To the Editor—During an evaluation for a solid organ transplant (SOT) procedure, we identified a rare, opportunistic nontuberculous (NTM) strain, Mycobacterium saskatchewanense. The patient, who suffered from chronic renal disease, was tested for several pathogens, Mycobacterium tuberculosis included. Transplantation procedures rely on immunosuppressive therapies, which increase the success of the surgery and the survival of the patient.Reference Allison1 However, they also render the patients more susceptible to microbial infections, such as multidrug-resistant bacterial species like Klebsiella pneumoniae.Reference Di Mento, Cuscino, Carcione, Cardinale, Conaldi and Douradinha2, Reference Monaco, Di Mento, Cuscino, Conaldi and Douradinha3 Thus, patients require rigorous evaluations to confirm that they are not infected with pathogenic microorganisms that might compromise their health or lead to the rejection of the transplanted organ. One of the most opportunistic pathogens that can cause complications in SOT is M. tuberculosis, which infects up to 6.4% of transplant recipients in developed countries and up to 15% in areas where tuberculosis is highly endemic.Reference Aguado, Silva, Samanta and Singh4 Thus, exhaustive investigations, including the use of classical diagnostic microbiological and molecular assays are essential to ensuring that patients are fit for a SOT procedure or that they receive the correct antimicrobial therapy.
At the Istituto Mediterraneo per i Trapianti e Terapie ad Alta Specializzazione (IRCCS-ISMETT), in Palermo, Italy, a 46-year-old man with hereditary chronic renal disease requiring dialysis for the previous 2 years was evaluated for renal transplantation. He had a positive interferon-γ release assay, suggesting prior exposure to M. tuberculosis. This patient had no previous history of tuberculosis infection, and his chest imaging did not show any signal of M. tuberculosis infection in the lungs. To confirm the presence of mycobacterial antigens in the blood, an ELISpot and T-SPOT.TB (Oxford Immunotec, Oxford, UK) were performed according to the manufacturer’s instructions. Samples of sputum and urine were also inoculated into liquid BBL mycobacteria growth indicator tubes (Becton Dickinson, Milan, Italy) according to the supplier’s instructions to identify potential Mycobacteria. We obtained positive results for the presence of M. tuberculosis antigens in the blood and observed mycobacterial growth in the urine sample. Sputum samples were negative.
Because no M. tuberculosis was present in the patient’s sputum and because this particular ELISpot has previously shown some limitations,Reference Zhu, Liu, Li, Mei and Hu5 we decided to extract the genomic material of this strain for sequencing. Total mycobacterial DNA was extracted using a QIAamp UCP Pathogen Mini Kit (Qiagen, Venlo, Netherlands), as specified by the kit’s manual. Next-generation sequencing (NGS, full genome) and Sanger sequencing (16S rRNA gene, internal transcribed spacer (ITS1) 16S-23S and hsp65) were performed as published elsewhere.Reference Di Mento, Cuscino, Carcione, Cardinale, Conaldi and Douradinha2, Reference Turenne, Thibert and Williams6 The sequences derived from both techniques showed a homology >99% with the publicly available sequences of previously identified NTM M. saskatchewanense, from the complex M. simiae,Reference Fedrizzi, Meehan and Grottola7 previously only reported in the United StatesReference Springer, Stockman, Teschner, Roberts and Bo8 and Canada.Reference Turenne, Thibert and Williams6 This particular NTM had been isolated from a bronchiectasis patient, both from sputum and from thoracenthesis fluid, and was thought to have contributed to the deterioration of the patient.Reference Turenne, Thibert and Williams6 Genomic sequences of the M. saskatchewanense ISMETT strain were deposited in the NCBI public database (accession no. SRP149411).
The ELISpot was repeated 3 weeks later, yielding a negative result. Thus, and since we had confirmed that he was not infected with M. tuberculosis, the patient was not subjected to any antimycobacterial therapy. Because these interferon-γ release assays do not allow proper distinction between colonization and infection,Reference Hermansen, Thomsen, Lillebaek and Ravn9 we can only assume, due to its brevity, that the patient underwent a self-resolving episode of M. saskatchewanense colonization.
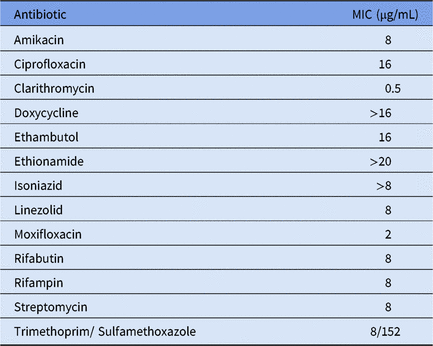
To the best of our knowledge, this is the first time this particular NTM has been detected outside North America. We assessed its antimicrobial sensitivity to several antibiotics. The minimal inhibitory concentrations (MIC) were determined using SlowMyco Sensititre plates (Thermo Fisher Scientific, Waltham, MA), according to the manufacturer’s instructions (Table 1). Our results show an antibiotic resistance profile similar to those previously observed in Canada with their local M. saskatchewanense strains.Reference Turenne, Thibert and Williams6 Some differences were also noted; for example, the MIC for rifampicin for all Canadian strains is 0.06 µg/mL, whereas our strain displayed a value of 8 µg/mL for the same antibiotic (Table 1). Although we cannot exclude the possibility that the differences observed may be due to the use of different assays, they may also indicate that the Italian M. saskatchewanense has either undergone mutations since it left North America or that its origin lies elsewhere. However, no epidemiological data about this NTM are currently available, and we can only speculate about whether it derives from the North American strains or from some other geographical location.
Table 1. Minimal inhibitory concentrations (MIC) of M. saskatchewanense ISMETT strain
To the best of our knowledge, we report for the first time the detection of a M. saskatchewanense clinical isolate in a European health facility. Our results highlight the fact that screening assays for TB detection in blood can produce misleading results and could lead to incorrect antimicrobial therapy. Careful evaluation for mycobacterial infection must be performed, and the organism must be identified and coupled with highly discriminating techniques such as NGS, as necessary.
Author ORCIDs
Bruno Douradinha, 0000-0002-9980-4505
Acknowledgments
We thank the Molecular Diagnostics at ISMETT team members for technical support and helpful insight. Informed consent of all inhouse patients is required by internal regulations before any experiments can be performed with their clinical samples.
Financial support
This study was supported by internal funding.
Conflicts of interest
All authors report no conflicts of interest relevant to this article.



