Introduction
On March 11, 2020, a pandemic was declared by the World Health Organization (WHO) due to a novel respiratory viral infection, the likes of which had not been witnessed in generations [1]. COVID-19, the illness caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused a range of symptoms from totally undetectable (i.e., “asymptomatic”) to flu-like effects to severe health issues that necessitated emergency medical care, hospitalization, and intensive care. Two years after the first documented case in the USA [2], over 68 million individuals across the country had been diagnosed with COVID-19 and over 1 million deaths have been attributed to the virus. In the same time frame, case counts around the world approached half a billion, with over six million reported deaths [Reference Elflein3,4]. One of the most significant early challenges in managing this outbreak was the lack of accurate viral prevalence and infection rates – especially among asymptomatic carriers.
Recognizing its pressing scientific and public health obligations, investigators at the National Institutes of Health (NIH) initiated a study to examine the presence of antibodies (seroprevalence) to SARS-CoV-2 in blood specimens collected from participants without a known diagnosis or symptoms of COVID-19. The study was initially designed to enroll a geographically constrained population that could physically visit the recruitment site on the NIH campus. Given the rapid spread of the disease, the NIH team soon realized that the study needed to scale across the country to have the greatest impact on the development of diagnostics that were reliable and generalizable and to inform national policies to mitigate the life-threatening impact of the pandemic in the USA. Therefore, they needed to expand the research sites and teams to other locations.
In 2006, the NIH Clinical and Translational Science Award (CTSAs) program established a consortium of research-intensive hubs to realize a vision for the biomedical research enterprise of the future. The program is empowered with the flexibility and support to innovate clinical and translational research workflows, develop critical scientific capacities and expertise, assemble a knowledgeable workforce, and engage contributors in an integrated approach to accelerating the translation of discoveries to improve health and health equity of individuals and communities [Reference Zerhouni5,Reference Austin6]. This ethos uniquely positions National Center for Advancing Translational Sciences (NCATS)-supported CTSA sites to pivot and scale in response to scientific opportunity. By virtue of their established research capacities, expertise and experience in large, multisite projects, two CTSA hubs (University of Pittsburgh (Pitt CTSI) and University of Alabama at Birmingham (UAB CCTS)) proposed a collaboration with the NIH in April 2020 to rapidly expand their seroprevalence study to a national scale.
The aim of the collaboration was to study COVID-19 positivity rates among asymptomatic individuals to inform national and local policy initiatives to combat the virus’ spread. The team recognized early on the critical need for having diverse representation in study participation to assess the potential impact of health disparities. By leveraging and pivoting existing resources at the CTSA hubs, the partnership succeeded in quickly developing and implementing comprehensive and harmonized processes that enabled recruitment of participants from all 50 states, data collection, specimen acquisition, and analyses (Fig. 1). Herein, the group details several lessons learned about creating a national research program, which may inform the conduct of future multisite investigation and pandemic preparedness.
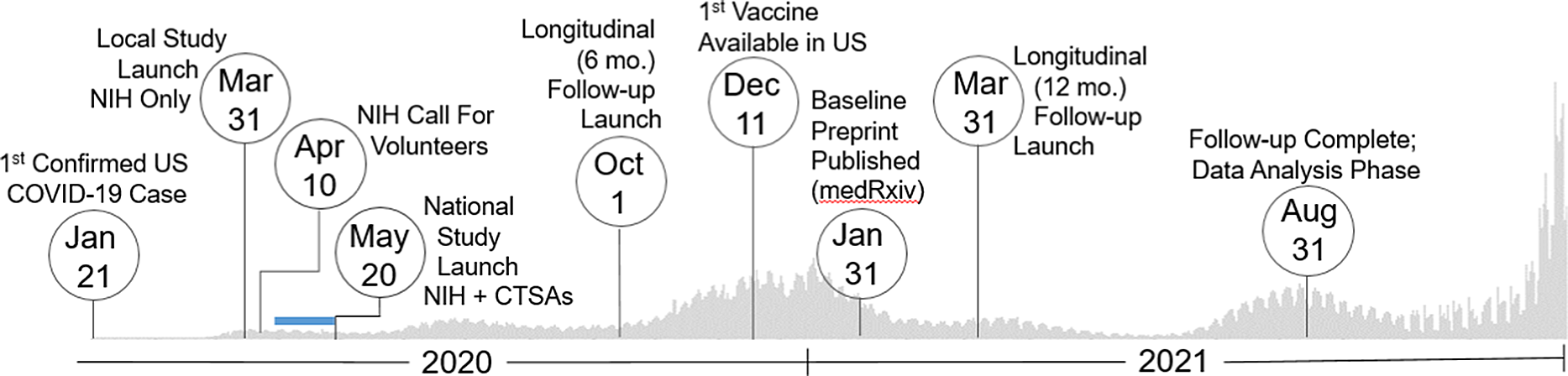
