1 Introduction
1.1 Overview of this Volume
In recent decades, the number of archaeometric investigations that make use of physical‒chemical techniques for the analysis of the composition of various archaeological materials continues to grow, as evidenced by the increasing number of publications in this area. One example of this type of studies is provenance analysis, which tries to relate archaeological materials to their original natural sources by discriminating their characteristic chemical fingerprint. In brief, it tries to determine the geological or natural origin of materials found in different archaeological contexts to establish the places of acquisition and production of the raw materials. We have chosen to approach this complex subject in two different ways, both based on very similar datasets.
In this Element, we take an applied, practical approach, allowing the reader to experiment with the provided datasets and scripts to be used in the R software package. In Statistical Processing of Quantitative Data of Archaeological Materials, we take a more theoretical and mathematical avenue, allowing the reader to amend and apply the discussed methods freely. These two Volumes can be used independently as well as complementary, throughout both ample cross-references are provided to facilitate the latter. As an introduction to the subject, let us first remember that the methods, basic principles and when to apply different statistical processing depends on three data scenarios: (1) when dealing with high-dimensional spectral data, (2) when employing compositional data, and (3) when managing a combination of compositional and spectral data.
Case 1 considers high-dimensionality data (n ≪ p, where n relates to the number of observations and p are the number of variables) using full spectrum readings, such as those obtained with Fourier transform infrared spectroscopy (FT-IR), Raman spectroscopy, or X-ray fluorescence (XRF) spectroscopy. For this type of data, the suggested approach is to apply chemometric techniques and unsupervised machine learning methods. First, the spectra are preprocessed by filtering the additive and multiplicative noise, correcting misaligned peaks, and detecting outliers by robust methods. Afterwards, the data are clustered using a parametric Bayesian model that simultaneously conducts the tasks of variable selection and clustering. The variable selection employs mixture priors with a spike and slab component, which make use of the Bernoulli distributions and the Bayes factor method to quantify the importance of each variable in the clustering.
Case 2 contemplates low-dimensional data (n > p) where the recorded data have been converted to chemical compositions. For this case, the recommended approach is to adopt the methodology proposed by Reference AitchisonAitchison (1986), which discusses some of the algebraic–geometric properties of the sample space of this type of data and implements log-ratio transformations. Respecting adequate preprocessing of compositional data, such as robust normalization and outlier detection, the use of model-based clustering that fits a mixture model of multivariate Gaussian components with an unknown number of components is proposed. This allows choosing the optimal number of groups as part of the selection problem for the statistical model. Mixture models have the advantage of not depending on the distance matrix used in traditional clustering analyses. Instead, the key point of the model-based clustering is that each data point is assigned to a cluster from several possible k-groups according to its posterior probabilities, thus determining the membership of each of the observations to one of the groups.
For Case 3, if reliable calibrations are available to obtain compositional data, this information can be combined with the spectra to obtain groups. For handling the data, a combination of chemometric techniques is used. In this case, a dependent variable y (or compositional values) is related to the independent variables x (or spectral values). The preprocessing is performed similarly as in Case 2; this allows calibrating a model of predictive purposes that can discriminate those variables that provide significant information to the analysis and eliminating the redundancy of information as well as collinearity. Once the selection of variables has been made, a new methodology called Databionic Swarm (DBS; implemented by Reference Thrun, Ultsch and HüllermeierThrun, 2018) is applied for clustering the data.
To fully understand how the proposed methods work and how to apply them to your own data, these are exemplified in this Element with different case studies using quantitative data acquired from archaeological materials. The datasets used in the examples are provided in the electronic format of this Element as worksheet files with the “csv” extension. To process the data according to the exercises, the selected dataset must be imported and the source code executed in the R environment (R Development Core Team, 2011); we used version 3.6.1 on a 64-bit Windows system, although more recent versions of R are now available. R is a programming language for statistical analysis and data modeling that is used as a computational environment for the construction of predictive, classification, and clustering models. R allows you to give instructions sequentially to manipulate, process, and visualize the data. The instructions or scripts employed for each part of the process are detailed in the case studies. To learn how to employ the scripts in each step of the data processing, we encourage the reader to consult the videos associated with this Element in the electronic format of the Element.
1.2 Introduction to R
R is a public domain language and environment managed by the R Foundation for Statistical Computing (© 2016 The R Foundation) that has the virtue of being an exceptional tool for data statistical analysis and projection. This project contains a large collection of software, codes, applications, documentation, libraries, and development tools that users are free to copy, study, modify, and run. Therefore, it can be seen as a collaborative project in which anyone is invited to contribute. Although initially written by Robert Gentleman and Ross Ihaka, since 1997, it has been operated by the R Development Core Team. From early 2000 until now, it has become a kind of “standard of the scientific community.” There are many publications and tutorials on its use aimed at all levels of different areas and specializations, some of which focused on the most technical and computational aspects of the language.
1.2.1 Getting Started
First, search online for CRAN R (the Comprehensive R Archive Network) or follow the link http://cran.r-project.org/ that will direct you to the web page and the instructions to download and install the latest version of R in various platforms (Linux, MacOS and Windows).
1.2.2 Data Import
Once R is installed, the next step is to call our data in the R window to be able to process them. In the screen, the indicator “>” appears and is where we must define what task we want to perform. The most commonly used configurations to perform data analysis in R are data frames, which are two-dimensional (rectangular) data structures. As in this case, we deal with datasets of low or high dimensionality, which must be arranged so that the rows in a data frame represent the cases, individuals or observations, and the columns represent the attributes or variables (see example in Table 1). These data frames must be prepared in a folder available for import and analysis.
Table 1 Example of a data frame of chemical concentrations of obsidian samples.
| ID | Mn | Fe | Zn | Ga | Th | Rb | Sr | Y | Zr | Nb | Source |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ahuisculco | 378 | 7468 | 47 | 18 | 10 | 109 | 45 | 20 | 143 | 19 | 1 |
| Ahuisculco | 379 | 7889 | 166 | 19 | 9 | 113 | 50 | 19 | 146 | 21 | 1 |
| … | … | … | … | … | … | … | … | … | … | … | …. |
| … | … | … | … | … | … | … | … | … | … | … | …. |
| El Chayal | 476 | 8162 | 33 | 17 | 11 | 146 | 154 | 20 | 100 | 9 | 12 |
| El Chayal | 486 | 8799 | 45 | 18 | 11 | 151 | 158 | 20 | 104 | 8 | 12 |
| El Chayal | 545 | 8309 | 36 | 17 | 10 | 140 | 151 | 19 | 101 | 9 | 12 |
The working directory is the place on our computer where the files we are working with are located. You can find what the working directory is with the function “> getwd()”. You only have to write down the function and execute it. You can change the working directory using the function “setwd()”, defining the path of the directory you want to use [e.g., setwd(”C:\obsidian”)]. Although there is extensive documentation on how to import/export data to R, we use the traditional method, as our data are usually in a spreadsheet with a csv extension. In R, it is sufficient to use the following command:
> data <- read.csv(”C:\\obsidian\\mydataset.csv”, header=T)

This command line provides the path to the folder where the data are found; the command “read.csv” indicates that a file with a “csv” extension is read from the “obsidian” folder located in the C root directory, and it is indicated that the data contain a name for each variable with the command “header=T”. The symbol “<-” is only used for assignment; in the previous example, the file name “mydataset.csv” refers to the name of the data frame that you are going to work with in R. To see the structure of the data, write down the next command.
str (data)

Another way is by selecting the option “Change directory” from the File menu and navigating to the folder where our file is located. Once this path is established, you must go to the folder where your dataset is, select it and drag it with the mouse to the R console and, later, copy that path from the console and paste it into the command “read.csv”. To see the dimensions of the dataset, you can write the function “dim (data)”; “names()” shows the names of the columns. In R, the “summary” function shows a general summary of the data frame variables (minimum, maximum, mean, median, first, and third quartile).
To perform an algebraic operation on a data frame such as the one exemplified in Table 1, the first column containing the identifier of each sample would have to be excluded; this is achieved by typing the following command:
data1 <- data [, 2:11]

In this way, only numerical variables are considered. Now you can transform all the data in the data frame, such as base 10 logarithms:
data2 <- log10 (data1)

If you also would like to transform negative values with logarithms you can use DataVisualizations::SignedLog(). Depending on the application this can be meaningful (cf, Reference Aubert, Thrun, Breuer and UltschAubert et al., 2016), even if in a strictly mathematical sense it is not allowed. If you want to see the values of a specific variable, you can do it with the following command:
data2$Na.

Some analyses require that the data be recognized as a matrix. In R, a matrix is a data structure that stores objects of the same type, conversely to a data frame, which is a rectangular array of data consisting only of numeric values. To convert a data frame to a matrix, you can use the following command:
newdata <- as.matrix(data2).

To save a file that has been transformed, just type the following command:
write.csv (newdata, file=”my_data.csv”).

This will save the file named “my_data” with a “.csv” extension to the working folder.
1.2.3 Functions
Once a dataset has been loaded, a large number of operations can be performed on it. If, for example, you have a variable “Na” from which you want to produce a histogram, it is enough to write the “hist (Na)” function to produce a bar graph of the variable. In R, the “plot()” function is generally used to create graphs. This function always asks for an argument for the axis of the abscissa (x) and another for the ordered (y). If two variables, x and y are available, say Sr and Zr, and you want to see how these variables relate, “plot(Data2$Sr, Data2$Zr)” is enough to get the graph. The “plot()” function allows you to customize the graph by entering titles or changing the size of the dots, the color, etc.
In addition to the “plot()” function, there are other functions that generate specific types of graphs. In Windows, right clicking the graph can be copied to the clipboard, either as a bitmap or as a metafile. There are a variety of graphic packages for R that extend their functionality or are intended to optimize things for the user. If you have any questions about any other function, you can always access Help documentation using the “help()” command. For example, “> help(mean)” directs us to a web page where we can obtain information about the concept “mean” that corresponds to the arithmetic average.
1.2.4 Packages
R has a large number of packages that offer different statistical and graphical tools. Each package is a collection of features designed to meet a specific task. For example, there are specialized packages for data grouping, others for visualization or for data mining. These packages are hosted on CRAN [https://cran.r-project.org/]. A small set of these packets is loaded into the processor’s memory when R is initialized. You can install packages using the “install.packages()” function, and typing in quotation marks the name of the package you want to install. They can also be installed directly from the console by going to the R menu and then selecting “Packages.” For example, to install the “cluster” package, type the following:
install.packages(”cluster”).

After you complete the installation of a package, you can use its functions by calling the package with the “library(cluster)” command. Every time an operation is performed in R, it is important to use the “rm(list = ls())” command to delete all objects in the session and to be able to start without any remaining objects stored in the program memory. Additionally, when calling any file, it is recommended to use the “str ()” command to know the structure of the data object.
1.2.5 Scripts
Scripts are text documents with the “.R” file extension. The scripts are the same as any text documents, but R can read and execute the code they contain. Although R allows interactive use, it is advisable to save the code used in an R file; this way, it can be used as many times as necessary. In this Element, we made use of the scripts published by the authors of the packages available for R. An advantage of these scripts and packages is that you can make use of the tutorials available for each. For example, to transform the data to the isometric log-ratio (ilr), go to CRAN [https://cran.r-project.org/] and look for the “compositions” package of Reference van den Boogaart, Tolosana-Delgado and Brenvan den Boogaart, Tolosana-Delgado and Bren (2023). You will find that the related script is the following:
## log-ratio analysis # transformation of the data to the ilr log-ratio library(compositions) xxat1 <- acomp(data) # “acomp” represents one closed composition; with this #command the dataset is now closed xxat2 <- ilr(xxat1) # isometric log-ratio transformation of the data str(xxat2) write.csv(xxat2, file=”ilr-transformation.csv”)

In this script, the command “acomp” tells the system to consider the argument as a set of compositional values, implicitly forcing the data to close to 1. Subsequently, following Reference Egozcue, Pawlowsky-Glahn, Mateu-Figueras and Barceló-VidalEgozcue et al. (2003), the “ilr” command is used to transform the data to the isometric log-ratio, which produces compositions that are represented in Cartesian coordinates.
A more complex task can be done with R. For example, let us say that you want to implement a Principal Component Analysis with the idea of exploring the data and seeing if the first components can reveal the existence of a pattern in the data. By installing the “ggfortify” package (Reference Horikoshi, Tang and DickeyHorikoshi et al., 2023), it is easy to perform the analysis and graphical display of the data. For example, suppose we have a matrix of n x p with untransformed data and whose eleventh column indicates the natural source from which some obsidian samples come, such as the one in Table 1. The first step is to call the package “ggfortify” and read the data.
library(ggfortify) data <- read.csv(”Sources.csv”, header=T) ## Sources.csv is an example file name str(data)autoplot(stats::prcomp(data[-11])) ## PCA without labels; the 11th column ##is deleted

In this script, the function “prcomp” will perform a principal component analysis of the data matrix and return the results as a class object; in turn, the “autoplot” function will provide the graph of the first two components. To produce a plot of the first two components that includes the provenance label for each sample and a color assignation to each group, use the following command line:
autoplot(stats::prcomp(data[-11]), data1 = data, colour = ‘Source’) ## ‘ Source’ specifies column name keyword in your dataset

A biplot of the components that explain most of the variance of the data using the loading vectors and the PC scores is obtained through the following command line:
autoplot(stats::prcomp(data[-11]),label=TRUE,loadings=TRUE,loadings.label=TRUE)

Assuming the grouping variable is available, the following command line automatically locates the centroid of each group and performs a PCA with 95 percent confidence ellipses:
autoplot(stats::prcomp(datos[-11]),data1=data,frame=TRUE, frame.type=‘t’,frame.colour=‘ Source’) ## ‘ Source’ specifies column name keyword in your dataset

In R, there are numerous clustering algorithms ranging from distance-based algorithms (e.g., to determine whether the data present a clustering structure) to more formal statistical methods based on probabilities, such as Bayesian methods or model-based clustering. For instance, for conventional clustering, the package “cluster” can be used (Reference Maechler, Rousseeuw, Struyf, Hubert and HornikMaechler et al., 2022). With this package, several classical classifications can be performed by selecting both the metric used to calculate the differences between the observations and the grouping method, among which are average, single, weighted, ward, and others.
This Element provides the scripts to perform all the proposed preprocessing and clustering techniques so that the user can easily execute the commands by copying and pasting them into the R environment. For example, in Section 3, we worked with model-based clustering; for this, we employed the R libraries “Rmixmod” (Reference Lebret, Lovleff and LangrognetLebret et al., 2015) and “ClusVis” (Reference Biernacki, Marbac and VandewalleBiernacki et al., 2021):
library (Rmixmod) out_data<-mixmodCluster(data2,nbCluster=2:8) summary(out_data) plot(out_data) library(ClusVis) clusvisu<-clusvisMixmod(out_data) plotDensityClusVisu(clusvisu)

