte cytoplasm implied that PUM3 might be an important regulator (Figure 1D), so we questioned that whether Pum3 might affect early embryo development after fertilization. Therefore, we obtained a large number of zygotes by in vitro fertilization (IVF), and then knocked down Pum3 in the fertilized oocytes by siRNA injection at day 0.5 (two-pronuclear stage, 2PN), and tracked the developmental progress of embryos in each group. Our RT-qPCR results showed that the efficiency of Pum3 knockdown in blastocysts reached 96% (Figure 3A). Next the zygote underwent cleavage to form the blastula. At 1.5 days, 96.9% of the control and 100.0% of siPum3 embryos developed to the 2-cell stage; at 2.5 days, 89.7% of the control and 92.5% of siPum3 embryos developed to the 4-cell stage; at 3.5 days, 91.8% of the control and 94.1% of siPum3 embryos developed to the compaction stage; at 4.5 days, 91.8% of the control and 90.3% of siPum3 embryos developed to the blastocyst stage. These data showed that siPum3 may slightly promote zygote cleavage and development from the 2-cell stage to the compaction stage, but we also finally obtained a similar blastocyst rate to the control group at 4.5 days (Figure 3B). Overall, these data suggested that Pum3 knockdown had little effect on early embryonic development in vitro.
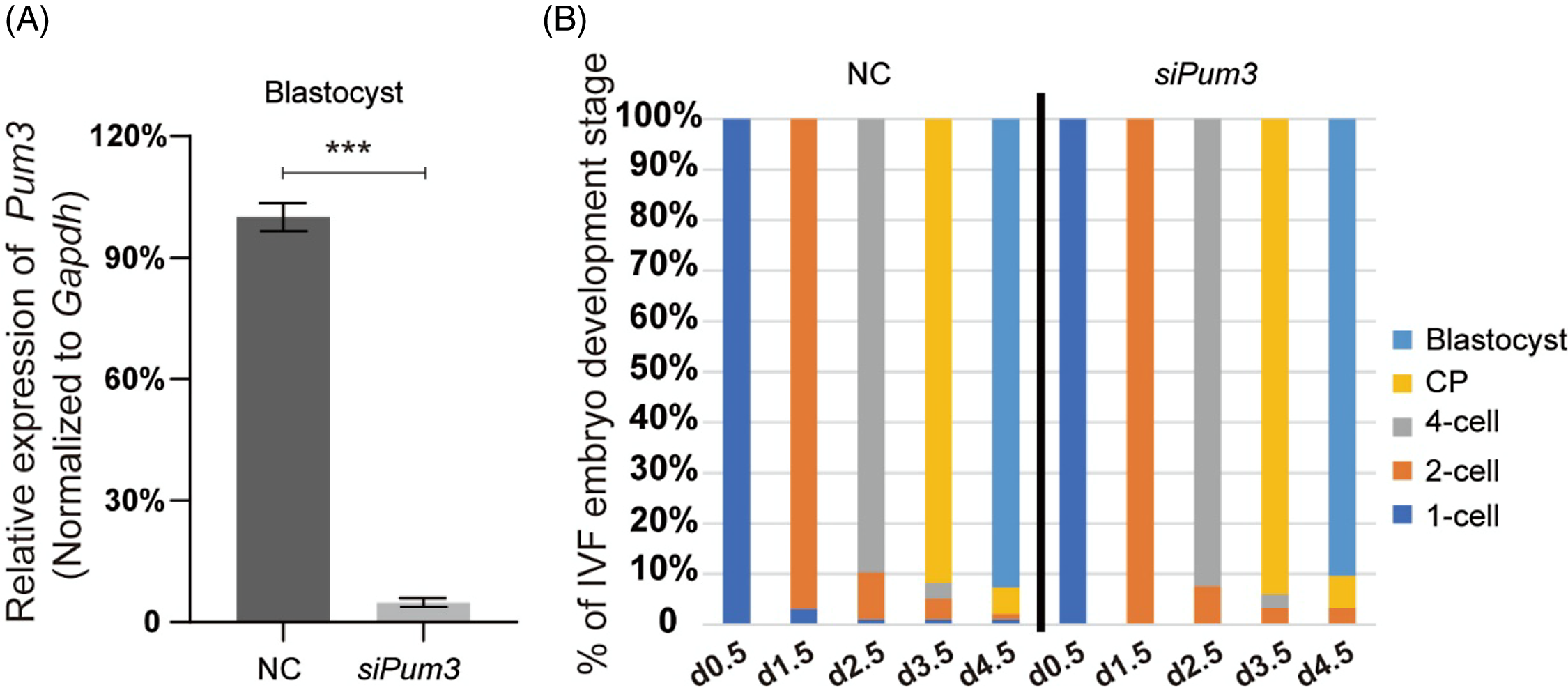
Figure 3. Effect of Pum3 knockdown on preimplantation embryo development in vitro. (A) RT-qPCR detection of Pum3 knockdown in the IVF blastocysts, ***P < 0.001. (B) Percentage of IVF embryos developed to the 2-cell, 4-cell, compaction and blastocyst stages in the siPum3 and control groups.
Discussion
In this study, we knocked down Pum3 by injecting siRNA into mouse GV-stage oocytes and obtained MII oocytes using IVM culture. We found that, although PUM3 was expressed at a high level in the oocyte at multiple stages of the follicle, deletion of PUM3 did not have significant negative effects on the GVBD and PBE of oocyte IVM. In addition, the knockdown of Pum3 in zygotes at the 2PN stage did not seem to cause significant defects in early embryonic development in vitro, and these embryos developed normally to blastocysts. These results suggested that PUM3 is not essential for oocyte maturation and early embryonic development in vitro.