Fig. 1. Study timeline. Set against the backdrop of actual US COVID-19 reported case counts [3,4], the collaborative team launched and regularly adapted an immune surveillance study across the nation that included three specimen and data collection time points (0, 6, and 12 months). The serosurvey project began as a single-site study based at the intramural NIH campus in March 2020. Over 400,000 persons nationally responded to an NIH press release calling for volunteers in mid-April 2020. As a result, the initiative quickly pivoted to a multisite study by leveraging two CTSA sites based at the University of Pittsburgh and the University of Alabama at Birmingham (UAB), which launched recruitment May 20, 2020. Over the next 15 months, the team succeeded to enroll over 10,000 individuals representative of the geographic and demographic diversity of the country. Blue Bar: Based on previously established capacity, trained workforce, and scientific agility, the two CTSA Hubs were quickly able to submit grants, establish IRB reliance, implement a comprehensive communications strategy, staff recruitment teams (during a pandemic work stoppage), receive and store 15000 kits, standardize a shipping protocol, and tailor informatics tools for rigorous data management (REDCap, Salesforce) to go live with recruitment ∼ 1 month later. Legend: CTSAs, Clinical and Translational Science Awards; mo., month; NIH, National Institutes of Health; US, United States.
Methods
Case Study: Undiagnosed SARS-CoV-2 Seropositivity in the USA
As the public health implications of the COVID-19 pandemic became apparent in 2020, the scientific community rapidly responded to an unprecedented public health threat and research challenge. Scientific projects focused on the biology of a novel coronavirus, the effects of the virus on human physiology, and the epidemiology and clinical impact of COVID-19. This knowledge was essential to the development of mitigation strategies, diagnostics, clinical care paradigms, and therapeutics to aid in the treatment of the disease and to reduce the risk of severe illness, hospitalization, and death.
During this early phase of the pandemic, very little was known about the transmission of SARS-CoV-2 infection. Those who were infected experienced a wide spectrum of symptoms, from very minor sniffles or manageable flu-like symptoms to life-threatening respiratory failure. Efforts to control the spread were hampered by a lack of information about presumed asymptomatic case rates, which made it very difficult to contain local outbreaks as these individuals were unaware if they were infected. Investigators at the National Institutes of Health (NIH) launched a study to evaluate the prevalence of immunity in the surrounding community by assaying the presence of antibodies to the virus in individuals who were not previously diagnosed with or recently experienced COVID-19 symptoms.
In a NIH press release dated April 10, 2020, Dr Anthony Fauci, Director of the NIH National Institute of Allergy and Infectious Diseases (NIAID), promoted the importance of the study [7]. Within 1 week of this announcement, NIAID received more than 400,000 emails from people across the country offering to participate in the research endeavor. The research team, which had launched the study by enrolling individuals onsite at the NIH, quickly pivoted to adapt their study design to a national-scale, remote, observational study.
In parallel, leaders of the CTSA hubs based at the University of Pittsburgh (Pitt CTSI) and the University of Alabama at Birmingham (UAB CCTS) acknowledged that CTSAs are uniquely equipped to rapidly leverage research capacities and mobilize well-trained teams to advance clinical and translational investigation at any scale. These two CTSA hubs, which support large-scale national research projects (e.g., All of Us Research Program, PCORnet, ACTIV), were ideally positioned to urgently apply their experiences to assist researchers at NIAID to scale their study across the USA. This agility to engage partners to quickly and efficiently enable clinical investigation and to provide a national resource responsive to public health needs is a hallmark of CTSA programs. In 1 month, the NIAID LID Clinical Studies Unit and the Pitt and UAB CTSA hubs partnered to assemble the expertise, regulatory approvals, technology, and engagement strategy to launch a national longitudinal observational seroprevalence study during the COVID-19 pandemic (Fig. 2).