By using the command “mixmodCluster”, an unsupervised classification based on Gaussian models with a list of clusters (from two to eight clusters) is performed, determining which model best fits the data according to the BIC information criterion. In turn, the “plot()” command provides a 2D representation with isodensities, data points, and partitioning. . Alternatively, two-dimensional density-based structures can be visualized with “ScatterDensity” (Reference Brinkmann, Stier and ThrunBrinkmann et al., 2023). Similarly, the “ClusVis” package (Reference Biernacki, Marbac and VandewalleBiernacki et al., 2021) is used to obtain a graph of Gaussian components that supplies an entropic measure on the quality of the drawn overlay compared to the Gaussian clustering of the initial space. For this, only two commands are needed, “clusvisMixmod” and “plotDensityClusVisu”, which are provided by the package authors.
Thus, the user has free access to the tutorials and scripts of each of the algorithms used in this Element. In many cases, the only thing that needs to be done is to replace the author’s data with your own. You can also experiment with other strategies for the analysis by changing the parameters, such as the number of iterations, the initialization method, and the model selection criteria. The instructions to do so, as well as a variety of examples that the same user can reproduce, can be consulted in the documentation associated with each R package or script. It is very important to remember that the theoretical assumptions of each model must be respected; unfortunately, the data do not always conform to these. That is why it is recommended that the reader pay close attention to the theory of each method and to the behavior of his data, since a violation of the theoretical assumptions can lead to an incorrect interpretation of the data.
2 Processing Spectral Data
2.1 Applications and Case Studies
This section presents the numerical experiments conducted on archaeological materials using spectral data and the Bayesian approach. Although only examples of X-ray fluorescence data are used in this Element, the proposed methods can be applied in the same way to any other spectrometric technique such as Raman or FT-IR. To exemplify the performance of the proposed methods, two analyses were carried out, one with 156 obsidian geological samples that served as a control test (matrix available in the supplementary material as file ‘Obsidian_sources.csv’) and a second one using 185 ceramic fragments of archaeological interest (matrix available in the supplementary material as file ‘NaranjaTH_YAcim.csv’). For the analysis of all samples, we employed a TRACER III-SD XRF portable analyzer manufactured by Bruker Corporation, with an Rh tube at an angle of 52°, a drift silicon detector and a 7.5 μm Be detector window.
The instrument was set with a voltage of 40 kV, a current of 30 μA, and a measurement time of 200 live seconds. Only for the case of the obsidian samples was employed a factory filter composed of 6 μm Cu, 1 μm Tl and 12 μm Al. A spectrum of each sample was obtained by measuring the photon emissions in 2048 channel intervals (corresponding to the energy range of 0.019 to 40 keV of the detector resolution). Is important to remember that portable XRF is more effective for detecting Na to U elemental concentrations, which is the range that corresponds to 1.040 (in the K-alpha layer) to 13.614 keV (in the L-alpha layer). Therefore, any peaks outside of this range supply no useful information. That is the reason why the original 2,048 channels were reduced by cutting the tails of the spectra that corresponded to noninformative regions, such as low Z elements (lower than Na), the Compton peak, and the palladium and rhodium peaks (K-alpha and K-beta). For example, in the obsidian exercise, the data that were not in the range from channel 38 to channel 900 were manually deleted, leaving only the central 862 channels that correspond to the energy range of 0.74 to 17.57 keV of the detector resolution (Figure 1).
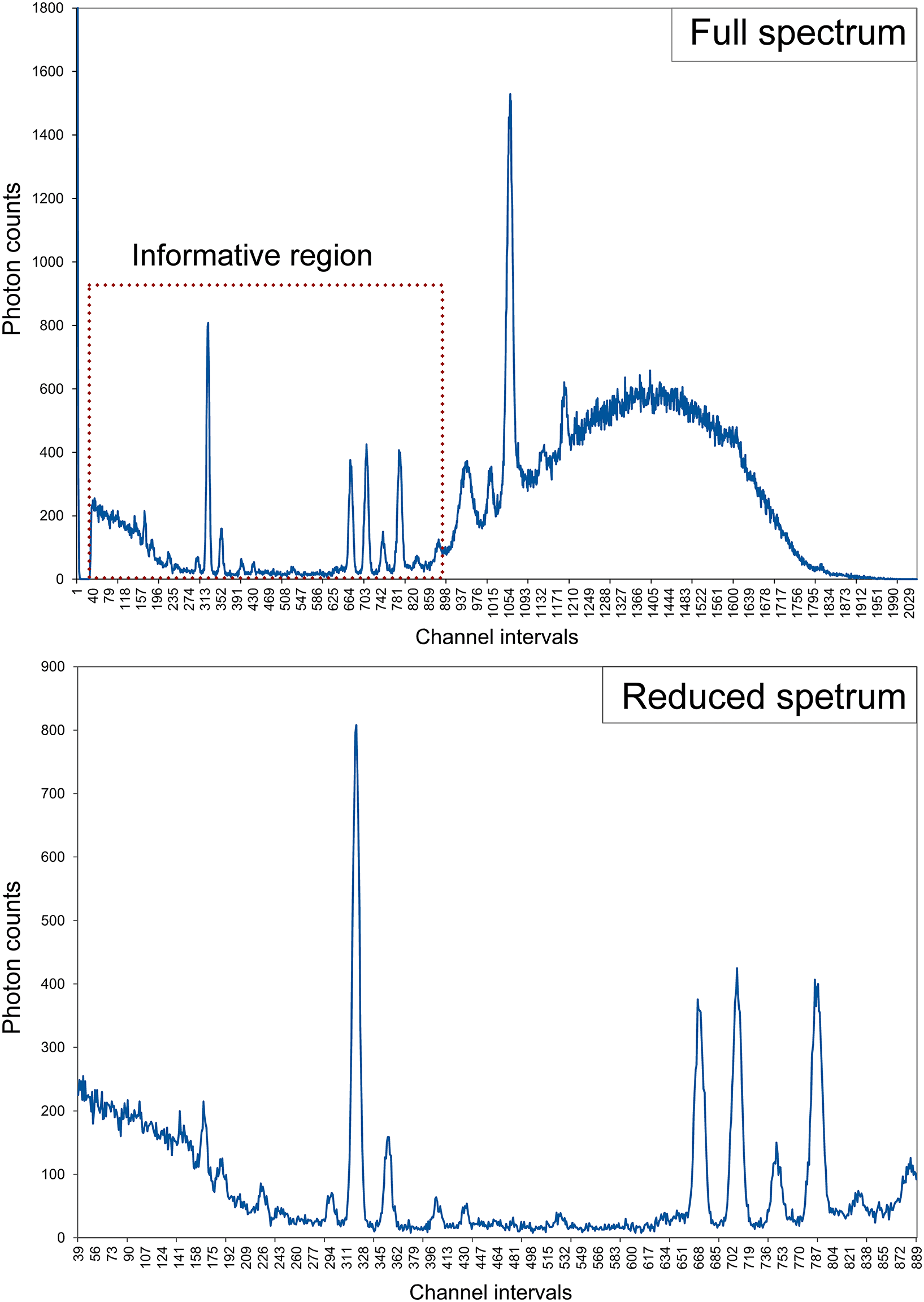
Figure 1 Comparison between the full spectrum of an obsidian sample (top) and a reduced spectrum containing only the informational region (bottom). Color version available at www.cambridge.org/argote_machine-learning
Using the numerical values obtained from the photon counts in each energy interval or channel, two n x p matrices were constructed (where n refers to the samples and p to the channel count interval):
1. An n = 149 x p = 862 matrix for the obsidians (available in the supplementary material as file ‘Obsidian_Sources_38_900.csv’).
2. An n = 185 x p = 791 matrix for the pottery fragments (available in the supplementary material as file ‘NaranjaTH_YAcim40_830.csv’).
The spectral intensities (photon counts) sampled at the given intervals (channels) represent the quantitative data employed in the statistical analysis instead of the major and trace element concentrations traditionally used for this purpose. Both datasets were preprocessed the same way. First, the EMSC algorithm, to filter the dispersion effects, and the smoothing procedure with the Savitzky‒Golay algorithm were applied. With this methodology, it is not necessary to standardize each variable before using the model-based clustering since applying the EMSC filter to the data are equivalent to normalizing it. If your own spectra show any peak displacement, you should apply at this point the CluPa algorithm for peak alignment. Before developing the classification model, the atypical values were detected, removing from the matrix the samples that recorded high values in their orthogonal and score distances with the ROBPCA algorithm. It is important to note that the clustering model presupposes that all the variable-wise centers equal zero.
2.2 Exercise 1: Obsidian Samples
As mentioned in Section 2.1, the control set consisted of 149 obsidian samples of known origin and p = 862 energy intervals (channels). These obsidian samples were collected from 12 Central Mexico (Figure 2). The number of samples analyzed from each source is specified in Table 2. A full description of the geological setting of the obsidian sources can be found in Reference Argote-Espino, Solé, Sterpone and López GarcíaArgote Espino et al. (2010), Reference Argote-Espino, Solé, López-García and SterponeArgote-Espino et al. (2012), Reference CobeanCobean (2002), and Reference López-García, Argote and BeirnaertLópez-García et al. (2019). These samples served as a controlled experiment for assessing the efficiency of the proposed method. The procedure is described step by step in the supplementary video “Video 1.”

Figure 2 Geographical location of the obsidian deposits. The numbers correspond to the following sources: (1) El Chayal, (2) Ixtepeque, and (3) San Martín Jilotepeque in Guatemala; (4) La Esperanza in Honduras; and (5–6) Otumba volcanic complex, Edo. México; (7) Ahuisculco, Jalisco; (8) El Paredón, Puebla; (9) El Pizarrín-Tulancingo, Hidalgo; (10) Sierra de Pachuca, Hidalgo; (11) Zacualtipán, Hidalgo; (12) Zinapécuaro, Michoacán. Color version available at www.cambridge.org/argote_machine-learning
Table 2 Number of obsidian samples per location.
| Source name | Sample ID | n | |
|---|---|---|---|
| 1 | El Chayal (Guatemala) | 1−17 | 17 |
| 2 | La Esperanza (Honduras) | 18−33 | 16 |
| 3 | Ixtepeque (Guatemala) | 34−50 | 17 |
| 4 | San Martin Jilotepeque (Guatemala) | 51−67 | 17 |
| 5 | Ahuisculco (Jalisco) | 68−76 | 9 |
| 6 | Otumba (Soltepec) | 77−86 | 10 |
| 7 | Otumba (Ixtepec-Pacheco-Malpaís) | 87−110 | 24 |
| 8 | El Paredon (Puebla) | 111−117 | 7 |
| 9 | El Pizarrin-Tulancingo (Hidalgo) | 118−122 | 5 |
| 10 | Sierra de Pachuca (Hidalgo) | 123−132 | 10 |
| 11 | Zacualtipán (Hidalgo) | 133−142 | 10 |
| 12 | Zinapecuaro (Michoacán) | 143−149 | 7 |
| Total: | 149 |
Step-by-step video on how to process spectral data of obsidian samples used in Video 1. Video files available at www.cambridge.org/argote_machine-learning
The reduced obsidian sample spectra were filtered using a combination of the EMSC + Savitzky-Golay filters; the script to perform this task is presented below.
## Script to filter with the EMSC algorithm version 0.9.2 (Reference Liland and IndahlLiland and Indahl, 2020) rm(list = ls()) library(EMSC) #Package EMSC (Performs model-based background correction and # normalisation of the spectra) dat <- read.csv(”Obsidian_source_38_900.csv”, header=T) # To call the spectral data file str(dat) # to see the data structure dat1 <- dat[2:863] # To eliminate the first column related to the sample identifier str(dat1) Obsidian.poly6 <- EMSC(dat1, degree = 6) #Filters the spectra with a 6th order #polynomial str(Obsidian.poly6) write.csv(Obsidian.poly6$corrected, file=”Obsidian_EMSC.csv”) # to save the data file # filtered with the EMSC. The User can choose other file names #To filter the spectra with the SG filter, use the ‘prospectr’ package (Stevens and #Reference Stevens and Ramirez-LopezRamirez–Lopez (2015) library(prospectr)## Miscellaneous functions for processing and sample selection of ## spectroscopic data (Reference Stevens, Ramirez-Lopez and HansStevens et al., 2022) dat2 <- read.csv(”Obsidian_EMSC.csv”, header=T) # Calls the file with EMSC filtered #data str(dat2) sg <- savitzkyGolay(Obsidian.poly6$corrected, p = 3, w = 11, m = 0) write.csv(sg, file=”Obsidian_EMSC_SG.csv”) # The user can choose another file name

Figure 3 shows the result of the filtered spectra compared to the untransformed raw data. Notice that the information was not altered. It is important to note that because we determined a polynomial of the sixth order for the SG filter, the initial matrix with p = 862 was reduced to p = 852, eliminating five channel intervals from each extreme of the data matrix.

Figure 3 Raw data (above) and EMSC + Savitzky‒Golay filtered spectra (below). Color version available at www.cambridge.org/argote_machine-learning
Because the original spectra were not shifted or the intensity peaks were misaligned, it was not necessary to apply the CluPa algorithm. Nevertheless, if anyone finds it necessary, the peaks can be aligned with the following script (published by Reference López-García, Argote and BeirnaertLópez-García et al., 2019):
# Run the whole script at one time devtools::install_github(”Beirnaert/speaq”) # download latest “speaq” package once! library(speaq) # Change file folder your_file_path = “/Users/” # Get the data (spectra in matrix format) matrix3 = read.csv2(file.path(your_file_path, “your_file.csv”), header = F, sep = “,”,colClasses = “numeric”, dec = ”.”) spectra.matrix = as.matrix(matrix3) index.vector = seq(1, ncol(spectra.matrix)) # Plot the spectra

speaq::drawSpec(X = spectra.matrix, main = ‘mexico’, xlab = “index”) # Peak detection

peaks <- speaq::getWaveletPeaks(Y.spec = spectra.matrix, X.ppm = index.vector, nCPU = 1, raw_peakheight = TRUE) # Grouping groups<- PeakGrouper(Y.peaks = peaks) # If the peaks are well formed and the peak detection threshold is set low, the filling step #is not necessary and can be omitted peakfill <- PeakFilling(groups_rawIntensity, spectra.matrix, max.index.shift = 5, window.width = “small”, nCPU = 1) Features <- BuildFeatureMatrix(Y.data = peakfill, var = “peakValue”, impute = “zero”, delete.below.threshold = FALSE, baselineThresh = 1, snrThres = 0)

# Aligning the raw spectra
peakList = list()
for(s in 1:length(unique(peaks$Sample))){
peakList[[s]] = peaks$peakIndex[peaks$Sample == s]
}
resFindRef<- findRef(peakList);
refInd <- resFindRef$refInd; Aligned.spectra <- dohCluster(spectra.matrix, peakList = peakList, refInd = refInd, maxShift = 5, acceptLostPeak = TRUE, verbose=TRUE); drawSpec(Aligned.spectra) write.csv (Aligned.spectra, file =” aligned.csv”)

After preprocessing the spectra, it is important to diagnose the data and detect outliers. For this task, use the following script extracted from the “rrcov” package (Reference TodorovTodorov, 2020):
## Script to diagnose outliers rm(list=ls()) library(rrcov) dat <- read.csv(File_X, header=T) str(dat) pca <- PcaHubert(dat, alpha = 0.90, mcd = FALSE, scale = FALSE) pca print(pca, print.x=TRUE) plot(pca) summary(pca)

The results can be observed in Figure 4. Reference Hubert, Rousseeuw and Vanden BrandenHubert et al. (2005) define this figure as a diagnostic plot based on the ROBPCA algorithm; it allows distinguishing regular observations and different types of outliers under the assumption that the relevant information is stored in the variance of the data (Reference López-García, Argote and ThrunLópez-García et al., 2020; Reference Thrun, Märte and StierThrun et al., 2023). In Figure 4, a group of orthogonal outliers (located at the top left quadrant of the graphic) that correspond to El Pizarrin-Tulancingo and Zinapecuaro source samples can be discriminated. According to this figure, there are ten observations with distances beyond the threshold of X2 that can be considered bad leverage points or outliers; the rest are regular observations. The bad leverage points correspond to the samples from Sierra de Pachuca, which have contrasting higher chemical concentrations of Zr, Zn, and Fe than the rest of the sources (Reference Argote-Espino, Solé, Sterpone and López GarcíaArgote-Espino et al., 2010). Therefore, they cannot be considered properly as outliers, but observations with a different behavior should not be deleted.

Figure 4 Diagnostic plot of the obsidian samples based on the ROBPCA algorithm. Color version available at www.cambridge.org/argote_machine-learning
Once the earlier steps were concluded, we can now classify the samples. In the Bayesian paradigm, the allocation of the samples in a cluster is regarded as a statistical parameter (Reference Partovi Nia and DavisonPartovi Nia and Davison, 2012). In general, it is better to work with the Gaussian distribution and set the default values of the hyperparameters given by the “bclust” algorithm. For this step, use the following script for R:
## bclust algorithm rm(list = ls()) library(bclust) # Reference Partovi Nia and DavisonPartovi Nia and. Davison (2015) datx <- read.csv(File_X, header=T) str(datx) dat2x <- as.matrix(datx) str(dat2x) Obsidian.bclust<-bclust(x=dat2x, transformed.par=c(-1.84,-0.99,1.63,0.08,-0.16,-1.68)) par(mfrow=c(2,1)) plot(as.dendrogram(Obsidian.bclust)) abline(h=Obsidian.bclust$cut) plot(Obsidian.bclust$clust.number,Obsidian.bclust$logposterior, xlab=”Number of clusters”,ylab=”Log posterior”,type=”b”) abline(h=max(Obsidian.bclust$logposterior)) str(Obsidian.bclust) Obsidian.bclust$optim.alloc # optimal partition of the sample Obsidian.bclust$order # produces teeth plot useful for demonstrating a grouping on clustered subjects Obsidian.bclust<-bclust(dat2x, transformed.par=c(-1.84,-0.99,1.63,0.08,-0.16,-1.68)) dptplot(Obsidian.bclust,scale=10,varimp=imp(Obsidian.bclust)$var, horizbar.plot=TRUE,plot.width=5,horizbar.size=0.2,ylab.mar=4) #unreplicated clustering wildtype<-rep(1,55) #initiate a vector wildtype[c(1:3,48:51,40:43)]<-2 #associate 2 to wildtypes dptplot(Obsidian.bclust,scale=10,varimp=imp(Obsidian.bclust)$var, horizbar.plot=TRUE,plot.width=5,horizbar.size=0.2,vertbar=wildtype, vertbar.col=c(”white”,”violet”),ylab.mar=4)

The result is displayed in the dendrogram of Figure 5, as well as in the partition of the sample space of Table 3. As expected, the Bayesian method clustered the obsidian samples into twelve groups (going from bottom to top in the dendrogram), all related to their geological sources: [1] Zacualtipan (ID. 133–142), which, according to the dendrogram, is subdivided into two subsources: [2] Zinapecuaro (ID. 143–149); [3] Ahuisculco (ID. 68–76); [4] El Paredon (ID. 111–117); [5] Otumba-Ixtepec, Pacheco, and Malpais (ID. 87–110); [6] Otumba-Soltepec (ID. 77–86); [7] El Pizarrin (ID.118–122); [8] Sierra de Pachuca (ID. 123–132), [9] El Chayal (ID. 1–17), which is also subdivided into two subsources: [10] La Esperanza, Honduras (ID.18–33); [11] San Martín Jilotepec (ID. 51–67), also subdivided into two subsources; and [12] Ixtepeque (ID. 34–50).