RNA-binding proteins are a large group of proteins that mediate post-transcriptional regulation by binding to RNA, regulating the fate of RNA and affecting related gene expression and the corresponding cellular functions (Matoulkova et al., Reference Matoulkova, Michalova, Vojtesek and Hrstka2012). The PUF family are evolutionarily conserved and widely distributed RNA-binding proteins in eukaryotes, such as yeast, green plants, invertebrates and vertebrates (Zhang et al., Reference Zhang, Chen, Xia, Han, Cui, Neuenkirchen, Hermes, Sestan and Lin2017). PUF family proteins possess a crescent-shaped RNA-binding domain called the Pumilio homology domain (PUM-HD), which is formed from eight PUF repeats (Jenkins et al., Reference Jenkins, Baker-Wilding and Edwards2009; Wang et al., Reference Wang, Zamore and Hall2001). This PUM-HD structure can specifically recognize the PUM-binding site in the 3′UTR of target RNAs, and this mode of binding is conserved and can be engineered for accurate identification of the designated RNA sequence (Lu et al., Reference Lu, Dolgner and Hall2009).
PUF family genes generally have multiple copies or variants in different species, which may be related to the number of target mRNAs that need to be regulated in different organisms (Najdrová et al., Reference Najdrová, Stairs, Vinopalová, Voleman and Doležal2020). The PUF family has two evolutionarily distant homologous proteins, PUM3 and NOP9. It is worth noting that PUM3 and NOP9 are also widely distributed in eukaryotes, but there is only one copy on average in different species, which is different from the gene copy number of the PUF family (Najdrová et al., Reference Najdrová, Stairs, Vinopalová, Voleman and Doležal2020). Both proteins contain a non-canonical PUM-HD domain consisting of 11 repeats, a U-shaped domain in NOP9 and an L-shaped domain in PUM3 (Wang et al., Reference Wang, Ogé, Perez-Garcia, Hamama and Sakr2018). In eukaryotes, NOP9 has the function of RNA binding and is mainly involved in the processing and folding of 18S rRNA in the nucleolus; PUM3 can bind to double-stranded DNA or RNA without sequence specificity and participate in the occurrence of large ribosomal subunits. In addition, PUM3 and PUF family proteins are more similar in sequence than NOP9 (Najdrová et al., Reference Najdrová, Stairs, Vinopalová, Voleman and Doležal2020). Therefore, PUM3 and NOP9 are evolutionarily orthologous proteins of the PUF family. Although they have similar PUM-HD domains and are capable of binding RNA, they have their own role in the biogenesis of ribosomal RNA. This may be completely different in function from the classical PUF family proteins.