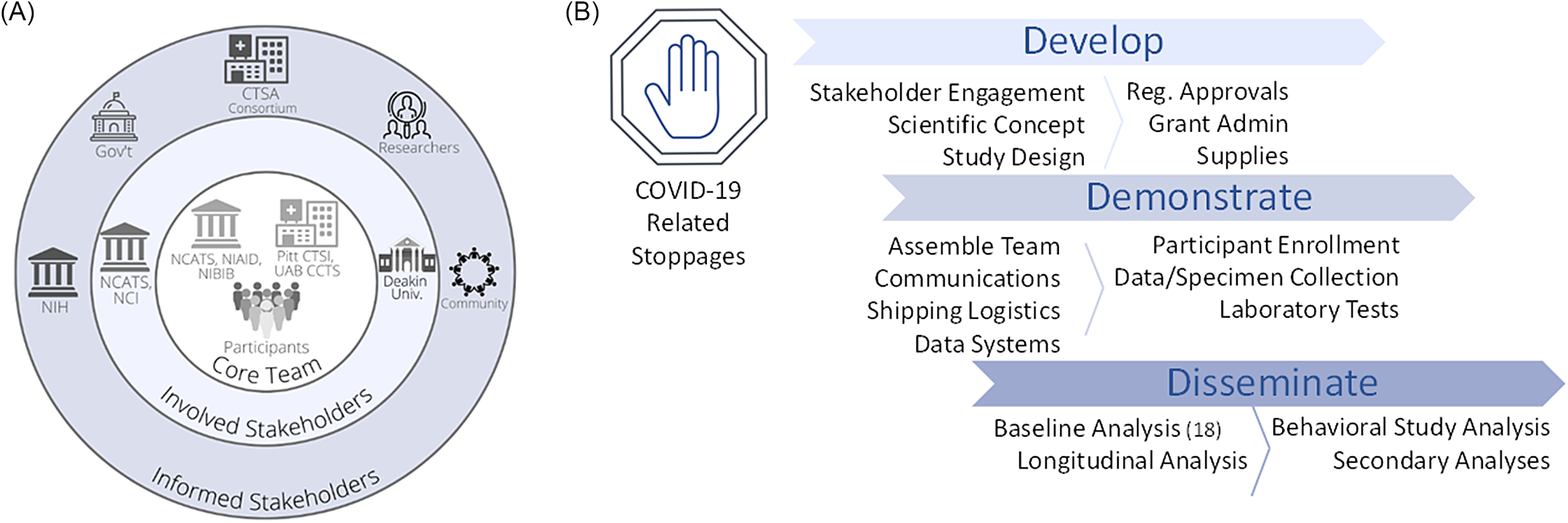
Fig. 2. Stakeholder and study map. NIH and two NCATS-sponsored CTSA hubs (Pitt CTSI and UAB CCTS) partnered to assemble the expertise, regulatory approvals, technology, and engagement strategy to launch a national longitudinal observational seroprevalence study during the COVID-19 pandemic. A. This project engaged stakeholders on multiple levels, including project drivers involved in the concept and implementation (Core Team), sponsors (Involved Stakeholders), and several areas with vested interest (Informed Stakeholders). B. Overview of study implementation. Following widespread, COVID-19-related research stoppages that went into effect in March 2020, the team of CTSAs and NIH were able to develop the project, quickly pivoting to the scientific opportunity and scaling to a national reach. Over the next 1.5 months, the team put in place the necessary study components to allow collaborative enrollment to begin as part of the Demonstration phase. Dissemination of initial findings succeeded within ∼ 8 months. Legend: CTSA, Clinical and Translational Science Award; Gov’t, Government; NIH, National Institutes of Health; NCATS, National Center for Advancing Translational Sciences; NCI, National Cancer Institute; NIAID, National Institute of Allergy and Infectious Diseases; NIBIB, National Institute of Biomedical Imaging and Bioengineering; Pitt CTSI, University of Pittsburgh Clinical and Translational Science Institute; Reg., Regulatory; UAB CCTS, University of Alabama at Birmingham Center for Clinical and Translational Science; Univ., University
Assembling the Team
The rapid implementation of this large, multisite, remote seroprevalence study required a transdisciplinary team of clinical investigators, laboratory scientists, biostatisticians, immunologists, infectious disease experts, epidemiologists, informaticists, clinical research coordinators, and award managers at the two CTSA hubs. This group had extensive and complementary training and skills in methodologic rigor and reproducibility, data and specimen management, regulatory knowledge, good clinical practice, and team science. The project, by design, assembled systems thinkers, communicators, domain experts, process innovators, “boundary crossers,” and team players. As a direct result of the learning opportunities available through Pitt CTSI & UAB CCTS, this team represented the characterization of translational scientists in action [Reference Gilliland, White and Gee8]. Additionally, given the wide range of experiences of each CTSA Hub (e.g., in supporting the All of Us Research Program [Reference Denny and Rutter9]), their leadership was immediately able to plan for staffing needs, budget requirements, computing power, communication equipment, IT platforms, and space necessary for this large complex project. This shared standard of excellence in workforce development and training precedent across CTSA-based institutions was also reflected in facile communications and standard operating procedures using Salesforce Customer Relationship Management (CRM) software, Microsoft Teams, Box file storage systems, and weekly video meetings in an effort to streamline efficient and consistent support of the project at all sites.
