INTRODUCTION
Tuberculosis (TB) is a major infectious disease in developing countries [Reference Maartens and Wilkinson1]. It remains a considerable global public health concern as every year there are more than 9 million new cases [Reference Lönnroth2].
The direct and indirect costs of TB, and the social consequences, are often catastrophic for the individual patient and for the community [Reference Hanson, Floyd, Weil and Raviglione3]. Therefore, the rapid and precise detection of Mycobacterium tuberculosis is of primary importance for the administration of empirical antibiotic therapy and for the appropriate implementation of public health measures to effectively cut transmission and prevent the disease from spreading [Reference Griffith4]. In fact, an intensive effort is needed to develop new medical technologies for prevention, diagnosis, and treatment of TB [Reference Abu-Raddad5–Reference Bark8].
Polymerase chain reaction (PCR) has been an increasingly used tool for a more sensitive and rapid diagnostic of many infectious diseases, including TB. Over the years, a significant improvement in PCR technologies has been achieved with the development of real-time PCR (rtPCR). The main advantages of rtPCR are a shortened turnaround times; automation of the procedure, which reduces hands-on time; and a decrease in the risk of cross-contamination [Reference Espy9].
There has been a plethora of studies published over almost 20 years which have shown a considerable variation in sensitivity (Se) and specificity (Sp) of PCR compared to culture for M. tuberculosis and direct detection of acid-fast bacilli (AFB) [Reference Greco10, Reference Murray11]. The microbiological culture of M. tuberculosis is considered the ‘gold-standard’ for the diagnosis of TB, although a negative culture result does not necessarily exclude the disease. In clinical practice, many cases of TB are diagnosed based only on clinical and radiological findings, without confirmation either by direct AFB or culture. In fact, there are many parameters that might be evaluated to define TB [12] and, therefore, it is important to consider the clinical aspects in addition to the results of laboratories assays to establish the appropriate Se and Sp of a new methodology, such as PCR assay.
Reports from the US Centers for Disease Control and Prevention and the American Thoracic Society have shown that nucleic acid amplification tests (NAATs) improve diagnostic certainty, especially in TB culture-negative cases [Reference Lönnroth2]. However, to the best of our knowledge, there is no study comparing the efficacy of NAATs for M. tuberculosis detection in clinical samples compared to the classical ‘gold-standard’ (culture for M. tuberculosis), plus clinical criteria of TB.
In this study, we assessed the performance of two NAATs, an in-house nested PCR (nPCR) and rtPCR, for the diagnosis of pulmonary TB, considering the results of the M. tuberculosis culture as well as clinical criteria for the disease.
MATERIALS AND METHODS
Patients and isolates
This was a retrospective diagnostic method study performed from February to June 2011 at Hospital de Clínicas de Porto Alegre, a tertiary-care teaching hospital in southern Brazil. Respiratory samples were collected from hospitalized patients at the discretion of the attendant physician. Only one specimen per patient was selected for this study. Patients were eligible if they had a respiratory sample submitted for M. tuberculosis culture for suspected TB at the discretion of the attendant physician. They were excluded if they have been taking any anti-TB drugs at the time of sample collection. All patients with positive cultures with no exclusion criteria were analysed and, in order to increase statistical power, two patients with negative cultures were randomly selected for each culture-positive case.
After collection, respiratory samples were immediately transported to the laboratory, where they were stored at 2–8°C and were processed within the next 12 h. All samples were submitted for direct detection of AFB, using the Ziehl–Neelsen stain, and culture for mycobacteria. Samples were decontaminated with N-acetyl-l-cysteine-sodium hydroxide and concentrated by centrifugation. Part of the sediment was inoculated onto Ogawa medium (Laborclin, Brazil) and in Middlebrook 7H9 broth (Difco, Oxford, UK). The Ogawa medium was incubated at 37°C for 8 weeks and visually monitored, once a week, for colony growth. The Middlebrook 7H9 broth was cultured in a BACTEC™ MGIT™ 960 Mycobacterial Detection (BD Diagnostics, USA) automated system. The sediments were also aliquoted and frozen at −20°C for subsequent PCR assay.