The earliest discovered physiological functions of the PUF family are the regulation of early embryo and germline stem cell development in invertebrates (Nüsslein-Volhard, Reference Nüsslein-Volhard1987; Lin and Spradling, Reference Lin and Spradling1997; Zhang et al., Reference Zhang, Gallegos, Puoti, Durkin, Fields, Kimble and Wickens1997; Forbes and Lehmann, Reference Forbes and Lehmann1998). This raised the question of whether the PUF family, a conserved family of RNA-binding proteins, could also play an equally important role in mammals. The subsequent studies in mice found that PUM1 deletion can reduce mouse fertility (Chen et al., Reference Chen, Zheng, Lin, Uyhazi, Zhao and Lin2012), whereas PUM2 deletion did not affect mouse fertility (Xu et al., Reference Xu, Chang, Salmon and Reijo Pera2007). In the absence of both PUM1 and PUM2, mouse embryos died at 8.5 days due to defective gastrulation differentiation (Lin et al., Reference Lin, Zhang, Shi, Zhu, Gao, Xia, Geng, Zheng and Xu2018), probably because there would be some complementation between two highly conserved paralogous proteins of the PUF family in mammals. This also showed that their functions were also conserved to a certain extent, except for the evolutionary conservation of the protein structure of the PUF family. Moreover, large numbers of studies have shown that the PUF family also has important functions in the nervous system of invertebrates and mammals (Chen et al., Reference Chen, Li, Zhang, Regulski, Sinha, Barditch, Tully, Krainer, Zhang and Dubnau2008; Vessey et al., Reference Vessey, Schoderboeck, Gingl, Luzi, Riefler, Di Leva, Karra, Thomas, Kiebler and Macchi2010; Gennarino et al., Reference Gennarino, Singh, White, De Maio, Han, Kim, Jafar-Nejad, di Ronza, Kang, Sayegh, Cooper, Orr, Sillitoe and Zoghbi2015). It has been previously reported that PUF-A plays an important role in the development of eyes and primordial germ cells in fish (Kuo et al., Reference Kuo, Wang, Chang, Chang, Huang, Lin, Yu, Li and Yu2009; Ko et al., Reference Ko, Chang, Cho and Yu2022), but it remains unclear whether PUM3 plays a role in mammalian germ cells. Recently, it has been reported that the PUF family members PUM1/2 may play an important role in the occurrence and development of multiple tumours, and their expression is significantly upregulated in cancer cells. PUM protein can bind to the mitosis-related cycle inhibitor cdkn1b through post-transcriptional regulation, thereby promoting the proliferation of tumour cells (Li et al., Reference Li, Zhu, Zang, Cao, Xie, Liang, Bian, Zhao, Hu and Xu2022; Silva et al., Reference Silva, Kohata and Shigunov2022). At the same time, it has also been reported that PUM3 can promote the growth of tumour cells (Fan et al., Reference Fan, Lee, Yu, Tzen, Chou and Chang2013; Cho et al., Reference Cho, Huang, Hung, Hung, Cheng, Liu, Kuo, Wang, Yu and Yu2022; Silva et al., Reference Silva, Kohata and Shigunov2022), but the current research is relatively limited and this remains to be studied.
During female oocyte development, shortly after the resumption of meiosis at the GV stage in vitro, including those artificially maintained in an arrest, the nucleolus (or nucleoli) rapidly disappears as does detectable rRNA and tRNA transcription (Kresoja-Rakic and Santoro, Reference Kresoja-Rakic and Santoro2019), as would be expected. Nucleolar dissolution is followed by GVBD and chromatin condensation into nascent bivalent chromosomes. Condensation is accompanied by a cessation of transcription that is not resumed at a significant level until approximately the 2-cell stage in mouse and is not fully recovered until full embryo genomic activation a few cleavage divisions later.