Figure 5 Dendrogram and optimal grouping found by the Gaussian model for the obsidian source data. The dendrogram visualizes the ultrametric portion of the selected distance (Reference MurtaghMurtagh, 2004). The method proposed twelve groups. Color version available at www.cambridge.org/argote_machine-learning
Table 3 Partition of the sample space

The Bayes factor (B10) can be regarded as a measure of the importance that each variable holds in the classification. To determine which oligo-elements are important, the algorithm provides a list of the potentially important variables that contribute to the grouping. The following script is used for this purpose:
# This function plots variable importance using a barplot Obsidian.bclust<-bclust(dat2x, transformed.par=c(-1.84,-0.99,1.63,0.08,-0.16,-1.68), var.select=TRUE) Obsidian.imp<-imp(Obsidian.bclust) #plots the variable importance par(mfrow=c(1,1)) #retrieve graphic defaults mycolor<-Obsidian.imp$var mycolor<-c() mycolor[Obsidian.imp$var>0]<-”black” mycolor[Obsidian.imp$var<=0]<-”white” viplot(var=Obsidian.imp$var,xlab=Obsidian.imp$labels,col=mycolor)

#plots important variables in black viplot(var=Obsidian.imp$var,xlab=Obsidian.imp$labels, sort=TRUE,col=heat.colors(length(Obsidian.imp$var)), xlab.mar=10,ylab.mar=4) mtext(1, text = “Obsidian”, line = 7,cex=1.5)# add X axis label mtext(2, text = “Log Bayes Factor”, line = 3,cex=1.2) # adds Y axis labels #Sorts the importance and uses heat colors; adds some labels to the X and Y axes str(Obsidian.imp) Obsidian.imp$var Obsidian.imp$order

The relevant variables can be separated from the nonrelevant variables by looking for the inflection point in the Gaussian variable selection model, such as the one observed in Figure 6, that is, the point in the distribution curve where the factor value of the variables stabilizes or stays more constant. For this case, the inflection point is presented at Log B10> 1.57E+07; therefore, values of B10 > 1.57E+07 are considered relevant variables. The higher Bayes factors correspond to the positions of Zr, Nb, and Sr peaks (Figure 7), indicating the relative importance of these elements in the classification task. The rest of the elements had negligible and negative Bayes factors (B10 <1.57E+07) and hence were irrelevant.

Figure 6 Log Bayes factor of variables (logB10) for the Gaussian variable selection model of the obsidian data. Color version available at www.cambridge.org/argote_machine-learning

Figure 7 XRF spectrum of an obsidian sample. Color version available at www.cambridge.org/argote_machine-learning
Although discriminating groups of samples with a similar spectral profile is not a simple task, the results obtained with this clustering algorithm leave no doubt of its accuracy. First, the structure of the dendrogram was clear, and atypical values were not observed. Second, it is possible to observe that the samples are not mixed with other groups and that each group remains characterized by its place of origin. Third, the number of groups calculated by the algorithm is correct, corresponding to the number of geological sources introduced. This allows us to conclude that each geological source has its own spectral signature, which is different from those of the other sources. It should be noted that to accurately identify the groups, it is necessary to always refer to the observations that serve as control samples (e.g., known sources).
2.3 Exercise 2: Thin Orange Pottery Samples
Thin orange ware, as its name says, is a light orange ceramic with very thin walls that became one of the main interchange products of the Classic period in Central Mexico. Its distribution over a large expanse of Mesoamerica has been considered to be closely related to the strong cultural dominion of Teotihuacan. Its wide geographical circulation has been documented in many places far from Teotihuacan (Reference Kolb and SandersKolb, 1973; Reference López Luján, Neff, Sugiyama, Carrasco, Jones and SessionsLópez Luján et al., 2000; Reference RattrayRattray, 1979), including Western Mexico, Oaxaca, and the Mayan Highlands (i.e., Kaminaljuyú, Tikal, and Copán). The use of this ware type had a broad extension over time, starting in the Tzacualli phase (ca. 50–150 AD), peaking at the Late Tlamimilolpa and Early Xolalpan phases (350–550 AD), and declining at approximately 700 AD (Reference Kolb and SandersKolb, 1973; Reference MüllerMüller, 1978).
According to Reference RattrayRattray (2001), the suggested chronology of the different ceramic forms of the Thin orange ware is the following. In the Tzacualli–Miccaotli phase (ca. 1–200 AD), some of the common forms are vessels with composite silhouettes, everted rims and rounded bases, vases with straight walls, pedestal base vessels, pots and a few miniatures; simple incised lines and red color decorations are present in some pieces. Some sherds found in archaeological contexts from the Miccaotli phase suggest that hemispherical forms were present. Nevertheless, the hemispherical bowls with ring bases, the most representative form in Thin orange, occur in the Early Tlamililolpan phase (ca. 200–300 AD) and continue until the end of the Metepec phase (ca. 650 AD).
Archaeological and petrographic studies performed in the 1930s (Reference LinnéLinné, 2003) found that the components of Thin orange pastes were homogeneous and of a nonvolcanic origin. Therefore, if Teotihuacan city was settled within a volcanic region, then the production center (or at least the raw material source) should be somewhere else. These findings opened the discussion about why the most distinctive ware of Teotihuacan culture was not produced there. In the 1950s, Cook de Leonard proposed that the natural clay deposits were located south of the state of Puebla based on the material excavated from some tombs in an archaeological site near Ixcaquixtla (Reference Brambila, Puche and NavarreteBrambila, 1988; Reference Cook de LeonardCook de Leonard, 1953).
Rattray and Harbottle performed neutron activation and petrographic analyses on samples classified as fine Thin orange ware and a coarse version of this ware called San Martin orange (or Tlajinga), the last one locally produced in Acatlán de Osorio, south of Puebla state (Reference Rattray, Harbottle and NeffRattray and Harbottle, 1992). In their conclusions, they proposed that the clay deposits and the production centers of Thin orange pottery were in the region of Río Carnero, 8 km south of Tepeji de Rodriguez town, south of Puebla state. Summarizing several former investigations about the compositional pattern of Thin orange ware, the following groups have been established:
1. A main ‘Core’ group, with clay and temper of homogeneous characteristics. Reference Rattray, Harbottle and NeffRattray and Harbottle (1992) and Reference López Luján, Neff, Sugiyama, Carrasco, Jones and SessionsLópez Luján et al. (2000) mentioned that its chemical profile is characterized mainly by high concentrations of Rb, Cs, Th, and K. This group was acknowledged as “Core Thin orange” by Reference AbascalAbascal (1974), “Thin Orange” by Reference Shepard, Kidder, Jenning and ShookShepard (1946), “group Alfa” by Reference Kolb and SandersKolb (1973), and “group A” by Reference Sotomayor and CastilloSotomayor and Castillo (1963).
2. A coarser second group, used for utilitarian purposes (domestic ware), is characterized by having different percentages of the minerals present in the first group. This group corresponds to the “group Beta” (Reference Kolb and SandersKolb, 1973) and the “Coarse Thin orange” group (Reference AbascalAbascal, 1974). Reference Rattray, Harbottle and NeffRattray and Harbottle (1992) assume that this group is formed by local imitations of the original Thin orange ware.
In this exercise, the purpose of this application is to determine possible differences in the manufacturing techniques of the Thin Orange pottery, providing a better understanding of the underlying production processes. The focus is on identifying natural groups with homogeneous chemical compositions within the data, leading to the determination of whether this ceramic type was crafted following a unique recipe (clay and temper) or if there were several ways to produce it. By comparing our results with those obtained by other researchers on the conformation of a single ‘core’ group (Reference AbascalAbascal, 1974; Reference Harbottle, Sayre and AbascalHarbottle et al., 1976; Reference Rattray, Harbottle and NeffRattray and Harbottle, 1992; Reference Shepard, Kidder, Jenning and ShookShepard, 1946), new evidence could be provided that might help refine the current classification of this significant ware.
The procedure is shown step by step in the supplementary video “Video 2”. The archaeological pottery set consisted of 176 ceramic fragments and 9 clay samples (extracted from a natural deposit near the Rio Carnero area). Both sets of materials were analyzed with a portable X-ray fluorescence spectrometer. To conduct the comparative analysis with adequate variability, it was necessary to collect several samples of the same ceramic type from different locations and contexts. Therefore, the pottery samples were provided by different research projects that performed systematic excavations at various Central Mexico archaeological sites (Figure 8; Table 4).
Step-by-step video on how to process spectral data of ceramic samples used in Video 2. Video files available at www.cambridge.org/argote_machine-learning

Figure 8 Geographical location of the archaeological sites from which the Thin orange pottery samples were collected. Color version available at www.cambridge.org/argote_machine-learning
Table 4 Number of Thin orange pottery samples and locations
| Archaeological site | Mexican state | N |
|---|---|---|
| Teteles de Santo Nombre | Puebla | 23 |
| Izote, Mimiahuapan, Mapache, and other sites | Puebla | 22 |
| Huejotzingo | Puebla | 6 |
| Xalasco | Tlaxcala | 53 |
| Teotihuacan | Mexico | 72 |
| Clay source | Puebla | 9 |
| Total: | 185 |
Pottery is a very heterogeneous material; therefore, the ceramic samples were treated differently. Following the recommendations of Reference Hunt and SpeakmanHunt and Speakman (2015), the ceramic samples were prepared as pressed powder pellets. From each pottery sherd, a fragment weighing approximately 2 g was cut. The external surfaces of each fragment were abraded with a tungsten carbide handheld drill, reducing the possibility of contamination from depositional processes. The residual dust was removed with pressurized air, and the fragments were pulverized in an agate mortar. After grinding and homogenizing, the powder was compacted into a 2-cm diameter pellet by a cylindrical steel plunger with a manually operated hydraulic press. No binding agent was added. These pellets provided samples that were more homogeneous and with a uniformly flat analytical surface.
Figure 9 displays an example XRF spectrum of one Thin orange pottery sample. The main elements in the ceramic matrix are iron (Fe), followed by calcium (Ca), potassium (K), silicon (Si), and titanium (Ti); aluminum (Al), manganese (Mn), nickel (Ni), rubidium (Rb), and strontium (Sr) are also present at lower intensities. Sulfur (S), chromium (Cr), copper (Cu), and zinc (Zn) can be considered trace elements.

Figure 9 XRF spectrum of a representative sample of Thin orange pottery. Color version available at www.cambridge.org/argote_machine-learning
In this case, the matrix has many more variables than observations (n ≪ p), with p = 2,048; thus, much of the information contained was irrelevant for clustering. As mentioned at the beginning of this section, it was decided to manually cut some readings as they contained values close to zero or corresponded to undesirable effects, such as light elements below detection limits, the Compton peak, Raleigh scattering and palladium and rhodium peaks (produced by the instrument). The cuts were made at the beginning (from channel 1 to 39) and end (from channel 831 to 2,048) of the spectrum, retaining the elemental information corresponding to the analytes between channels 40 and 830, related to the energy range of 0.78 to 16.21 keV of the detector resolution. In this way, only p = 791 channel intervals were kept. This matrix can be found in the supplementary material as file ‘NaranjaTH_YAcim40_830’. It should be noted that because there were no displacements in the spectra, it was not necessary to use the peak alignment algorithm.
The next step was to filter the spectra as in the previous study case, using only the EMSC, as the spectrum did not contain scattering effects. For this purpose, use the following script:
## Script to filter with the EMSC algorithm rm(list = ls()) library(EMSC) #Package EMSC (Performs model-based background correction and # normalization of the spectra) dat <- read.csv(“C:\\NaranjaTH_YAcim40_830.csv”, header=T) # Calls the spectral data #file str(dat) # to see the structure of the data dat1 <- dat[2:792] # Eliminates the first column related to the sample identifier str(dat1) pottery.poly6 <- EMSC(dat1, degree = 6) #Filter spectra with a 6th-order polynomial str(pottery.poly6) write.csv(pottery.poly6$corrected, file=”pottery_EMSC.csv”) # to save the data file # filtered with the EMSC

Once filtering was performed, the diagnosis of outliers was performed with the following script:
## Script to diagnose outliers (Reference TodorovTodorov, 2020) rm(list=ls()) library(rrcov) dat <- read.csv(File_X, header=T) str(dat) pca <- PcaHubert(dat, alpha = 0.90, mcd = FALSE, scale = FALSE) pca print(pca, print.x=TRUE) plot(pca) summary(pca)

In the outlier detection with the ROBCA algorithm (Figure 10), most of the ceramic sample data vectors have regular patterns with normal punctuation and orthogonal distances. We can distinguish seventeen orthogonal outliers in the upper left quadrant of the graph, one observation with an extreme orthogonal distance (observation no. 126), and a small group of ten bad leverage points in the upper right quadrant. The robust PCA high-breakdown method treats this last group as one set of outliers. An interesting fact about the set of detected bad leverage points is that nine of them correspond to clay deposits (cases no. 177 to 185); only one sample (case no. 115) is related to an archaeological site in Puebla that shows a low amount of manganese content. Therefore, instead of just being measurement errors, outliers can also be seen as data points that have a different origin from regular observations, such as the case of the pure clay samples. According to this, no observations were removed from the analysis.

Figure 10 Outlier map of the Thin Orange pottery dataset computed with ROBPCA based on five principal components. Color version available at www.cambridge.org/argote_machine-learning
For the Bayesian clustering, the model parameters were set the same way as in the obsidian case. The algorithm provides a list of the potentially important variables that contribute to the clustering. In this case, the variable selection extension of the Gaussian model (Figure 11) selected 22 of the 791 initial variables as the most important ones. These twenty-two variables (channels) corresponded to the energy ranges of Fe (6.3 to 6.55 and 7 to 7.11 KeV) and Ca (3.7 to 3.75 KeV) chemical elements. Calcium and iron oxides (such as hematite) are two components that are commonly found in pottery and mudrock composition at variable concentrations depending on the parental material (Reference Callaghan, Pierce, Kovacevich and GlascockCallaghan et al., 2017; Reference Minc, Sherman and ElsonMinc et al., 2016; Reference Ruvalcaba-Sil, Ontalba Salamanca and ManzanillaRuvalcaba et al, 1999; Reference StonerStoner, 2016). These results are different from Reference Rattray, Harbottle and NeffRattray and Harbottle (1992) analysis in which the pottery was mainly determined by high concentrations of Rb, Cs, Th and K. On the other hand, Reference Kolb and SandersKolb (1973) found that Fe and Ti were important elements present in his Alpha and Beta groups.

Figure 11 Log Bayes factor (logB10) for the Gaussian variable selection model of Thin orange data. The Bayes factors are computed for the optimal grouping found by agglomerative clustering using the Gaussian model. Color version available at www.cambridge.org/argote_machine-learning
The resulting dendrogram (Figure 12) grouped the data into two main clusters (I and II) subdivided into six (1 to 6) and two subgroups (7 and 8), respectively. Subgroups 7 and 8 showed chemical differences that distinctively separated them from the rest of the groups. The number of samples (n) assigned to each subgroup were as follows: Group 1 = 23, Group 2 = 51, Group 3 = 16, Group 4 = 36, Group 5 = 13, Group 6 = 12, Group 7 = 18, and Group 8 = 16. Samples from group 1 come mostly from the archaeological site of Xalasco. Samples from Groups 2, 3, 6, 7, and 8 come from Teotihuacan, Xalasco, and several Puebla sites. Group 4 contains samples from some sites in Puebla State and the northeastern sector of Teotihuacan city. Group 5 has samples mainly from Teteles del Santo Nombre and a few from Xalasco and Teotihuacan. Groups 4 and 6 contain the clay samples from the Rio Carnero area. Table 5 summarizes the ceramic shapes included in each group, showing a great variability of forms in each group.