Regulatory Oversight
NIH served as the IRB of record to oversee all human subjects activities, adapting the original site-specific regulatory approval to a single IRB model, with Pitt CTSI and UAB CCTS establishing reliance agreements that ceded protocol review. Though the NIH IRB is not typically a resource for extramural projects, the process of rapid adaptation of site-specific protocols to shared regulatory models is a generalizable model with feasibility for any additional sites. All protocol modifications were approved by the NIH IRB (e.g., participant communications used during the recruitment, prescreening, enrollment, and sample collection phases of the project). The well-established regulatory expertise at the CTSA hubs allowed the efficient launch of the study within a few days of protocol submission and rapid approval of all amended documents throughout the project implementation. The team was also well positioned to leverage observed increases in IRB productivity and accelerated reviews of COVID human subjects protocols [Reference Ford, Johnson, Nichols, Rothwell, Dubinett and Naeim10]. The synergies enabled by prior training and process know-how combined with active communication with the Office of the IRB as a partner in the process were instrumental in this accelerated time to activation.
Study Design
Statisticians based at NIAID collaborated with colleagues in the Pitt CTSI Biostatistics, Epidemiology and Research Design (BERD) Core to revise their study design. The initial single-site recruitment model based at the NIH was insufficient for a national study design. Therefore, they adopted a quota sampling approach with algorithms to adapt recruitment targets in real time to more effectively reflect the representative diversity of the country. Their challenge, however, was how to maintain rigorous and reproducible capacities for consistent outreach, screening, enrollment, home blood sampling, and data collection of this volume.
Information Technology Support
As part of a mission-aligned implementation strategy, CTSA organizations are developing innovative informatics solutions to use digital assets to improve health. NIH, Pitt CTSI, and UAB CCTS have established a range of expertise for the capture, management, and analysis of biological and clinical data to advance scientific discovery, following collaborative standards for interoperability and data sharing. This project used an instance of Salesforce (Salesforce, Inc.) licensed to Pitt CTSI and accessible to UAB CCTS for research purposes to track participant information, contact (both email and phone), and shipping information for the test kits. The team developed an application programming interface (API) to link Salesforce to the NIAID REDCap instance to register participants and document informed consent [Reference Harris, Taylor, Thielke, Payne, Gonzalez and Conde11,Reference Harris, Taylor and Minor12]. The REDCap instance was used to track consent/withdrawal status and collect patient-reported data (medical, geographic, demographic, and socioeconomic information) via electronic questionnaire. The Pitt CTSI Enterprise Relationship Management team configured the Pardot (email) and Lightning Dialer (telephone) components of Salesforce for the Pitt CTSI and UAB CCTS staff to manage the IRB-approved communication and phone outreach plans for this study. Salesforce relationship management capabilities were extended to seek and record consent from participants. Pitt CTSI wrote automated services to query the UPS and FedEx APIs, to retrieve shipping status for the kits that have gone out, and to automatically update tracking fields in Salesforce to provide a real-time picture of where participant kits were. The dedicated clinical research informatics and information technology expertise in place at the CTSA hubs and NIH allowed the facile adoption and integration of systems with the requisite security measures and rigorous data management procedures to serve a multisite clinical study.