This early embryonic period is largely characterized by the degradation of inherited maternal transcripts and their replacement by those originating from the embryonic genome, when the PUF family might be expected to function. PUM1, as the highly conserved PUF family protein, is considered to be an important maternal factor. Maternal deletion of PUM1 can lead to poor quality oocytes. Loss of maternal and paternal PUM1 results in severe preimplantation embryonic lethality (Mak et al., Reference Mak, Xia, Cheng, Lowther and Lin2018). As PUM3 is a non-canonical member of the PUF family, it is expressed at high levels during the development of mouse oocytes. We wondered whether PUM3 was also involved in mammalian oocyte development and early embryonic development. However, our data implied that PUM3 was not essential for mouse oocyte maturation and preimplantation embryo development in vitro.
It would appear that Pum3 does not play a regulatory role in reactivating transcription and would not be expected to have such a role from the GV stage until early cleavage, as translation is driven primarily by maternal RNAs and the post-translational modifications of existing proteins. Although PUM3 is thought to have the ability to bind DNA and RNA, fluorescence staining actually revealed that PUM3 is mainly localized in the cytoplasm of GV-phase eggs, with extremely low levels in the nucleus and no signal in the nucleolus (Figure 1D). This might explain why PUM3 may not be mainly involved in RNA transcriptional activities in the GV phase. Therefore, knockdown of PUM3 in the GV phase did not affect two significant biological events (GVBD and PBE) during oocyte maturation.
What is surprising is that PUM3 is seemingly not involved in RNA processing and expression during the preimplantation stages, implying that if this processing is occurring other RNA regulatory elements may also have this function, including other members of RNA-binding proteins and/or the PUF family. The redundant function of the PUM1 and PUM2 on embryogenesis, body size, and testis development has been reported previously (Lin et al., Reference Lin, Zhang, Shi, Zhu, Gao, Xia, Geng, Zheng and Xu2018, Reference Lin, Qiang, Zhu, Ding, Shi, Chen, Zsiros, Wang, Yang, Kurita and Xu2019; Zhao et al., Reference Zhao, Xiao, Cao, Xia, Gao, Cheng, Zang, Li and Xu2022). Furthermore, earlier studies of preimplantation mouse embryogenesis have suggested that there may be long-lived maternal mRNAs that presumably occur at the GV stage and persist through to the blastocyst stage (Schultz et al., Reference Schultz, Stein and Svoboda2018). If true, siRNA knockout at the GV stage may not have evident downstream developmental consequences, as these transcripts, suggested by some to be stored in a subcortical complex, already existed prior to siRNA treatment.
As oocytes originate from primordial germ cells in the fetal period, it takes a long time for female germ-cell growth during folliculogenesis and eventually maturation in the ovary. It would be useful to know whether implantation occurs, and if fetal development is normal in blastocysts derived from oocytes from Pum3 null mice or injected with siRNA at the GV stage prior to the cessation of transcription. Therefore, a Pum3 knockout animal model is still required to more clearly and accurately display the physiological functions of Pum3 in germ-cell and embryonic development in the future.
Acknowledgements
This research acknowledges the effort of all authors in planning the experimental design and interpreting the results.
Financial support
This work was supported by research grants from the Young Scientists Fund of the National Natural Science Foundation of China (Grant No. 32000599); the Hainan Provincial Natural Science Foundation of China (Grant No. 821RC686); and the Shanghai Ninth People’s Hospital (Grant No. JC201806).
Conflict of interest
The authors declare none.
Ethical standards
All animal experiments were approved by the Animal Care and Use Committee (ACUC) of Nanjing Medical University and performed in accordance with institutional guidelines.