Figure 12 Dendrogram and optimal grouping found by the Gaussian model for the Thin Orange samples. The horizontal bar at the bottom refers to the optimal grouping found by the Gaussian model. Color version available at www.cambridge.org/argote_machine-learning
Table 5 Number of samples (n) and ceramic forms included in each subgroup:
| Group 1 | Group 2 | Group 3 | Group 4 |
|---|---|---|---|
| (n = 23) | (n = 51) | (n = 16) | (n = 36) |
| Hemispherical bowl with ring base (13 sherds) | Hemispherical bowl with ring base (12 sherds) | Bowl with ring base (4 sherds) | Bowl with ring base (11 sherds) |
| Vase with incised exterior decoration (1 sherd) | Hemispherical bowl (6 sherds) | Cylindrical vase (4 sherds) | Hemispherical bowl (3 sherds) |
| Tripod vase with nubbin supports ( 1 sherd) | Vessel with convex wall (4 sherds) | Jar with incised exterior decoration (2 sherds) | Cylindrical vase (4 sherds) |
| Jar with incurved rim and incised simple double-line (1 sherd) | Cylindrical vase (5 sherds) | Tripod vase with nubbin supports ( 1 sherd) | Incense burner (1 sherd) |
| Jar with incised exterior decoration (1 sherd) | Tripod vase with nubbin supports ( 2 sherds) | Undetermined shape (5 sherds) | Basin (1 sherd) |
| Pot (1 sherd) | Vase with small appliqués (1 sherd) | Vessel with pedestal base (1 sherd) | |
| Undetermined shape (5 sherds) | Tripod vessel with deep parallel grooves (1 sherd) | Vessel with incised exterior decoration (1 sherd) | |
| Vessel with pedestal base (1 sherd) | Tzacualli phase Pot (1 sherd) | ||
| Miniature vessel (1 sherd) | Undetermined shape (12 sherds) | ||
| Undetermined shape (18 sherds) | Natural clay deposit (1 sample) | ||
| Group 5 | Group 6 | Group 7 | Group 8 |
| (n = 13 ) | (n = 12 ) | (n = 18 ) | (n = 16 ) |
| Bowl with ring base (4 sherds) | Hemispherical bowl with ring base (3 sherds) | Hemispherical bowl with ring base (7 sherds) | Hemispherical bowl with ring base (5 sherds) |
| Vessel with pedestal base (1 sherd) | Jar with incised exterior decoration (1 sherd) | Hemispherical vessel (2 sherds) | Annular-based hemispherical bowls and incised exterior decoration (1 sherd) |
| Vessel with incised exterior decoration (1 sherd) | Natural clay deposit (8 samples) | Vessel with recurved composite wall (1 sherd) | Tripod vessel with everted rim (1 sherd) |
| Cylindrical vase (3 sherds) | Vessel with exterior punctate decoration (1 sherd) | Vessel with red pigment and incised exterior decoration (1 sherd) | |
| Vessel with convex wall and flat-convex base (1 sherd) | Pot (1 sherd) | Jar with incised exterior decoration (1 sherd) | |
| Undetermined shape (3 sherds) | Undetermined shape (6 sherds) |
The results obtained in this spectral analysis revealed the existence of two large groups subdivided into several subgroups that exhibit a certain degree of chemical differentiation, indicating that different raw materials were used to produce the Thin Orange ware. Pottery is produced by mixing clays and aplastic particles or temper, with the clay predominating over the temper. In this case, the clay deposit samples were classified inside the main group I (in subgroups 1 and 6); thus, it can be assumed that, for manufacturing the pieces related to this main group, clay banks from the same geological region were used. The Rio Carnero region is shaped by a set of deep barrancas (canyons) predominantly from the Acatlán Complex, the geological vestige of a Paleozoic ocean formed between the Cambro-Ordovician and late Permian periods (Reference Nance, Mille, Keppie, Murphy and DostalNance et al., 2006). This region contains banks of schists rich in hematite located in the ravines of Barranca Tecomaxuchitl and Rio Axamilpa. On the other hand, the chemical differences of main group II (with only 34 samples) indicate the extraction of clay from a different region.
According to the results, an interpretation can be as follows. The division of the samples into two main groups seems to be associated with two different clay deposit regions from which the raw material was exploited. The internal differences in their chemical composition, probably related to differences in temper, influenced the clustering algorithm to classify them into separate subgroups. This could mean that the aplastic particles used in the mixture for manufacturing the ceramic pieces did not naturally occur in the clay and were added by the artisans. The last observation is consistent with Reference Kolb and SandersKolb (1973), who stated that the temper was deliberately added and is not found in situ in the natural clay deposit.
The considerable variety of patterns presented by each of the eight subgroups suggests that the recipe for manufacturing the pieces was not used uniformly and that multiple ceramic production centers existed, employing their own and specific production recipes. In other words, each center would have produced its version of the Thin orange pottery with a standardized composition, and this was different to some extent from the ceramic made in other workshop centers. The results also support the idea that there was compositional continuity through time, despite the different shapes of the analyzed pieces.
3 Processing Compositional Data
3.1 Applications and Case Studies
In this section, data from published case studies were used to illustrate the techniques for processing compositional data. In summary, the steps for handling all datasets are as follows. First, the data are rescaled in such a way that the sum of the elemental concentrations row is equal to 100 percent. Afterwards, the data are transformed to log-ratios using the ilr transformation, translating the geometry of the Simplex into a real multivariate space. Once the ilr coordinates are obtained, data are standardized using a robust min/max-standardization. Any observation with a value equal to zero is imputed. As a diagnosis method, an MCD estimator is applied to identify the presence of outliers in the data. Then, model-based clustering is employed for the classification and visualization of the data. Finally, an SBP is calculated and graphically represented using the CoDa-dendogram for understanding differences in the composition of the clusters.
3.2 Exercise 1: Unsupervised Classification of Central Mexico Obsidian Sources
In this case study, the compositional values of obsidian samples collected from different natural sources (Table 6) located in Central Mexico, published by Reference López-García, Argote and BeirnaertLopez-Garcia et al. (2019), and Guatemala, retrieved from Reference CarrCarr (2015), were processed. The procedure is described step by step in the supplementary video “Video 3”. The intention of this analysis is to demonstrate the performance of the model-based clustering and an unbiased visualization system in a controlled environment. The dataset was ideal for the analysis because the correct number of clusters and the cluster to which each of the observations belonged were known.
Table 6 Samples collected from Mesoamerican obsidian sources.Footnote a
| Source name | Geographic region | Sample ID no. | N | ||
|---|---|---|---|---|---|
| Ahuisculco | Jalisco | 1−9 | 9 | ||
| El Chayal | Guatemala | 10−25 | 17 | ||
| San Martin Jilotepeque | Guatemala | 26−39 | 14 | ||
| Ixtepeque | Guatemala | 40−55 | 16 | ||
| Otumba (Soltepec) | Edo. de México | 56−65 | 10 | ||
| Otumba (Ixtepec-Malpais) | Edo. de México | 66−88 | 23 | ||
| Oyameles | Puebla | 89−95 | 7 | ||
| Paredon | Puebla | 96−102 | 7 | ||
| Tulancingo-El Pizarrin | Hidalgo | 103−107 | 5 | ||
| Sierra de Pachuca | Hidalgo | 108−117 | 10 | ||
| Zacualtipan | Hidalgo | 118−127 | 10 | ||
| Zinapécuaro | Michoacán | 128−134 | 8 |
a The compositional values of samples from El Chayal, San Martin Jilotepeque, and Ixtepeque were retrieved from Reference CarrCarr (2015).
Step-by-step video on how to process compositional data of the obsidian samples used in Video 3. Video files available at www.cambridge.org/argote_machine-learning
The dataset consisted of n = 136 samples with p = 10 variables containing the elemental composition of the samples (Mn, Fe, Zn, Ga, Th, Rb, Sr, Y, Zr, and Nb), obtained with a portable X-ray fluorescence (pXRF) instrument. This matrix is available in the supplementary material as file “Mayas_sources_onc.csv”. In this example, we considered the problem of determining the structure of the data without prior knowledge of the group membership. The estimation of the parameters was performed with the maximum likelihood, and the best model was selected using the ICL criterion, resulting in K = 12 components. Compositional data are constrained data and therefore must be translated into the appropriate geometric space. To convert the data to completely compositional data, the following code from the “compositions” package in R is used (Reference van den Boogaart, Tolosana-Delgado and Brenvan den Boogaart et al., 2023):
rm(list=ls()) # Delete all objects in R session ## log-ratio analysis # load quantitative dataset. The name of the file for this example is Sources.csv. You can #change it for the location and name of your own data file. data <- read.csv(”C:\\obsidian\\Mayas_sources_onc.csv”, header=T) str(data) # displays the internal structure of the file, including the format of each column dat2 <- data[2:11] # delete data identification column str(dat2) # transformation of the data to the ilr log-ratio library(compositions) ## Reference van den Boogaart, Tolosana-Delgado and Brenvan den Boogaart, Tolosana-Delgado and Bren (2023) xxat1 <- acomp(dat2) # the function “acomp” representing one closed composition. # With this command, the dataset is now closed. xxat2 <- ilr(xxat1) # isometric log-ratio transformation of the data str(xxat2) write.csv(xxat2, file=”ilt-transformation.csv”) #You can choose a personalized file name

In the isometric transformation output file, the geometric space is D – 1. Once the data have been taken to the Simplex geometry, it is important to normalize them robustly so that there are no variables with greater weight. This is achieved by loading the clusterSim package in R with the normalization option = na3 using the code presented below:
# Robust normalization # In our case, the file “xxat2” that contains the isometric transformation of the #compositional data was normalized with the robust equation presented in the #transformations section. library(clusterSim) ## Reference Walesiak and DudekWalesiak and Dudek (2020) z11<- data.Normalization(xxat2,type=”n3a”,normalization=”column”,na.rm=FALSE) # This corresponds to the robust normalization described in ‘Compositional and #Completely compositional data’ Section of Volume I. # n3a positional unitization ((x-median)/min(x)- max(x)) z12 <- data.frame(z11) # After the previous operations, it is necessary to convert the #data output to a data frame to tell the program that the observations are in the #rows and columns represent the attributes (variables) str(z12)

After transforming the data through robust normalization, it is convenient to verify that there are no values equal to 0. In this example, no zero values were present; thus, no imputation was needed. In the case that your data have values equal to zero, it is recommended to employ an imputation algorithm such as Amelia II:
# Imputation of data # Loads the user interface to perform the imputation of values Library (Amelis) AmeliaView()

Afterwards, to identify the presence of outliers in the data, a diagnosis is performed through the MCD estimator of the rrcov package (Reference Todorov and FilzmoserTodorov and Filzmoser, 2009):
# Parameters of the model # kmax maximal number of principal components to compute. The default is kmax=10. # Default k= 0; if we do not provide an explicit number of components, the algorithm # chooses the optimal number. alpha: 0.7500; this parameter measures the fraction # of outliers the algorithm should resist (default). # The matrix dimension in this example is n = 136 and p = 10 library(rrcov) ## Reference TodorovTodorov (2020) MCD_1D <- data.matrix(z12[, 1:9]) cv <- CovClassic(MCD_1D) plot(cv) rcv <- CovMest(MCD_1D) plot(rcv) summary(MCD_1D)

Figure 13 shows the distance–distance plot, which displays the robust distances versus the classical Mahalanobis distances (Reference Rousseeuw and van ZomerenRousseeuw and van Zomeren, 1990), allowing us to classify the observations and to identify the potential outliers. Note that the choice of appropriate distance metric is essential (Reference ThrunThrun, 2021a). The dotted line represents points for which the classical and robust distances are equal. Vertical and horizontal lines are plotted in values x = y =
![]() ; points beyond this threshold are considered outliers. While the robust estimation detects many observations whose robust distance is above the threshold, the Mahalanobis Distance classifies all points as regular observations. Observations with large robust distances are not candidates for outliers because they do not have an impact on the estimates. The only two observations that exceed the values of X2 are those with ID numbers 9 and 136, which are convenient to eliminate from the estimate and leave only n = 134 samples in the dataset.
; points beyond this threshold are considered outliers. While the robust estimation detects many observations whose robust distance is above the threshold, the Mahalanobis Distance classifies all points as regular observations. Observations with large robust distances are not candidates for outliers because they do not have an impact on the estimates. The only two observations that exceed the values of X2 are those with ID numbers 9 and 136, which are convenient to eliminate from the estimate and leave only n = 134 samples in the dataset.

Figure 13 Distance–distance plot of the samples from obsidian sources. Color version available at www.cambridge.org/argote_machine-learning
Once the preprocessing of the data has concluded, the mixed model of multivariate Gaussian components is adjusted for clustering purposes using the Rmixmod and Clusvis packages:
# Classification with Mixture Modeling: Clustering in Gaussian case # To fit the mixture models to the data and for the classification, two additionaL #programs must be loaded. rm(list=ls()) library(Rmixmod) ## Reference Lebret, Lovleff and LangrognetLebret et al., 2015) library(ClusVis) ## (Reference Biernacki, Marbac and VandewalleBiernacki et al., 2021) fammodel <- mixmodGaussianModel(family=”general”,equal. proportions=FALSE) Mod1<-mixmodCluster(data,12, strategy = mixmodStrategy(algo = “EM”, nbTryInInit = 50, nbTry=25)) #EM (Expectation Maximization) algorithm # nbTryInInit: integer defining the number of tries in the initMethod algorithm. # nbTry: integer defining the number of tries summary(Mod1) Mod1[”partition”] # partition output made by mixing model ## Gaussian-Based Visualization of Gaussian and Non-Gaussian Model-Based Clustering library(ClusVis) ## (Reference Biernacki, Marbac and VandewalleBiernacki et al., 2021) resvisu <- clusvisMixmod(Mod1) # Gaussian-Based Visualization of Gaussian Model-Based #Clustering plotDensityClusVisu(resvisu) # probabilities of classification are generated by using the model #parameters.

In this script, the command mixmodGaussianModel is an object defining the list of models to run. The function mixmodCluster computes an optimal mixture model according to the criteria supplied and the list of models defined in [Model] using the algorithm specified in the command ‘strategy’ [strategy = mixmodStrategy (algo = “CEM”, nbTryInInit = 50, nbTry=25)] (Reference Lebret, Lovleff and LangrognetLebret et al., 2015). The estimation of the mixture parameters can be carried out with a maximum likelihood using the EM algorithm (Expectation Maximization), the SEM (Stochastic EM), or by maximum likelihood classification using the CEM algorithm (Clustering EM). In this example, we use the EM algorithm. With the general family command, it is possible to give more flexibility to the model that best fits the data by allowing the volumes, shapes, and orientations of the groups to vary (Reference Lebret, Lovleff and LangrognetLebret et al., 2015).
Because our groups had different proportions (each group had a different sample size), the FALSE command was established as Z12 in the data frame. As an output of this estimation step, the program provides a partition and other parameters, including the proportions of the mixed model in each group, their averages, variances and likelihood, and the associated source of each group obtained for this example. Table 7 shows the output of the algorithm; in this table, it can be observed that although the analysis was carried out without labeling the observations, they were correctly assigned to their corresponding source group, except for a single observation of the Otumba (Ixtepec-Mailpais) subsource that was assigned to Otumba (Soltepec).
Table 7 Assignation of the mixing proportions with Rmixmod (z-partition of the obsidian sources).
| Cluster | Proportion | Group assigned | n | Source |
|---|---|---|---|---|
| 1 | 0.1269 | 6,6,6,6,6,6,6,6 | 8 | Ahuisculco |
| 2 | 0.0746 | 1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1 | 17 | El Chayal |
| 3 | 0.0522 | 4,4,4,4,4,4,4,4,4,4,4,4,4,4 | 14 | San Martin Jilotepeque |
| 4 | 0.1045 | 12,12,12,12,12,12,12,12,12,12,12,12, 12,12,12,12 | 16 | Ixtepeque |
| 5 | 0.0522 | 8,8,8,8,8,8,8,8,8,8,8 | 11 | Otumba (Soltepec) |
| 6 | 0.0597 | 7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7 | 22 | Otumba (Ixtepec-Malpais) |
| 7 | 0.1642 | 5,5,5,5,5,5,5 | 7 | Oyameles |
| 8 | 0.0821 | 11,11,11,11,11,11,11 | 7 | Paredon |
| 9 | 0.0746 | 10,10,10,10,10 | 5 | Tulancingo-El Pizarrin |
| 10 | 0.0373 | 2,2,2,2,2,2,2,2,2,2 | 10 | Sierra de Pachuca |
| 11 | 0.0522 | 9,9,9,9,9,9,9,9,9,9 | 10 | Zacualtipan |
| 12 | 0.1194 | 3,3,3,3,3,3,3 | 7 | Zinapécuaro |
In this case, the model that best fitted the data turned out to be “Gaussian_pk_Lk_C” with the following cluster properties: Volume = Free, Shape = Equal, and Orientation = Equal. It is also possible to analyze the clustering results graphically. It is important to note that the graphics produced by the “ClusVis” package may vary in the output. The authors state that, for some specific reproducibility purposes, the Rmixmod package allows the random seed to be exactly controlled by providing the optional seed argument (“set.seed: number”). However, despite having performed multiple tests with different seeds, the resulting graphics tend to vary. Figure 14 displays the bivariate spherical Gaussian visualization associated with the confidence areas; the size of the gray areas around the centers reflects the size of the components. The accuracy of this representation is given by the difference between entropies and the percentage of inertia of the axes.

Figure 14 Component interpretation graph of the obsidian sources. Color version available at www.cambridge.org/argote_machine-learning
In the graph, it can be observed that the mapping of f is accurate because the difference between entropies is zero:
![]() . The first dimension provided by the LDA mapping was the most discriminative, with 55.7 percent of the discriminant power; the sum of the inertia of the first two axes was 55.7 + 23.22 = 78.92 percent of the discriminant power; thus, most of the discriminant information was present on this two-dimensional mapping. Components 12, 7, 4, and 2 contain most of the observations. The components that show the greatest difference in their chemical composition are the samples from Components 1 and 6 (El Chayal and Ahuisculco, respectively) and Components 2 and 10 (Pachuca and Pizarrin) in the other extreme. Components 5 and 11 (Oyameles and El Paredon) are the ones that are closest to each other (regarding their mean vectors) and slightly join without meaning that the observations are mixed, as seen in the partition results of Table 7. An important observation about these results is that there are no overlaps between any of the sources used in the classification, hence fulfilling all the conditions of a good classification.
. The first dimension provided by the LDA mapping was the most discriminative, with 55.7 percent of the discriminant power; the sum of the inertia of the first two axes was 55.7 + 23.22 = 78.92 percent of the discriminant power; thus, most of the discriminant information was present on this two-dimensional mapping. Components 12, 7, 4, and 2 contain most of the observations. The components that show the greatest difference in their chemical composition are the samples from Components 1 and 6 (El Chayal and Ahuisculco, respectively) and Components 2 and 10 (Pachuca and Pizarrin) in the other extreme. Components 5 and 11 (Oyameles and El Paredon) are the ones that are closest to each other (regarding their mean vectors) and slightly join without meaning that the observations are mixed, as seen in the partition results of Table 7. An important observation about these results is that there are no overlaps between any of the sources used in the classification, hence fulfilling all the conditions of a good classification.
3.3 Exercise 2: Obsidian Sources in Guatemala
Reference CarrCarr (2015) performed a study to identify obsidian sources and subsources in the Guatemala Valley and the surrounding region. In his project, he analyzed a total of 215 samples from El Chayal, San Martin Jilotepeque, and Ixtepeque geological deposits with pXRF spectrometry. Of these samples, n = 159 were collected from 36 different localities in El Chayal, n = 34 were gathered from eight sampling localities in San Martin Jilotepeque, and n = 22 were collected from four sampling locations in Ixtepeque. The data matrix for this example can be found in the supplementary file “Obs_maya.csv”. To discriminate between these three main obsidian sources, Reference CarrCarr (2015) used bivariate graphs, plotting the concentrations of Rb versus Fe (see the graphic reproduced from his data in Figure 15). In the figure, it is possible to discriminate three different groups, but it is difficult to distinguish between different subsources. In addition, there is a great dispersion of the points from the El Chayal and Ixtepeque sources.