Participant Screening, Enrollment, Specimen, and Data Collection
This project involved quota sampling from 462,949 volunteers with the goal of enrolling a convenience sample of over 11,000 individuals reflecting demographic characteristics of the US population (see [Reference Kalish, Klumpp-Thomas and Hunsberger13] for more detailed study description). The recruitment teams at Pitt CTSI and UAB CCTS followed approved telephone scripts to screen and enroll participants, which were documented in Salesforce and REDCap. Consented individuals were shipped a Mitra Home Collection Kit to provide an 80-µl blood specimen [Reference Hall, Luisi and Zlotorzynska14]. Each kit contains gauze, lancets, bandages, collection device, and all necessary shipping materials along with detailed instructions for participants and a return postage slip. Program managers at Pitt CTSI and UAB CCTS were able to package, label, and ship up to 200 kits/day. Tracking information was monitored to confirm delivery and unreturned kits, which in turn was used to trigger automated reminder messages (up to 3) and to inform the NIAID statistical team when necessary to adjust the planned consenting strategy. Follow-up email and phone messaging were facilitated by the Pitt CTSI Communications team, which also served to bolster individual participant understanding when needed (e.g., how to use the collection kit). Specimen kits were returned directly to the NIH for laboratory analysis.
Participant-reported health data and personality survey feedback were captured digitally using an emailed link to a REDCap questionnaire or facilitated by a study team member by telephone if necessary. While the project’s screening, recruitment, and collection components were customized to the study aims and did not exist “on the shelf,” the team was able to draw on experience in multimodal communication methods, near-peer mentoring, and design thinking – common topics in CTSA-based workforce development programs – to rapidly prototype and refine solutions to overcome barriers to shipping kits, calling participants in different time zones and other logistical challenges.
Research Dissemination
The vested interest of the public to participate in this seroprevalence study and to contribute to the understanding of a disease that affected everyone’s lives is exemplified by the initial response of individuals volunteering to participate in the study within hours of NIAID’s press release. While return of individual results was not possible, the team was committed to scientific transparency and, therefore, acquired regulatory approval to share summary data with participants by emailing them the initial published, peer-reviewed manuscript of baseline study results [Reference Kalish, Klumpp-Thomas and Hunsberger13]. Similarly, understanding the urgency to share COVID-19 results, the data were published on the medRxiv preprint server a few months earlier for rapid access by the scientific community [Reference Kalish, Klumpp-Thomas and Hunsberger15,Reference Else16].
Discussion, Lessons Learned, and Perspectives for the Future
Rapidly launching a large, multisite, decentralized clinical research study to enroll participants across the nation and collect biospecimens and data is a complex endeavor, even under traditional circumstances. Carrying out the study in the context of a global health crisis that led to hiring freezes and furloughs of research staff, virtual (socially distanced) work, and supply chain constraints added to the project management’s complexity. Yet, as is apparent by the present study, CTSA hubs are optimally positioned to rapidly implement high-impact translational research projects at local, regional, and national levels, even under the most challenging circumstances, as a direct result of their expertise, research capacities, infrastructure, and established collaborations established years before the pandemic (Fig. 3).

Fig. 3. Key advantages for rapid response. Legend: CTSA, Clinical and Translational Science Award; IRB, Institutional Review Board; IT, Information Technology; NCATS, National Center for Advancing Translational Sciences; NCI, National Cancer Institute; NIAID, National Institute of Allergy and Infectious Diseases; NIBIB, National Institute of Biomedical Imaging and Bioengineering.
CTSA Advantage for Large-Scale, Rapid Deployment
Transdisciplinary Teamwork
The advancement of scientific discovery to improve health is increasingly team science. The ability for diverse investigative teams to work together toward a shared goal is a strength in the conduct of science [17]. Sponsorship of this project relied on intra-agency collaboration at the NIH, bringing together NIAID, NCATS, NIBIB, and NCI to support the project. Conduct of the project relied on seamless integration of daily activities across NIH (NCATS, NIAID LID CSU, NIBIB) and CTSA organizations (Pitt CTSI and UAB CCTS). The team – comprised of faculty, students, and staff – represented clinical investigators, laboratory scientists, biostatisticians, epidemiologists, health economists, informaticists, data managers, research coordinators, and more. The success of this multidisciplinary team to rapidly launch and complete a high-impact study on a national scale is a direct result of each member’s training, skills, and experience in core principles of clinical and translational team science.
Translational Spectrum
The seroprevalence study collaboration integrated all phases of the translational research spectrum, which are supported by CTSA hubs [18]:
T1 – Basic Research: Use of ELISA to detect SARS-CoV-2-related antibodies in blood
T2 – Clinical Research: National population enrollment and specimen collection
T3 – Population Research: Outcomes investigation of seropositivity relative to gender, race/ethnicity, urban/rural, geography, etc.