Figure 15 Bivariate plot using Rb (ppm) and Fe (ppm) concentrations of El Chayal, San Martin Jilotepeque, and Ixtepeque obsidian source systems (graphic reproduced with data from Reference CarrCarr, 2015). Color version available at www.cambridge.org/argote_machine-learning
Reference CarrCarr (2015) also intended to examine the chemical variability of the samples to discriminate subsources within each of the main sources. For this purpose, the author analyzed the samples from each of the regions separately. For the El Chayal region, he used Rb versus Zr components in a bivariate display that resulted in five different subsources. Using this procedure, the author determined the existence of two distinct geochemical groups for the San Martin Jilotepeque obsidian source and two for Ixtepeque. To support the bivariate classification, Carr calculated the Mahalanobis distances (MD) to obtain the group membership probabilities of the observations, making use of the following set of variables: Sr/Zr, Rb/Zr, Y/Zr, Fe/Mn, Mn, Fe, Zn, Ga, Rb, Sr, Y, Zr, Nb, and Th. It should be noted that this was not possible for El Chayal Subgroups 4 and 5 because their sample size was too small, preventing the calculation of their probabilities.
From his study, several observations can be made. First, the data were processed without any transformation. Therefore, it is advisable to open the data to remove the constant sum constraint. Second, the author managed to establish a total of nine obsidian subsources using bivariate graphs for each of the regions separately. Third, the calculation of probabilities to determine group memberships using the MD was not possible in all cases due to the restrictions imposed by the sample size. Furthermore, if Reference CarrCarr (2015) data were processed according to one of the commonly established methodologies, that is, transforming the data to log10 and applying a PCA, the explained variance of the first two components would have been only 56.53 percent, and six PCs would have been needed to explain 95.58 percent of the variance.
By plotting all the data of the nine subsources together using a PCA (Figure 16), including the information about the origin of the samples, it can be observed that the overlap between the different groups is unavoidable. In this way, it can be concluded that this methodology is not able to differentiate the chemical characteristics of the samples. Therefore, the discrimination of sources using concentrations is a procedure that requires nonconventional methods. According to this, we can assume that many of the published classifications that follow preestablished methods have incurred serious classification errors.

Figure 16 Plot of the two principal components of the samples from nine subsources in Guatemala valley. Color version available at www.cambridge.org/argote_machine-learning
Employing the same data contained in Reference CarrCarr (2015), it was applied the methodology described in the associated section in Statistical Processing of Quantitative Data of Archaeological Materials www.cambridge.org/Argote. The entire procedure is described step by step in the supplementary video “Video 4”. First, as part of the preprocessing, the data were transformed to the isometric log-ratio and then robustly standardized. Afterward, and as an essential step, the diagnosis of the data was made with the robust MCD estimator (see the scripts for this part of the procedure in the previous exercise). Figure 17 shows the distance-distance plot in which eleven outliers were detected (92, 115, 142, 150, 166, 189, 190, 191, 192, 193, 204), displayed in the right half of the graph. In this case, it was decided not to exclude them from the analysis because part of these observations (190 to 193) corresponded to the samples identified by Reference CarrCarr (2015) as belonging to the Jilotepeque 2 subsource (FP01, SAI01, SAI03, SAI04).
Step-by-step video on how to compositional data of the obsidian samples used in Video 4. Video files available at www.cambridge.org/argote_machine-learning

Figure 17 Distance–Distance plot of the samples of obsidian sources. Color version available at www.cambridge.org/argote_machine-learning
Once the diagnosis was performed, unsupervised classification was carried out using the Gaussian mixture model with the Rmixmod and clusVis packages. The data matrix for this example consisted of n = 215 observations and D = 10 parts representing the Simplex. If the number of components is unknown and is to be estimated from the data, Rmixmod includes the parameter nbCluster to run a cluster analysis with a list of clusters (from 2 to n clusters). In this example, this parameter was set to “1” for the minimum number of components and “9” as the maximum number. Using the Gaussian mixture model, the Maximum likelihood inference was performed, and model selection was performed by the ICL criterion, detecting nine components. The output of the Rmixmod algorithm can be found in Table 8. The model that best fitted the data turned out to be Gaussian_pk_Lk_C, with clusters with the following properties: Volume = Free, Shape = Equal, and Orientation = Equal. The script used is the following:
Table 8 Output of the Rmixmod algorithm.
| ******************************************************* |
| * Number of samples = 215 |
| * Problem dimension = 9 |
| ******************************************************* |
| * Number of cluster = 9 |
| * Model Type = Gaussian_pk_Lk_C |
| * Criterion = ICL(−3126.1685) |
| * Parameters = list by cluster |
| truncated output |
| z-partition |
| 8 8 8 8 8 8 8 8 8 8 8 8 8 8 8 8 8 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 5 9 5 9 5 5 5 5 5 5 5 5 5 9 9 9 9 5 9 9 9 9 9 9 9 9 5 5 5 5 5 5 9 5 5 5 9 9 5 5 5 5 5 9 5 5 5 5 9 5 5 9 5 5 9 9 9 9 5 5 9 9 9 9 9 9 9 9 9 9 9 9 9 5 5 5 5 5 5 5 5 5 5 5 5 5 5 2 2 2 2 4 2 2 2 2 2 2 4 2 5 2 2 4 4 4 4 4 4 4 4 4 5 9 9 9 1 1 1 1 3 1 3 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 3 1 1 3 7 7 7 7 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 6 |
| ICL(−3117.1847) |
| Log-likelihood = 1963.1254 |
rm(list=ls()) library(Rmixmod) library(ClusVis) data <- read.csv(”C:\\Cobean\\Nueve_grupos\\robust_n-imp5. csv”,header=T) str(data) fammodel <- mixmodGaussianModel(family=”general”,equal.proportions=FALSE) Mod1<-mixmodCluster(data, 1:9, criterion = “ICL”, strategy = mixmodStrategy(algo = “CEM”, nbTryInInit = 50, nbTry=25)) summary(Mod1) Mod1[”partition”] resvisu <- clusvisMixmod(Mod1) plotDensityClusVisu(resvisu, add.obs = F, positionlegend = “topleft”)

The partition of the sample space of Figure 18 presents the Gaussian-like component overlap on the most discriminative map, where the difference between entropies is almost zero [
![]() ]. The sum of the inertia of the first two axes is 61.62 + 18.93 = 80.55 percent of the discriminant power. The groups with more observations are numbers 8, 3, 1, and 6. The most isolated groups are 1, 6, and 7; their position on opposite sides suggests that their chemical signatures are entirely different with respect to the rest of the subsources, with Component 7 differing the most. This component corresponds to Jilotepeque 2.
]. The sum of the inertia of the first two axes is 61.62 + 18.93 = 80.55 percent of the discriminant power. The groups with more observations are numbers 8, 3, 1, and 6. The most isolated groups are 1, 6, and 7; their position on opposite sides suggests that their chemical signatures are entirely different with respect to the rest of the subsources, with Component 7 differing the most. This component corresponds to Jilotepeque 2.

Figure 18 Component interpretation graph of the Mayan obsidian sources. The component numbers indicate the following subsources: [1] Jilotepeque 1, [2] El Chayal 3, [3] El Chayal 2*, [4] El Chayal 4, [5] Ixtepeque 2, [6] Ixtepeque 1, [7] Jilotepeque 2, [8] El Chayal 2, and [9] El Chayal 1. Color version available at www.cambridge.org/argote_machine-learning
The first thing that we can observe in these results is that the algorithm estimates the optimal number of groups as K = 9 components, which would correspond to the 9 subsources registered for the region. Therefore, it can be concluded that the chemical variability of each subsource is sufficiently different from the others, allowing recognition of its origin with adequate accuracy. However, the assignments of the samples to the components are different than the assignment made by Carr, as seen in the assignment of the units to each one of the components or subsources displayed in Table 9. This is because the mixture model is more robust for modeling group analysis.
Table 9 Assignation of the mixture proportions with Rmixmod (z-partition of Guatemala obsidian sources and subsources).
| Reference CarrCarr (2015) Subsource | Reference CarrCarr (2015) sample size (n) | Rmixmod component group ID | Rmixmod sample assignation | Proportion | z-partition |
|---|---|---|---|---|---|
| El Chayal 1 | 17 | 8 | 17 | 0.0791 | El Chayal 1 |
| El Chayal 2 | 113 | 5 | 78 | 0.3628 | El Chayal 2 |
| 9 | 40 | 0.1860 | El Chayal 2* | ||
| El Chayal 3 | 16 | 2 | 13 | 0.0605 | El Chayal 3 |
| El Chayal 4 | 10 | 4 | 11 | 0.0512 | El Chayal 4 |
| El Chayal 5 | 3 | ||||
| Jilotepeque 1 | 30 | 1 | 26 | 0.1209 | Jilotepeque 1 |
| 3 | 4 | 0.0186 | Jilotepeque 1* | ||
| Jilotepeque 2 | 4 | 7 | 4 | 0.0186 | Jilotepeque 2 |
| Ixtepeque 1 | 17 | 6 | 22 | 0.1023 | Ixtepeque 1 |
| Ixtepeque 2 | 5 | ||||
| Total = | 215 | 215 | 1.0000 |
Table 9 shows the z-partition of the Gaussian mixture model. In the first column are the nine subsources classified by Reference CarrCarr (2015). In the second column, the sample size of each subsource estimated by the author is tabulated. In the third column, the z-partition of nine components of the sample can be observed. The number of observations assigned to each component is recorded in the fourth column, and the proportions of each component are recorded in the fifth column. In the last column are the differences between the algorithm assignation and the classification provided by Carr.
In this example, the first component coincides with Carr’s classification; that is, the same 17 sample units were assigned to the El Chayal 1 subsource. For the El Chayal 2 subsource, Carr classified 113 samples to this subsource, while the mixture model assigns only 78 observations to this component and 40 observations to component 9, now marked as El Chayal 2*. In other words, the original El Chayal 2 subsource was divided into two subsources by the mixture model, separating 35 samples from the original El Chayal 2 and transferring two samples from El Chayal 3 and three from El Chayal 5 to the new El Chayal 2* subgroup. Carr classified 16 observations in the El Chayal 3 group and 10 samples in the El Chayal 4 group, but the mixing model classification assigned only 13 samples to the El Chayal 3 group and 11 to the El Chayal 4 group. Due to the new assignations, the original El Chayal 5 subsource in Carr’s classification was eliminated.
For Jilotepeque, Carr identified two subsources: Jilotepeque 1 and Jilotepeque 2, with 30 and 4 observations, respectively. The mixture model also identified two subsources but with slight differences in sample size for the case of Jilotepeque 1, with 26 observations in one component and 4 observations in another subsource marked as Jilotepeque 1*. The case of Jilotepeque 2 still consisted of four samples. For the Ixtepeque subsources, Carr identified two subsources: Ixtepeque 1 with 17 observations and Ixtepeque 2 with five observations. Conversely, the mixture model did not find significant differences to divide this source into two subsources, so the 22 original observations were assigned to a single component (Ixtepeque 1).
The Mahalanobis distance (MD) has been widely used as a classification technique in archaeometry to estimate relative probabilities of group membership. However, as discussed in the introduction of Statistical Processing of Quantitative Data of Archaeological Materials www.cambridge.org/Argote, this statistic presents several drawbacks that can cause serious problems in the calculation of group memberships. For example, Reference Glascock, Braswell, Cobean and ShackleyGlascock et al. (1998) analyzed the provenance of obsidian samples from Central Mexico using the first three principal components, which explained approximately 92 percent of the variance. The author used the MD to calculate relative probabilities for the assignment of samples to the groups and obtained erroneous results for the sources of Santa Elena (Hidalgo) and the subsources of Pachuca. This exhibits that using a technique such as MD to make assignments can force some observations to belong to one of the groups. Furthermore, it was shown that multimodal distance distributions are preferable (Reference ThrunThrun, 2021b), which is a property that MD rarely possesses.
In contrast, model-based clustering produces an association weight based on a formal probability, called the posterior probability of each observation belonging to each group. The partition is derived from a maximum likelihood estimator using the MAP procedure (see the section on model-based clustering in Statistical Processing of Quantitative Data of Archaeological Materials www.cambridge.org/Argote). This procedure is carried out through the EM algorithm or one of its variants. Parameter estimation using the EM algorithm calculates the weights for each observation, given the parameter values of the mixture components and the overall mixture weights (Reference KesslerKessler, 2019). These weights are used as measures of the strength of the association between each observation and the groups in the model in such a way that, in the model-based method, the observations in the same cluster are generated from the same probability distribution of the cluster (Reference Grün, Frühwirth-Schnatter, Celeux and RobertGrün, 2019).
To exemplify this procedure, the groups established by Reference CarrCarr (2015) were contrasted according to his scatter diagrams and the probabilities obtained from the calculation of the MD made by the same author and the probabilities calculated using the mixture model. The reader can directly contrast the results of the calculation of the probabilities of membership obtained with both methods by directly consulting the work of Reference CarrCarr (2015) and computing the model-based clustering probabilities with the following command:
(Mod1@bestResult@proba) # Calculation of probabilities with the mixture model

By making this comparison, one can appreciate that the memberships calculated with the MD do not coincide with the groups designated by Carr for each of the localities in the region. It is important to note that, in some cases, the MD underestimates and, in others, overestimates the probabilities of belonging to a group. The MD sometimes assigns the observations to the groups even when their probability of membership to a group is well below 50 percent. This fact has to do with the clear violation of the assumption of normality established by the MD method. On the other hand, the probabilities obtained by the mixture model are usually above 90 percent, except for three cases.
If the number of components is small and the sample size is large, the ClusVis program allows a pseudoscatter plot of the observation memberships to be obtained. In this plot, each observation is projected as colored dots representing the partition membership z; the information about the uncertainty of the classification is given by the curves of the iso-probability of classification, and information about the visualization accuracy is given by the difference between entropies and the percentages of inertia (Reference Biernacki, Marbac and VandewalleBiernacki et al., 2021). In this case, it was considered to use only the five subsources belonging to El Chayal, forming a data matrix with n = 159 and D = 10; the processing was done the same way as for the previous case.
In Figure 19, we can see that the difference between entropies is almost zero [
![]() ]. The sum of the inertia of the first two axes is 61.94 + 26.98 = 88.92 percent of the discriminant power. Three probability levels of classification were obtained for El Chayal samples (0.50, 0.80, and 0.95); the observations are represented with the label of the component maximizing the posterior probability of classification. Only four of the 159 observations were misclassified into component 4; in the rest of the components, there are no misclassified observations, and the membership of the observations is above 0.80 percent and 0.95 percent. To obtain the pseudoscatter plot of the observation memberships, the following command is used:
]. The sum of the inertia of the first two axes is 61.94 + 26.98 = 88.92 percent of the discriminant power. Three probability levels of classification were obtained for El Chayal samples (0.50, 0.80, and 0.95); the observations are represented with the label of the component maximizing the posterior probability of classification. Only four of the 159 observations were misclassified into component 4; in the rest of the components, there are no misclassified observations, and the membership of the observations is above 0.80 percent and 0.95 percent. To obtain the pseudoscatter plot of the observation memberships, the following command is used:

Figure 19 Bivariate scatter plot of the observation memberships of samples from El Chayal, Guatemala. Component 1 corresponds to El Chayal 3, component 2 to El Chayal 1, component 3 to El Chayal 2, component 4 to El Chayal 4, and component 5 to El Chayal 2 *. Color version available at www.cambridge.org/argote_machine-learning
plotDensityClusVisu(resvisu, add.obs = T, positionlegend = “ topleft”)
At times it is important to have an idea of the chemical variation of each of the existing subsources and, for this, it is not enough to compare the vectors of means or their standard deviations. To obtain a better notion of the variability between subsources, one can turn to the definition of Binary Sequential Partition (SBP) of parts that defines an orthonormal basis in the Simplex (see the section on Compositional and Completely Compositional Data in Statistical Processing of Quantitative Data of Archaeological Materials www.cambridge.org/Argote). The partition criterion must be based on a wide knowledge of the association and affinity between parts (Reference van den Boogaart and Tolosana-Delgadovan den Boogaart and Tolosana-Delgado, 2013) but can also be obtained through the variation matrix of the log-ratios or the implementation of compositional biplots obtained from the variance and covariance matrices using the clr transformation (Reference Pawlowsky-Glahn and EgozcuePawlowsky-Glahn and Egozcue, 2011).
Another option is to group the parts by means of a hierarchical cluster analysis, such as the Ward method, and use the variation matrix to calculate the distances between the parts. With this method, a signed matrix is built, containing the bases from which an ilr definition matrix is obtained. The employment of the SBP allows the bases to be obtained to generate a CoDa-dendogram (Reference Parent, Parent, Rozanne, Hernandes, Natale and IssakaParent et al., 2012). To calculate the SBP and the CoDa-dendogram, it is necessary to include in the data matrix a column that identifies the group to which each of the samples belongs. As an example, the matrix in Table 10 illustrates the ordering of the groups.
Table 10 Example of the matrix to obtain the SPB bases and the CoDa-dendogram.
| ID | Class | Mn | Fe | Zn | Ga | Th | Rb | Sr | Y | Zr | Nb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CH01 | group2 | 651 | 5844 | 30 | 17 | 9 | 137 | 133 | 17 | 101 | 9 |
| CH02 | group2 | 712 | 6075 | 32 | 17 | 10 | 140 | 139 | 20 | 105 | 9 |
| CH03 | group2 | 687 | 5943 | 33 | 17 | 10 | 138 | 138 | 20 | 106 | 10 |
| CH04 | group2 | 662 | 6009 | 34 | 17 | 9 | 136 | 135 | 19 | 105 | 9 |
| CH05 | group2 | 680 | 6029 | 37 | 18 | 10 | 136 | 139 | 21 | 107 | 9 |
| CH06 | group2 | 656 | 6084 | 34 | 17 | 12 | 140 | 140 | 19 | 106 | 9 |
| CH07 | group2 | 679 | 6115 | 33 | 17 | 10 | 137 | 138 | 18 | 106 | 10 |
| CH08 | group2 | 667 | 5994 | 30 | 17 | 9 | 135 | 137 | 19 | 108 | 10 |
| CH09 | group2 | 656 | 6042 | 33 | 17 | 10 | 138 | 134 | 19 | 105 | 9 |
| CH10 | group2 | 617 | 6155 | 34 | 17 | 10 | 139 | 140 | 20 | 103 | 10 |
| LM13 | group2 | 680 | 6399 | 31 | 17 | 8 | 132 | 132 | 18 | 105 | 9 |
| LJ01 | group5 | 703 | 7829 | 36 | 18 | 11 | 159 | 165 | 22 | 120 | 12 |
| LJ02 | group5 | 668 | 7920 | 35 | 18 | 12 | 152 | 161 | 20 | 116 | 10 |
| LJ03 | group5 | 651 | 8352 | 43 | 18 | 12 | 157 | 172 | 23 | 115 | 9 |
| AC13 | group5 | 516 | 6675 | 29 | 17 | 10 | 129 | 125 | 18 | 103 | 7 |
| KM01 | group5 | 597 | 6999 | 37 | 18 | 11 | 141 | 149 | 19 | 107 | 9 |
| KM02 | group5 | 712 | 7243 | 31 | 17 | 10 | 141 | 153 | 21 | 111 | 9 |
| KM03 | group5 | 669 | 8248 | 32 | 17 | 10 | 147 | 154 | 20 | 109 | 10 |
To obtain the SBP, use this script:
rm(list=ls())
dat1 <- read.csv(X,header=T) #uploads the data file X arranged by groups
str(dat1)#view the file structure
library(compositions)#uploads the package
x = acomp(dat1[,-c(1:2)]) #applies the closure operator only to numeric data
x#to see the closed data
gr = dat1[,2] #assigns the variable class to gr= to identify the obsidian groups (useful afterwards)
gr#to observe if the variable was correctly assigned
#Use an ilr basis coming from a clustering of parts
dd = dist(t(clr(x))) #computes the Euclidian distances of the variation matrix
hc1 = hclust(dd,method=”ward.D2”) #builds the dendrogram with the Ward method
plot(hc1)#dendogram
mergetree=hc1$merge#basis to use, described as a merging tree color=c(”green3”,”darkviolet”,”red”, “blue”, “orange”) CoDaDendrogram(X=acomp(x),mergetree=mergetree,col=”black”,range=c(-6,6),type=”l”) xsplit = split(x,gr) for(i in 1:5){ CoDaDendrogram(X=acomp(xsplit[[i]]),border=color[i], type=”box”,box.pos=i-2.5,box.space=1.5,add=TRUE) CoDaDendrogram(X=acomp(xsplit[[i]]),col=color[i], type=”line”,add=TRUE)
}
The CoDa-dendogram of the El Chayal dataset, following the SBP, is represented in Figure 20. In the electronic version of this Element, the colors assigned to the subsources can be appreciated, which were the following: El Chayal 1 = red, El Chayal 2* = green, El Chayal 2 = purple, El Chayal 3 = blue and El Chayal 4 = orange. The length of the vertical bars represents the variability of each ilr coordinate. In this case, we have nine balances with D = 10 parts (components). As seen, the third balance has the largest variance and involves the Fe (iron) part; this implies that this balance explains a great portion of the total variance. The location of the mean of an ilr-coordinate is determined by the intersection of the vertical segment with the horizontal segment (variance); when this intersection is not centered, it indicates a major or minor influence of one of the groups of the parts.

Figure 20 CoDa-dendrogram for the El Chayal dataset using SBP. The vertical bars correspond to the decomposition of the variance by balances. Color version available at www.cambridge.org/argote_machine-learning
For this example, we can see in the first balance (b1) that the median of the five groups does not coincide. In the first balance, the box plot shows a certain asymmetry and a greater dispersion than the second balance. This is due to the low variability given by Fe, Mn, Zr, Rb, and Sr concentrations in the second group of parts. In balance 1 (b1), El Chayal 4 (orange) deviates slightly from the other groups and shows a greater contribution in the Fe–Mn–Zr–Rb‒Sr parts. Conversely, El Chayal 2* (green) shows smaller amounts in the Th–Nb–Zn–Ga–Y parts. The other groups contain slight variations between the two major groups of the parts. In balance 2 (b2), El Chayal 1 (red) has a lower variance, and as it is loaded to the left, it would have smaller quantities of Th and Nb parts and a greater contribution of Zn–Ga–Y parts.
In balance 3 (b3), El Chayal 2 (purple) shows a lower content of Fe and minimal variations in Mn-Zr-Rb‒Sr with respect to the other groups. Balance 7 (b7) also displays a similar aspect, but El Chayal 1 shows a slight increase in the Zr, Rb, and Sr parts, followed by El Chayal 2* and El Chayal 4. The effect is null in the rest of the balances because there is good symmetry in the parts. El Chayal 4 registers a greater variance in balances b4 and b5, as does El Chayal 3, so these two subsources would present a greater dispersion in this group of parts (Th–Nb–Zn–Ga–Y). The parts that play a greater role in the classification are those to the right of the CoDa-dendogram (Fe–Mn–Zr–Rb‒Sr).
In summary, it can be said that the range of chemical variability of each of the subsources is different and that the discrimination between them is because the material does not correspond to the same lava flow, which would explain the variation in the chemical elements that compose the samples. In general, one can speak of the existence of five different systems that could correspond to different eruptions or volcanic events.
4 Processing a Combination of Spectral and Compositional Data
4.1 Application and Case Studies
Let us remember that chemometry is directly related to all those methods that transform relatively complex analytical signals and data to provide the maximum amount of relevant chemical information; it is strongly connected to multivariate quantitative analysis and pattern recognition. From the point of view of chemometry, spectral data can be represented in a matrix form (Figure 21) for further classification with multivariate methods. This matrix (i.e., spectral data) combined with a compositional data vector can be used for the classification of archaeological samples combining spectral preprocessing techniques (such as that described in Section 2 of this Element), variable selection methods, and projection-based clustering analysis. In this section, the proposed methods will be applied to data measured from obsidian samples and to a hypothetical “human-in-the-loop” cluster analysis

Figure 21 Matrix representation of the spectra. Color version available at www.cambridge.org/argote_machine-learning
4.2 Exercise 1: Mesoamerican Obsidian Deposits
In this example, geological samples from eight different obsidian sources (enlisted in Table 11) were analyzed with a pXRF spectrometer, employing a TRACER III-SD XRF portable analyzer manufactured by Bruker Corporation, with an Rh tube at an angle of 52°, a drift silicon detector, a 7.5 μm Be detector window and a factory filter composed of 6 μm Cu, 1 μm Tl, and 12 μm Al. The instrument was set with a voltage of 40 kV, a current of 30 μA, and a measurement time of 200 live seconds. The measured spectra were used to construct a matrix that consisted of n = 136 [samples] and p = 2,048 [channels], available in the supplementary material as file “Obsidian_chp4.csv”. The spectral and compositional data from these sources are similar to the data used in the first exercise of Sections 2 and 3 of this Element. The procedure is described step by step in the supplementary video “Video 5.”
Table 11 Number of samples collected from eight Mesoamerican obsidian sources.
| Source name | Geographic region | n |
|---|---|---|
| Ahuisculco | Jalisco | 9 |
| El Chayal | Guatemala | 34 |
| San Martin Jilotepeque | Guatemala | 17 |
| Ixtepeque | Guatemala | 17 |
| La Esperanza | Honduras | 16 |
| Otumba (Soltepec) | Edo. de México | 10 |
| Otumba (Ixtepec-Malpais) | Edo. de México | 23 |
| Sierra de Pachuca | Hidalgo | 10 |
| Total = | 136 |
Step-by-step video on how to process a combination of spectral and compositional data used in Video 5. Video files available at www.cambridge.org/argote_machine-learning
In this case, the spectra showed a high number of noisy signals that are more visible at higher keV values (Figure 22). Moreover, at the beginning and the end of the spectra are areas that contain no relevant information, either because they contain values close to zero or correspond to undesirable effects such as the Compton peak or Raleigh scattering. Therefore, columns 1 to 38 and 903 to 2,048 were manually deleted from the original matrix, reducing the dimension of the new matrix to n = 136 obsidian samples by p = 865 channel counts (matrix available in the supplementary material as file ‘Ixtq_2_38_865.csv’).

Figure 22 (Top) Full spectra of the obsidian samples. (Bottom) Selection of useful channels from the pXRF spectra. Color version available at www.cambridge.org/argote_machine-learning
The second step was to filter the spectra to maximize the quality of the information. The raw spectra were processed using the EMSC and SG filters separately; the two resulting matrices were saved in different files. Afterward, the EMSC-filtered data were processed using the Savitzky‒Golay filter, and the SG-filtered matrix was treated with the EMSC algorithm to obtain two other files, one with a combination of EMSC + SG and one with SG + EMSC filters. It is important to note that these procedures eliminate five columns from both extremes of the matrix, leaving only 855 variables in our final matrix. The filtering system with the best performance is chosen by evaluating the parameter values that are calculated later in the procedure. The script below allows filtering with the EMSC algorithm and then filtering with the SG algorithm. Note that to perform the inverse action, first use the script for the SG filter and afterward the code for the EMSC. For individual filtering systems, that is, only EMSC or only SG, the scripts are used separately. Just remember to update the file names to call and run the proper one.
## Script to filter with the EMSC algorithm rm(list = ls()) library(EMSC) #Package EMSC. Performs model-based background correction and # normalization of the spectra (Reference Liland and IndahlLiland and Indahl, 2020) dat <- read.csv(”C: \\Ixtq_2_38_865.csv’”, header=T) # To call the spectral data file str(dat) # to see the data structure dat1 <- dat[,2:866] # To eliminate the first column related to the sample identifier str(dat1) EMSC.basic <- EMSC(dat1) EMSC.poly6 <- EMSC(dat1, degree = 6) #Filters the spectra with a 6th order # polynomial str(EMSC.poly6) write.csv(EMSC.poly6$corrected, file=”FEmsc.csv”) # to save the data file # filtered with the EMSC. The user can choose other file names ##–––––––––––––––––––––––––––- ## Script to filter with the SG algorithm library(prospectr)## Miscellaneous Functions for Processing and Sample Selection of ## Spectroscopic Data (Reference Stevens, Ramirez-Lopez and HansStevens et al., 2022) data <- read.csv(”FEmsc.csv”, header=T) # Calls the file with EMSC filtered data # data <- read.csv(“C: \\Ixtq_2_38_865.csv’”, header=T) #Use this instead for only SG str(data) # data1 <- dat[,2:866] #Add this when applying only SG #str(data1) #Add this when applying only SG sg <- savitzkyGolay(data, p = 3, w = 11, m = 0) # sg <- savitzkyGolay(data1, p = 3, w = 11, m = 0) #Use this instead for only SG write.csv(sg, file=” FEmsc _SG.csv”) # or ‘SG.csv’. The user can choose other file names

In the third step, the data were processed using the iPLS algorithm to select the interval(s) within the data that would provide the most significant variables. In the model, the photon counts in each of the spectrum channels measured with the pXRF instrument were used as the explanatory variables (X). To obtain the response variable (y), the luminescent data were converted to concentration values according to the Empirical Coefficients method (Reference Rowe, Hughes and RobinsonRowe et al., 2012) using a variant of the Reference Lukas-Tooth and PriceLukas-Tooth and Price (1961) equation. Once the chemical concentrations were obtained, the resulting matrix contained the following components: Mn, Fe, Zn, Ga, Th, Rb, Sr, Y, Zr, and Nb; this matrix is provided in the supplementary material of the electronic version of this volume as file “analitos.csv”. The data were then moved to their native geometric space according to Aitchison’s theory (Reference AitchisonAitchison, 1986) employing the centered log-ratio (clr) transformation using the following script:
rm(list=ls()) data <- read.csv (”C:\\analitos.csv”, header=T) #You can use your own file str(data) # displays the internal structure of the file, which includes the format of each#column dat2 <- data[2:11] # delete data identification column str(dat2) # transformation of the data to the clr log-ratio library(compositions) ## Reference van den Boogaart, Tolosana-Delgado and Brenvan den Boogaart, Tolosana-Delgado and Bren (2023) xxat1 <- acomp(dat2) #the function “acomp” representing one closed composition. #With this command, the dataset is now closed. xxat2 <- clr(xxat1) # Centered logratio transformation str(xxat2) write.csv(xxat2, file=”Clr-transformation.csv”) ##From this resulting output file #(”Clr-transformation.csv”), extract Sr (strontium) vector and save it as a separate.cvs # file

From the resulting output file, the values obtained for the variable Sr (strontium) are copied to a separate .cvs file for its posterior use in the iPLS calibration model as the response variable (y = Sr). It is important to note that the selection of the response variable will depend on the material you are working with. For example, pottery, Fe or Ca could be relevant; for some Roman glasses, Na, Ca, Sb, and Pb can differentiate the real origin of the pieces (Reference López-García and ArgoteLópez-García and Argote, 2023); for obsidian, Sr is a discriminatory element, so it was chosen for the analysis. For this exercise, the file “Stroncio.csv” is provided in the supplementary material. Table 12 shows an example of the difference between the raw values of Sr composition of some samples and its clr transformed values.
Table 12 Example of the raw compositional data and the data after the clr transformation.
| ID | Raw data | Clr transformation |
|---|---|---|
| Sr | Sr | |
| 1 | 129.36 | 0.465173738 |
| 2 | 132.46 | 0.507479359 |
| 3 | 131.64 | 0.504697572 |
| 4 | 132.17 | 0.474821295 |
| 5 | 127.27 | 0.439785067 |
| 6 | 133.33 | 0.488857386 |
| 7 | 137.82 | 0.502847437 |
| ---------- | -------------- | |
| ---------- | -------------- | |
| ---------- | -------------- | |
| 134 | 4.61 | −3.303411956 |
| 135 | 5.60 | −3.137748288 |
| 136 | 7.23 | −2.938981779 |
For iPLS regression analysis, the full spectrum (1–855) was divided into 10 equidistant subintervals, each containing approximately 85 variables. Then, a PLS calibration model was developed for each subinterval. The iPLS algorithm was applied to the four filtering systems (EMSC, SG, ESMC+SG, and SG+EMSC), and the one with the best performance was chosen according to the values observed in the RMSECV, RMSE, and R2 parameters. The EMSC + SG combination turns out to be the best for filtering the data. To run the iPLS algorithm using the next script, the output was the following:
## Interval variable selection rm(list=ls()) ## To remove all objects from memory library(mdatools) # Reference KucheryavskiyKucheryavskiy (2020) X <- read.csv(”C:\\FEmsc_SG.csv”, header=T) #Call the filtered spectra data file str(X) # mean centering, in case you want to autoscale the spectrum #X1 = prep.autoscale(X, center = T, scale = F) # Call the concentration data file of Sr (“Stroncio.csv”) or the y variable you selected y <- read.csv(”C:\\Stroncio.csv”, header=T) str(y) # ipls model # for a model without mean centering, use X instead of X1 m = ipls(X, y, glob.ncomp = 4, int.num = 10) # Model parameters # glob.ncomp = maximum number of components for a global PLS model # int.num = number of intervals summary(m) plot(m) plotRMSE(m) show(m$int.selected) show(m$var.selected) par(mfrow = c(1, 2))

The output details information about the selected intervals, the number of variables at both ends, and the value of R2:
Model with all intervals: RMSECV = 0.107, nLV = 4 Iteration 1/ 10 … selected interval 9 (RMSECV = 0.125, nLV = 4) Iteration 2/ 10 … selected interval 8 (RMSECV = 0.105, nLV = 4) Iteration 3/ 10 … selected interval 2 (RMSECV = 0.105, nLV = 4) Iteration 4/ 10 … selected interval 5 (RMSECV = 0.105, nLV = 4) Iteration 5/ 10 … selected interval 3 (RMSECV = 0.105, nLV = 4) Iteration 6/ 10 … selected interval 1 (RMSECV = 0.105, nLV = 4) Iteration 7/ 10 … no improvements, stop.