T4 – Public Health Research: Value of seropositivity estimates in guiding pandemic response and understanding regional variability
Agile and Responsive Research Capacity
Like the NIH, the CTSA consortium is part of a national resource committed to the development of scientific and operational innovations that improve the efficiency and effectiveness of clinical and translational investigations while maintaining high standards of research quality, reproducibility, and safety. The CTSA hubs are highly adept at promoting partnerships and collaborations to accelerate these processes and in developing a diverse, skilled, and knowledgeable workforce capable of supporting the life cycle of a study. Accordingly, the hubs based at Pitt CTSI and UAB CCTS were able to rapidly respond to the call to expand a local project from NIH to a national scale [Reference Austin, Jonson and Kurilla19]. They leveraged their experiences with large research programs like All of Us and PCORnet to adapt to the tailored needs of the new project. They had well-established infrastructure, relationships, and expertise that could guide the startup and conduct of the project within each site and the communication platforms to harmonize standard operating procedures across sites. The similarities in training between CTSA hubs also allowed team members to relate, to mentor, and to collaborate with each other in support of the project. This cross-coverage between sites was especially helpful in both grants management and regulatory approvals as well as managing participant outreach to different time zones and appreciating variable work preferences (early risers and night owls).
While the study team was expeditious and flexible in implementation, the speed of the pandemic spread proved challenging. The rate of transmission, mutation rate of the virus, development and deployment of vaccines, and the adaptive sampling strategy necessitated considerations in data collection and rigorous analysis that delayed the availability of study insights to impact COVID-19 public health policy and management. While some other population-based serosurveys conducted in the USA were published more quickly [Reference Barzin, Schmitz and Rosin20-Reference Sutton, Cieslak and Linder28] or represented larger enrollments [Reference Rosenberg, Tesoriero and Rosenthal26,Reference DeSantis, León-Novelo and Swartz29,Reference Pathela, Crawley and Weiss30], they were all limited to a constrained geographic region (e.g., city, county/parish, state). This seroprevalence experiment is the only known project conducted on a national scale, recruiting over 11,000 individuals and publishing baseline data within 10 months of the first enrollment in March 2020 (Fig. 1) [Reference Kalish, Klumpp-Thomas and Hunsberger13]. The experience and established methods will be exceptionally valuable to future pandemic preparedness and response other health crises.
Conclusion – Preparing for the Future
Response to scientific opportunity, whether prompted by a global pandemic or not, requires greater speed and efficiency to launch studies, recruit participants, analyze data, and disseminate results. As evidenced in this use case, institutions and investigators are better equipped to address emerging research needs in an environment that has assembled resource components (informatics/IT, cohesive leadership, biostatistics, trained workforce, grants management, regulatory expertise/good clinical practice, etc.) and a track record in external partnerships. Experience with verbal informed consent, digital survey of participant information, and remote sample collection can yield greater enrollments, reach remote (e.g., rural) geographic areas, and increase the diversity and inclusion of groups underrepresented in biomedical research. The involvement of multiple sites expands this functionality and offers an understanding of cultural sensitivities, health literacy, and local attitudes. Challenges of harmonizing activities in multiple locations can be minimized with a proactive training, communication, and capacity building. This seroprevalence study is an important demonstration of how the complementary capabilities of CTSA hubs can come together to achieve programmatic synergies and rapidly conduct large-scale studies on a national level, an experience that will inform and enable collaborations and pandemic preparedness in the future.
Acknowledgements
We thank all the participants in this study (ClinicalTrials.gov NCT04334954; NIH IRB #20I0083; UAB IRB-300005253; Pitt IRB STUDY20110122). This research was supported, in part, by the Intramural Research Program of the National Institutes of Health, including the National Institute for Biomedical Imaging and Bioengineering, the National Institute of Allergy and Infectious Disease and the National Center for Advancing Translational Sciences (NCATS). This project has been funded, in part, with Federal funds from the National Cancer Institute, NIH, under contract number HHSN261200800001E, 75N91019D00024, Task Order No. 75N91019F00130, and Clinical and Translational Science Awards program grants UL1TR003096 (UAB) and UL1TR001857 (University of Pittsburgh). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government. The NIH, its officers, and employees do not recommend or endorse any company, product, or service. We thank the CCTS Clinical Research Support Program, the University of Pittsburgh CTSI staff and leadership, and University of Pittsburgh Information Technology for their contributions to this study. The authors also thank Michelle Kienholz for editorial assistance.
Disclosures
The authors have no conflicts of interest to declare.