In this case, the global model has an RMSECV = 0.107 with four components. Interval 9, with RMSECV = 0.125, gave the best performance for building local models with individual intervals. On the other hand, the second iteration selected interval 8 as the best local model with an RMSECV = 0.105, and according to the cross-validation, four components were optimal. In the subsequent iterations, more intervals were included; nevertheless, the RMSECV value did not vary at all, so it was no longer convenient to add more intervals. Intervals 8 and 9 present an R2 similar to the global PLS model using the entire spectrum, suggesting a meaningless variation of the RMSE or R2 values if any other intervals were included. In this way, it was possible to determine that intervals 8 and 9 contained the most informative variables of the spectrum; these intervals included the range of variables from columns 601 to 685 and from 686 to 770, respectively.
The iPLS variable selection results were as follows:
Validation: venetian blinds with 10 segments Number of intervals: 10 Number of selected intervals: 6 RMSECV for global model: 0.107508 (4 LVs) RMSECV for optimized model: 0.105035 (4 LVs) Summary for selection procedure: n start end selected nComp RMSE R2 1 0 1 855 FALSE 4 0.107 0.988 Global Model 2 9 686 770 TRUE 4 0.125 0.984 3 8 601 685 TRUE 4 0.105 0.988 4 2 87 172 TRUE 4 0.105 0.988 show(m$int.selected) [1] 9 8 2 5 3 1 Method: forward

Figure 23 shows the performance of individual models and the selected interval or intervals, interpreted as follows. The average spectrum can be appreciated along the bars. Green or dark gray bars are the local intervals selected by the iPLS model; the height of each bar corresponds to the RMSECV value for the local model using variables from this interval as predictors (X). The number within each bar is the number of PLS components used in the local model. A dashed horizontal line indicates the RMSECV obtained by using all variables, and the number 4 at the end of that line is the number of latent variables (LV).

Figure 23 iPLS model for 10 intervals. Color version available at www.cambridge.org/argote_machine-learning
Finally, it is convenient to check the existence of outliers using the ROBPCA algorithm of the ‘rrcov’ package:
rm(list=ls()) library(rrcov) ## Scalable Robust Estimators with High Breakdown Point (Reference TodorovTodorov, 2020) dat <- read.csv(”C:\\FEmsc_SG.csv”, header=T) str(dat) rpca <- PcaGrid(~., data=dat) rpca plot(PcaHubert(dat, k=0), sub=”data set: dat, k=4”) str(rpca) rpca$flag

In this case, to establish the optimal number of components to retain, we set k = 0 such that
![]() . E − 3 and
. E − 3 and
![]() . Refer to Reference Hubert, Rousseeuw and Vanden BrandenHubert et al. (2005) and the ‘rrcov’ package (Reference TodorovTodorov, 2020) in R for more information. In the diagnosis of the data (Figure 24), eleven observations exceeded the cutoff value: ten forming a small group of bad leverage points at the top of the quadrant and the isolated case 119 at the far right of the graph. The bad leverage points correspond to the Pachuca samples; their separation from the rest of the sources is because these samples register significantly higher concentrations of Fe, Zr, and Zn and lower amounts of Sr compared to the other deposits. Therefore, they cannot be considered outliers. Conversely, the isolated sample (no. 119) belongs to Otumba (Ixtepec-Malpais); it is possible that it has some measurement error or contamination because it completely departs from the group of normal observations; thus, it is convenient to eliminate it from the analysis.
. Refer to Reference Hubert, Rousseeuw and Vanden BrandenHubert et al. (2005) and the ‘rrcov’ package (Reference TodorovTodorov, 2020) in R for more information. In the diagnosis of the data (Figure 24), eleven observations exceeded the cutoff value: ten forming a small group of bad leverage points at the top of the quadrant and the isolated case 119 at the far right of the graph. The bad leverage points correspond to the Pachuca samples; their separation from the rest of the sources is because these samples register significantly higher concentrations of Fe, Zr, and Zn and lower amounts of Sr compared to the other deposits. Therefore, they cannot be considered outliers. Conversely, the isolated sample (no. 119) belongs to Otumba (Ixtepec-Malpais); it is possible that it has some measurement error or contamination because it completely departs from the group of normal observations; thus, it is convenient to eliminate it from the analysis.

Figure 24 Robust diagnostic plot for the obsidian data with k = 4. Color version available at www.cambridge.org/argote_machine-learning
Now that the preprocessing has concluded, the next stage of the analysis is to perform the projection-based clustering. For this, we used a reduced matrix of n = 135 by p = 170 that included only the range from 601 to 770 channel counts (intervals 8 and 9 selected by the iPLS) and eliminated sample 119. Although, in fact, the provenance of the sources is known, we considered this exercise as a nonsupervised classification, that is, where the data are unlabeled. The first thing we want to know is whether the data exhibit a clustering structure according to the information recorded in the variables. If this is true, then we will need to determine the optimal number of groups and the correct assignment of the observations using the projection-based clustering method (Reference Thrun, Ultsch and HüllermeierThrun, 2018).
The first module of the script (see “Introduction into the Usage of Projection-based clustering” in Statistical Processing of Quantitative Data of Archaeological Materials www.cambridge.org/Argote) allows us to visually appreciate the existence of groups and determine the optimal number of these through a topographic map (Reference Thrun, Lerch, Lötsch, Ultsch and SkalaThrun et al., 2016). The colors presented by the topographic elements depend on their elevation and are based on the U-matrix principle (Reference UltschUltsch, 2003; Reference Ultsch and SiemonUltsch and Siemon, 1990). The greater the spacing of the partitions in the high dimensional space, the higher the mountains on the topographic map. If two high-dimensional data points are in the same partition, both points end up in a blue lake or on a green meadow. Blue lakes indicate partitions with particularly high densities. Green grasslands represent homogeneous partitions. Conversely, if the two data points belong to different partitions, the landscape folds and a mountain range is created between the two points; depending on the real distance, the ranges can go from brown to white. Outliers land on volcanoes or mountaintops.
Here, we assume that the data was preprocessed appropriately and that the Euclidean distance is the best choice of similarity. Then the following code can be used:
rm(list = ls()) library(DatabionicSwarm) datos <- read.csv(”C:\\Two_intervals.csv”, header=T) str(datos) datos=as.matrix(datos)

library(DatabionicSwarm) InputDistances = as.matrix(dist(datos)) projection = Pswarm(InputDistances) library(DatabionicSwarm) library(GeneralizedUmatrix) genUmatrixList=GeneratePswarmVisualization( Data=datos, projection$ProjectedPoints, projection$LC)

GeneralizedUmatrix::plotTopographicMap( genUmatrixList$Umatrix, genUmatrixList$Bestmatches, NoLevels = 10)

Figure 25 displays the topographic map computed for the obsidian samples, where we can identify the existence of eight groups well separated by topographical barriers. The observations are clustered according to the position of the projected points. In this way, the points in each of the sections of the map correspond to observations that are similar between them and different from the rest of the groups according to their characteristics. In contrast to other multivariate methods, there is no overlap of the groups.

Figure 25 Topographic map of the DBS projection of the obsidian dataset with the Generalized U-matrix. Color version available at www.cambridge.org/argote_machine-learning
Once the existence of groups in the data is determined, the number of groups is specified in the script (with k = 8), and it is run again:
#Use previously loaded data library(DatabionicSwarm) ## (Reference ThrunThrun, 2021a) library(GeneralizedUmatrix) ## (Reference Thrun, Pape and UltschThrun et al., 2021a) Cls = DBSclustering( k = 8, InputDistances, genUmatrixList$Bestmatches, genUmatrixList$LC, PlotIt = FALSE ) GeneralizedUmatrix::plotTopographicMap(

genUmatrixList$Umatrix, genUmatrixList$Bestmatches, Cls, NoLevels = 10) library(DatabionicSwarm) library(ProjectionBasedClustering) library(GeneralizedUmatrix) Imx = ProjectionBasedClustering::interactiveGeneralizedUmatrixIsland( genUmatrixList$Umatrix, genUmatrixList$Bestmatches, Cls) GeneralizedUmatrix::plotTopographicMap( genUmatrixList$Umatrix, genUmatrixList$Bestmatches, Cls = Cls, Imx = Imx) Cls # provides the labels of the instances

In this first part of the analysis, it is important to remember that the data continue to be processed as a nonsupervised classification, where there is no a priori information of the data. By setting k = 8, the algorithm labels the observations according to the distance and the density of each data point, assigning a different color to each group. In this way, a group can be distinguished from the others by their high-dimensional distances from the original dataset on a map by hypsometric tints defined by the generalized U matrix (see Figure 26). What is important in this map is that the groups are clearly visible with a more compact structure defined by the valleys. Similarly, it is possible to perceive the absence of outliers in the data.

Figure 26 Topographic map of the DBS projection of the obsidian dataset with Generalized U-matrix using k = 8, interactively cropped. Color version available at www.cambridge.org/argote_machine-learning
In the last code, the “Cls” command allows you to see to which group was assigned each sample, as in Table 13.
Table 13 Assignation of samples to groups.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 |
| 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49 | 50 | 51 | 52 | 53 | 54 |
| 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 |
| 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 |
| 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 4 | 4 |
| 73 | 74 | 75 | 76 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 | 86 | 87 | 88 | 89 | 90 |
| 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 5 | 5 | 5 | 5 | 5 | 5 |
| 91 | 92 | 93 | 94 | 95 | 96 | 97 | 98 | 99 | 100 | 101 | 102 | 103 | 104 | 105 | 106 | 107 | 108 |
| 5 | 5 | 5 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 7 | 7 | 7 | 7 | 7 |
| 109 | 110 | 111 | 112 | 113 | 114 | 115 | 116 | 117 | 118 | 119 | 120 | 121 | 122 | 123 | 124 | 125 | 126 |
| 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 8 |
| 127 | 128 | 129 | 130 | 131 | 132 | 133 | 134 | 135 | |||||||||
| 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | |||||||||
In this example, all the samples were correctly assigned to their respective sources. As seen in Table 13, the first n = 34 samples were assigned to group 1 related to the El Chayal source, observations 35 to 50 (n = 16) were assigned to group 2 related to the La Esperanza source (Honduras), and so on. Therefore, it can be concluded that both the filtering system and the selection of the most informative intervals were appropriate procedures for partitioning the data into natural or significant groups. The final step is validating the model. As mentioned before in this Element, it is important to confirm the obtained results in a quantitative and objective manner to establish if the model fits the data well or if only represents a spurious solution. One way is to calculate the percentage of the accuracy, a supervised index defined by the ratio of the number of true positives to the number of cases (see Section 4 in Statistical Processing of Quantitative Data of Archaeological Materials www.cambridge.org/Argote). Another approach is to validate a clustering with the help of domain experts (e.g., Reference López-García, Argote and ThrunLópez-García et al., 2020; Reference Thrun, Ultsch and BreuerThrun et al., 2021b; Reference Thrun, Mack and NeubauerThrun et al., 2022). A third option is to evaluate if clustering is useful for a specific application (Reference ThrunThrun, 2022).
For simplicity, we compute the cluster accuracy in the dataset. For this, it is necessary to add a column in the data matrix with the header “Cls”, which refers to the class or group assigned by the algorithm to each of the observations (use the assignations of Table 13). Once this column has been added to the data matrix, the algorithm is rerun with the following script:
## with labeled data rm(list=ls()) library(DatabionicSwarm) ## Reference ThrunThrun (2021a) DataRaw <- read.csv(”C:\\Two_intervals_cls.csv”, header=T) ##Call the csv file with the #two intervals and add the Cls column. The user can choose a personalized file name str(DataRaw) Cls_prior=DataRaw$Cls Cls_prior # if you want the full unsupervised way

ind=which(colnames(DataRaw)!="Cls") Data=as.matrix(DataRaw[,ind]) library(DatabionicSwarm) projection = Pswarm(Data, Cls = Cls_prior, PlotIt = T, Silent = T)

library(DatabionicSwarm) library(GeneralizedUmatrix) visualization = GeneratePswarmVisualization(Data = Data, projection$ProjectedPoints, projection$LC)

GeneralizedUmatrix::plotTopographicMap(visualization$Umatrix, visualization$Bestmatches)

library(DatabionicSwarm) library(GeneralizedUmatrix) Cls = DBSclustering(k = 8, Data, visualization$Bestmatches, visualization$LC, PlotIt = FALSE) FCPS::ClusterCount(Cls) GeneralizedUmatrix::plotTopographicMap(visualization$Umatrix, visualization$Bestmatches, Cls) FCPS::ClusterAccuracy(PriorCls = Cls_prior, Cls)

library(DataVisualizations) #(Reference ThrunThrun, 2021a) Heatmap(as.matrix(dist(Data)),Cls = Cls) Silhouetteplot(Data,Cls =Cls)

In this case, the accuracy was 100 percent, so the assignments were made without error. Other exemplary validation indexes, known as nonsupervised indexes, used for evaluating the quality of the clustering are the silhouette index (Reference Kaufman and RousseeuwKaufman and Rousseeuw, 2005), the Dunn index (Reference DunnDunn, 1974), and the Davies Bouldin index (Reference Davies and BouldinDavies and Bouldin, 1979). Furthermore, visualization techniques like the heatmap (Reference Wilkinson and FriendlyWilkinson and Friendly, 2009) or the Silhouette plot can be used. Some of these are called in the last part of the previous script. For example, in the silhouette plot, the ideal number of clusters is displayed as separated silhouettes within a range of values that go from –1 to +1, where +1 indicates that the samples are correctly assigned to a cluster, 0 shows that the observations are very close to the decision limit between two neighboring clusters, and negative values indicate that the samples might have been assigned to the wrong cluster. The result of the silhouette plot (Figure 27, left image) clearly marks the presence of eight clusters as the optimal number, with no negative observations and no zero values. Therefore, we can sustain with confidence the existence of eight groups.

Figure 27 (Left) Silhouette plot of the obsidian dataset indicates a cluster structure. (Right) Heatmap of the obsidian dataset showing the existence of eight groups, where the intracluster distances are distinctively smaller than the intercluster distances. Color version available at www.cambridge.org/argote_machine-learning
Another graphical representation for visualizing high-dimensional data is the heatmap. A heatmap visualizes the distances ordered by the clustering through variations in color; this display simultaneously reveals row and column hierarchical cluster structure in a data matrix (Reference Wilkinson and FriendlyWilkinson and Friendly, 2009). In this example, the heatmap (Figure 27, right image) confirmed the DBS clustering of eight separated clusters; it also showed that this dataset was defined by discontinuities with small intracluster distances and large intercluster distances. Hence, the obsidian set is a high-dimensional dataset with natural clusters that are specified by the values represented in the two intervals of the spectra.
The clustering is also evaluated by means of a contingency table, whose rows are the source groups and whose columns are the results of the clustering. This contingency table is computed by the last part of the script of the projection-based clustering algorithm, copied below:
## With the following script, the contingency table is calculated
Cls_prior=DataRaw$Cls
Cls_prior
ind=which(colnames(DataRaw)!=“Cls”)
Data=as.matrix(DataRaw[,ind])
ContingencyTableSummary=function (RowCls, ColCls)
{
# contingency table of two Cls
# INPUT
# RowCls,bCls vector of class identifiers (i.e., integers or NaN’s) of the same length
# OUTPUT list with these elements:
# cTab cTab(i,j) contains the count of all instances where the i-th class in RowCls
#equals the j-th class inColCls
# rowID the different classes in RowCls, corresponding to the rows of cTab
# colID the different classes inColCls, corresponding to the columns of cTab
# RowClassCount, RowClassPercentages instance count and percentages of classes in #RowCls sorted according rowID # ColClassCount, ColClassPercentages instance count and percentages of classes #inColCls sorted according colID
RowID = length(unique(RowCls))
ColID = length(unique(ColCls))
Ctable = table(RowCls, ColCls)
AllinTab = sum(Ctable)
ColumnSum = colSums(Ctable)
ColPercentage = round(ColumnSum/AllinTab * 100, 2)
RowSum = rowSums(Ctable)
RowPercentage = round(RowSum/AllinTab * 100, 2)
Rows <- rbind(round(Ctable), ColumnSum, ColPercentage)
Xtable <- cbind(Rows, c(RowSum, AllinTab, 0), c(RowPercentage, 0, 100))
colnames(Xtable) = c(1:ColID, “RowSum”, “RowPercentage”)
return(Xtable)
} Table=ContingencyTableSummary(Cls_prior,Cls)
Table
Table 14 shows that all the elements of the main diagonal are well classified and that there are no misclassified observations. According to the overall results, it was shown that the analyzed obsidian samples had a clear cluster structure linked to their geological origin.
Table 14 Contingency table computed from the data of the obsidian sources.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Rowsum | RowPercentage | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 34.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 34 | 25.19 |
| 2 | 0.00 | 16.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 16 | 11.85 |
| 3 | 0.00 | 0.00 | 17.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 17 | 12.59 |
| 4 | 0.00 | 0.00 | 0.00 | 17.00 | 0.00 | 0.00 | 0.00 | 0.00 | 17 | 12.59 |
| 5 | 0.00 | 0.00 | 0.00 | 0.00 | 9.00 | 0.00 | 0.00 | 0.00 | 9 | 6.67 |
| 6 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 10.00 | 0.00 | 0.00 | 10 | 7.41 |
| 7 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 22.00 | 0.00 | 22 | 16.3 |
| 8 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 10.00 | 10 | 7.41 |
| Columsum | 34.00 | 16.00 | 17.00 | 17.00 | 9.00 | 8.00 | 24.00 | 10.00 | 135 | 0.00 |
| ColPercentage | 25.2 | 11.9 | 12.6 | 12.6 | 6.7 | 5.93 | 17.8 | 7.41 | 0 | 100 |
In sum, DBS is a nonlinear projection that displays the structure of the high-dimensional data into a low-dimensional space, preserving the cluster structure of the data. This model exploits the concepts of self-organization and emergence, game theory and swarm intelligence (Reference Thrun, Ultsch and HüllermeierThrun, 2018; Reference Thrun and UltschThrun and Ultsch, 2021). Pswarm does not require any input parameters other than the dataset of interest and is able to adapt itself to structures of high-dimensional data such as natural clusters characterized by distance and/or density-based structures in the data space. The result of the clustering consists of a 3D landscape with hypsometric tints, where observations with similar characteristics are represented as valleys while differences are represented as mountain ranges. In addition, the procedure can detect outliers that are represented as volcanoes on the 3D display and can be interactively marked on the display after the automated grouping process.
Another advantage of the method is that it is not necessary to have a priori knowledge of the classes to which the observations belong; the number of clusters and the cluster structure can be estimated by counting the valleys in the 3D topographic map and from the silhouette plot. Unlike other clustering algorithms, Pswarm does not impose any type of geometric structure in the formation of clusters, and the user does not need to specify any parameters. The results are evaluated using supervised and unsupervised validation indexes and visualization techniques, as well as a contingency table, confirming the goodness of the model to detect natural groups in the dataset. The example confirmed that the method proposed here is suitable for handling unbiased quantitative spectral analysis of archaeological materials.
4.3 Exercise 2: Human-in-the-Loop Cluster Analysis
The practical case study below serves as motivation to outline an alternative to investigate the detection and recognition of cluster structures using a human-in-the-loop. In this context, higher-level structures are detected in the data by enabling recognition of structures by the human-in-the-loop (HIL) at critical decision points. The authors thus follow the reasoning of Reference HolzingerHolzinger (2018) in that “the integration of an HIL’s knowledge, intuition, and experience can sometimes be indispensable, and the interaction of an HIL with the data can significantly improve the overall ML pipeline. Such interactive ML uses the HIL to make possible what neither a human nor a computer could do alone (Reference HolzingerHolzinger, 2018). The HIL is an agent that interacts with algorithms, allowing the algorithms to optimize their learning behavior (Reference Holzinger, Plass and Kickmeier-RustHolzinger et al., 2019). This perspective fundamentally integrates humans into an algorithmic loop with the goal of opportunistically and repeatedly using human knowledge and skills to improve the quality of ML systems (Reference Holzinger, Plass and Kickmeier-RustHolzinger et al., 2019; see also Reference Mac Aodha, Stathopoulos and BrostowMac Aodha et al., 2014; Reference ZanzottoZanzotto, 2019).
An HIL is usually necessary because automatic detection pipelines (see example in Reference Wiwie, Baumbach and RöttgerWiwie et al., 2015) have pitfalls and challenges that are systematically highlighted by Reference ThrunThrun (2021b). The work shows that parameter optimization on datasets without distance-based structures, algorithm selection using unsupervised quality measures on biomedical data, and benchmarking of detection algorithms with first-order statistics or box plots or a small number of repetitions of identical algorithm calls are biased and often not recommended (Reference ThrunThrun, 2021b). Hence, an alternative is proposed in Reference Thrun, Lerch, Lötsch, Ultsch and SkalaThrun et al. (2020; Reference Thrun, Pape and Ultsch2021a). Unlike typical approaches, the HIL is not overwhelmed with extensive parameter settings or evaluation of many complex quality measures (see examples in Reference Choo, Lee, Liu, Stasko and ParkChoo et al., 2013; Reference Müller, Assent, Krieger, Jansen and SeidlMüller et al., 2008; Reference Yang, Kandogan, Li, Sen and LaseckiYang et al., 2019; Reference Zhang, Tang and ShiZhang et al., 2002).
The coexistence of nonlinear projection methods and automatic detection of structures in projection-based clustering allows – apart from estimating whether there is a tendency for separable high-dimensional structures – estimating the number of partitions in the data as well as the correct choice of only one Boolean parameter for projection-based clustering. The HIL extension of the projection-based clustering incorporates user decisions in the detection process to visually discriminate structures. HIL-projection-based clustering is an open-source method that integrates the HIL at critical decision points through an interactive topographic map to detect separable structures.
Comparable interactive approaches fall into the category of visual analytics, which use visualizations to assist in manually searching for partitions in various types of datasets or to check the results of detection algorithms (e.g., Reference Cavallo and DemiralpCavallo and Demiralp, 2018; Reference Jeong, Ziemkiewicz, Fisher, Ribarsky and ChangJeong et al., 2009; Reference Kwon, Eysenbach and VermaKwon et al., 2017; Reference Müller, Assent, Krieger, Jansen and SeidlMüller et al., 2008; Reference Rasmussen and KarypisRasmussen and Karypis, 2004). However, the Johnson-Lindenstrauss lemma states that two-dimensional similarities in a scatter plot do not necessarily represent high-dimensional structures (Reference Dasgupta and GuptaDasgupta and Gupta, 2003; Reference Johnson and LindenstraussJohnson and Lindenstrauss, 1984). In praxis, projections of several datasets with distance- and density-based structures show a misleading interpretation of the underlying structures and unsupervised quality measures for dimensionality reduction are biased toward assumed underlying structures (Reference Thrun, Märte and StierThrun et al., 2023).
The HIL-projection-based clustering is proposed in a toroidal 2.5D representation, where a zoom out is preferred instead of other possible alternatives such as the four-tile of the toroidal projection. Toroid means that the boundaries of the topographic map are cyclically connected (Reference Ultsch, Oja and KaskiUltsch, 1999), which avoids problems of projections at the edges and therefore edge effects. In the four-tile representation, each projection point and structure would be represented four times. The HIL detects the number of partitions as the number of valleys. After the automatic detection phase, the HIL interactively rectifies the result on the topographic map via its own detection of high-dimensional structures. Using the 2.5D representation of the topographic map avoids the drawbacks of 3D representations and eliminates the challenge of the Johnson–Lindenstrauss lemma. The performance of HIL-projection-based clustering outperforms other accessible methods both qualitatively and quantitatively (Reference Thrun, Pape and UltschThrun et al., 2020, Reference Thrun2021b).
The GUI for the HIL-projection-based clustering is called with the following script:
rm(list=ls()) library(FCPS) library(ProjectionBasedClustering) ## (Reference Thrun, Pape and UltschThrun et al., 2020) data(”Chainlink”,package = “FCPS”) str(Chainlink) Data <- Chainlink$Data str(Data) V= IPBC (Data) #with prior classification Cls=Chainlink$Cls V=IPBC(Data,Cls)

The interfaces of the HIL-projection-based clustering algorithm (Reference Thrun, Pape and UltschThrun et al., 2021a) are presented in Figures 28 (‘Projection’ menu) and 29 (‘Clustering’ menu), in which every parameter setting is listed and numbered. In the ‘Projection’ menu, after loading the Chainlink dataset in listing (1), selecting the NeRV projection in (2), and clicking on the button in (4), the topographic map shown in (10) is obtained. The user can select another projection method in (2) and click in (4) to visualize a new topographic map. The user can automatically cluster the data with (15) by setting the number of clusters in (13) as the number of valleys. If the automatic clustering does not overlap with the valleys, the critical parameter for the clustering in (14) can be changed and a second trial of clustering can be performed. The user can frame points with the mouse either in the borderless view or the toroidal view (8); with “Add Cluster” in (12), a new cluster is assigned. Be aware that the assumption is that the data had been preprocessed properly and that the parameter of the selected projection method had been chosen wisely. If parameter setting of a projection method seems to be a challenge, the parameter-free projection method Pswarm can be selected.

Figure 28 Screenshot of the interface of the “Projection” menu of the IPBC method. Color version available at www.cambridge.org/argote_machine-learning

Figure 29 Screenshot of the interface of the “clustering” menu of the IPBC method. Color version available at www.cambridge.org/argote_machine-learning
5 Final Comments
One of the problems faced by archaeologists is that of data classification, which is defined by a series of attributes. Both clustering and classification have in common creating a model capable of recognizing instances according to their attributes by assigning them to different classes or groups. Both are complex tasks in archaeological data, since they involve the choice between many methods, the transformation and diagnosis of data, the selection of parameters and different metrics. Traditional unsupervised and supervised methods perform poorly at uncovering the underlying group structure in the data because they lack a formal statistical model. This Element compares the performance of different supervised and unsupervised classification methods that improve the outcome in clustering and classification of archaeological data.
If the data is high-dimensional, as in the case of pXRF spectral quantification, pre-processing is an essential part of comprehensive analysis to improve data quality. For example, spectra can show noise, displacements, or overlap between elements. To correct these problems in the related examples, a simple method for peak alignment was proposed using the hierarchical Cluster-based Peak Alignment (CluPA) algorithm. CluPa takes care of bringing all the peaks to the same origin, showing that the peaks from different origins are not misaligned. To eliminate unwanted interference in the spectra, model-based pre-processing techniques allow to quantify and separate different types of physical and chemical variations in the spectra. The recommended filtering was the use of a combination of the Savitzky–Golay and Extended Multiplicative Scatter Correction (EMSC) algorithms, which are filters that allow the preservation of the main characteristics of the function such as width and height of the spectral peaks.
The diagnosis of the data was another point to consider since extreme values can seriously distort the behavior of statistical estimators. To detect outliers, a robust estimator was proposed by replacing the covariance matrix obtained with the classical method with the covariance matrix obtained with the Minimum Covariance Determinant (MCD) robust method. On the other hand, a feature selection procedure is essential to separate variability related to relevant information from non-relevant information. In the high-dimensional (spectral) data scenarios, two variable selection methods were proposed. One approach based on Bayesian models and the other based on Partial Least Squares Interval (iPLS). The Bayesian approach allows for the selection of relevant variables and clustering simultaneously; at the same time is able to automatically determine the memberships of instances to their respective groups and to determine the optimal number of groups in the data. On the other hand, the iPLS method allows the selection of specific spectral regions for further analysis.
Continuing with the spectral analysis, Databionic swarm (DBS) algorithm propose an approach in which dimensionality reduction methods coexist with clustering algorithms, using a swarm-based AI system. The resulting groups define different generating processes related to the chemical composition of each group, which can be visualized with a topographic map of high-dimensional structures. The central problem in clustering is the correct estimation of the number of clusters; this is also addressed by the topographic map, which evaluates the optimal number of clusters. This is important because most clustering algorithms can detect clusters even if the distribution of the data is random. Unlike traditional methods, DBS is not going to force a cluster if there really are not natural clusters in the data.
When dealing with compositional data, first, it is important to consider that geometric space is different from real Euclidean space. In general, standard statistical methods are designed to work with classical Euclidean geometry (RD) or unconstrained p-dimensional spaces, so it is advisable to use a suitable transformation for compositional data whose geometric space is the simplex (SD). This is a bounded space with a constant-sum constraint, in which Aitchison’s geometry is applied; so only by working with the ratios between the parts do the problems of the constant-sum constraint disappear. There are many clustering methods in the literature without any specific method being uniformly better. To select the most suitable one, it is important to understand the intrinsic nature of the data and the strengths and weaknesses of the different algorithms.
To classify compositional data, an alternative is to model the data using components of the Gaussian mixture distributions, which assumes that the sample to be classified is divided into G groups or components. The estimation of parameters for each group is estimated by the maximum likelihood method, allowing us to estimate the probability that each of the instances has of belonging to one of the classes. To find the right number of groups, information criteria are considered, which are statistical criteria for evaluating models in terms of their posteriori probabilities. These statistics allow you to select from among the competing models and determine the optimal from a finite family of models.
Because of the excellent performance in partitioning the experimental datasets, the suggested algorithms and methodologies applied in this Element have proved to work well in the difficult task of clustering when the number of groups is unknown. The advantage of the proposed methods is that they allow the creation of a model for the determination of groups of well-defined characteristics that allows the optimization of the classification of the data. It is worth mentioning that the software for the implementation of the proposed methods is freely accessible, which allows the implementation to be established in an easy and simple way. We believe that these methods can be of great use to the archaeological community, as well as being applicable to a large number of cases beyond those described here. We encourage readers to practice with the proposed techniques and try new ways to solve their research problems.
Abbreviations
- alr
Additive Log-ratio
- BIC
Bayesian Information Criterion
- CA
Cluster Analysis
- CAIS
Center for Applied Isotope Studies
- CEM
Classification Expectation Maximization
- clr
Centered Log-ratio
- CluPA
Cluster-based Peak Alignment
- CRM
Certified Reference Material
- DA
Discriminant Analysis
- DBS
Databionic Swarm
- DR
Dimensionality Reduction
- EM
Expectation-Maximization
- EMSC
Extended Multiplicative Signal Correction
- FAST-MCD
Fast Minimum Covariance Determinant
- FCPS
Fundamental Clustering Problems Suite
- FT-IR
Fourier Transform Infrared Spectroscopy
- HIL
Human-in-the-Loop
- ICP‒MS
Inductively Coupled Plasma‒Mass Spectrometry
- ICL
Integrated Complete Likelihood
- ilr
Isometric Log-ratio
- iPLS
Interval Partial Least Squares
- LDA
Linear Discriminant Analysis
- LV
Latent Variables
- MAP
Maximum a Posteriori Probability
- MCD
Minimum Covariance Determinant
- MD
Mahalanobis Distance
- MD-plot
Mirrored Density plot
- ML
Maximization of Log-likelihood
- n
Number of observations or samples
- NAA
Neutron Activation Analysis
- NEC
Normalized Entropy Criterion
- NPE
Neighborhood Proportion Error
- ODi
Orthogonal Distance
- p
Number of variables or components
- PBC
Projection-based clustering
- PCA
Principal Component Analysis
- PC
Principal Component
- PDE
Pareto Density Estimation
Probability Density Function
- PLS
Partial Least Squares
- PP
Projection Pursuit
- pXRF
Portable X Ray Fluorescence
- QDA
Quadratic Discriminant Analysis
- RMSECV
Root Mean Squared Error of Cross-Validation
- RMSEP
Root Mean Square Error of Prediction
- ROBPCA
Robust Principal Component Analysis
- SBP
Sequential Binary Partition
- SDi
Score Distance
- SEM
Stochastic Expectation-Maximization
- SG
Savitzky‒Golay filter
- SNE
Stochastic Neighbor Embedding
- SVD
Singular Values Decomposition
- XRF
X Ray Fluorescence
Series Editors
Hans Barnard
Cotsen Institute of Archaeology
Hans Barnard was associate adjunct professor in the Department of Near Eastern Languages and Cultures as well as associate researcher at the Cotsen Institute of Archaeology, both at the University of California, Los Angeles. He currently works at the Roman site of Industria in northern Italy and previously participated in archaeological projects in Armenia, Chile, Egypt, Ethiopia, Italy, Iceland, Panama, Peru, Sudan, Syria, Tunisia, and Yemen.
Willeke Wendrich
Polytechnic University of Turin
Willeke Wendrich is Professor of Cultural Heritage and Digital Humanities at the Politecnico di Torino (Turin, Italy). Until 2023 she was Professor of Egyptian Archaeology and Digital Humanities at the University of California, Los Angeles, and the first holder of the Joan Silsbee Chair in African Cultural Archaeology. Between 2015 and 2023 she was Director of the Cotsen Institute of Archaeology, with which she remains affiliated. She managed archaeological projects in Egypt, Ethiopia, Italy, and Yemen, and is on the board of the International Association of Egyptologists, Museo Egizio (Turin, Italy), the Institute for Field Research, and the online UCLA Encyclopedia of Egyptology.
About the Series
Cambridge University Press and the Cotsen Institute of Archaeology at UCLA collaborate on this series of Elements, which aims to facilitate deployment of specific techniques by archaeologists in the field and in the laboratory. It provides readers with a basic understanding of selected techniques, followed by clear instructions how to implement them, or how to collect samples to be analyzed by a third party, and how to approach interpretation of the results.



















