DNA extraction
The sediments of the clinical samples were thawed and a 140-μl aliquot was treated with 10 μl proteinase K (Invitrogen, USA) at 56°C for 20 min. The DNA was extracted using a commercial extraction kit (QiaAmp Mini kit 250, Qiagen, Germany) according to the manufacturer's instructions.
nPCR
Nested PCR was used to detect M. tuberculosis complex DNA, as described previously [Reference Dora13] with a few modifications. The external primers TB290 (5′-GGC GGG ACA ACG CCG AAT TGC GAA-3′) and TB856 (5′-CGA GCG TAG GCG TCG GTG ACA AAG-3′) were used to generate a fragment of 600 bp. Ten microlitres of DNA were amplified in a 40-μl reaction mixture containing 1·25 U Taq polymerase (Super-Therm, JMR Holdings, UK), 1·5 mm MgCl2 buffer, 2·5 mm deoxynucleotide triphosphates (ABgene, UK), 25 μ m of external primers (Invitrogen) and distilled water, totalling 50 μl. The mixture was submitted to a denaturation period of 100 s at 94°C, followed by 33 cycles of amplification (each cycle consisted of 94 °C for 30 s, 65°C for 30 s, 72°C for 30 s). In the second reaction, the amplicon of the first DNA reaction was amplified to generate a DNA fragment of 170 bp, using the internal primers TB500 (5′-TAC TAC GAC CAG ATC-3′) and TB607 (5′-TTG GTG ATC AGC CGT-3′). Two microlitres of the amplicon from the first reaction were amplified in a 24-μl reaction mixture containing 1·25 U Taq polymerase (Super-Therm, JMR Holdings), 1·5 mm MgCl2 buffer, 2·5 mm deoxynucleoside triphosphates (ABgene), 25 μ m of internal primers (Invitrogen) and distilled water, totalling 26 μl. The mixture was submitted to a denaturation period of 45 s at 94°C, followed by 33 cycles of amplification (each cycle consisted of 94°C for 20 s, 52°C for 20 s, 72°C for 30 s).
We used distilled water as a negative control and the M. tuberculosis strain H37RV as positive control for all reactions. Visualization under ultraviolet light of the amplification products was done using 10 μl of the final volume after gel electrophoresis in 2% agarose with 0·5% ethidium bromide.
rtPCR
The primers and probes were selected from the gene sequence encoding the IS6110 (NCBI accession no. X17348). The probe was synthesized by Applied Biosystems (USA) and labelled with 6-carboxyfluorescein (6-FAM) attached to the 5′ end and MGB quencher linked to the 3′ end, as described previously [Reference Inoue14].
An internal control (IC) single-strand DNA was added to detect inhibition of amplification in all samples. The primers and probe for the IC have been described previously [Reference Inoue14]. The probe was synthesized with a VIC reporter fluorescein dye attached to the 5′ end and a TAMRA quencher linked to the 3′ end. Sequences of the primers, probes and IC are shown in Table 1.
Table 1. Sequences of the primers, probes and IC used in the real-time PCR assay

IC, Internal control, PCR, polymerase chain reaction.
The PCR mixture was performed with the Reagents DyNAmo™ Flash Probe qPCR kit (Finnzymes, USA). Two separate reactions were prepared for amplification, one for the template and another for the IC. The reactions consisted of 7 μl target DNA, 0·3 μ m forward and reverse primers, and 0·1 μ m probe, in a final reaction volume of 25 μl. A concentration of 1 pmol of the IC was used for the amplification procedure. The amplification was done in a ABI PRISM 7500 (Applied Biosystems) under the following conditions: 50°C for 2 min, 95°C for 10 min, followed by 45 cycles of amplification at 95°C for 15 s and 64°C for 45 s. PCR was repeated on all samples containing inhibitory substances that were determined by lack of amplification of the IC. The M. tuberculosis H37RV was used as a positive control in all rtPCR assays.
Clinical criteria
Medical records were reviewed and patients with suspected TB, according to the attendant physician, received an independent and blind evaluation by investigators and were further classified as having a high, undetermined or very low probability of pulmonary TB, according to the following criteria:
-
(1) High probability. Presence of cavitary lesion and/or consolidation, or miliary infiltrate in HIV-positive patients plus treatment for TB with no other antimicrobial drug associated, or cavitary or consolidative lesion at the upper pulmonary lobes or at the upper segment of inferior lobes, or miliary lesions with no confirmed fungal infection, or any pulmonary lesion with a positive AFB examination, regardless of culture results, plus treatment for TB with no other antimicrobial drug associated.
-
(2) Undetermined probability. Patients with the same radiological characteristics of ‘high probability’ but with empirical antimicrobial treatment for other suspected infections, or patients treated for TB with no radiological pulmonary lesions.
-
(3) Very low probability. Patients who had not received treatment for TB regardless of the presence and characteristic of pulmonary lesions, after exclusion of the diagnosis by the attendant physician, by other diagnostic methods (including diagnosis of neoplasia, other infections such as invasive mycosis, other non-TB mycobacteria (NTM) organisms, among others.
Sample size
Based on a rtPCR Se of 85% and a Sp of 95% [Reference Inoue14] with a precision of 10% and allocating positive and negative tests in a 1:1 ratio, the sample size was estimated in 100 patients (50 positives, 50 negatives).
Statistical analysis
The Se and Sp, were calculated using two distinct ‘gold-standards’ for pulmonary TB: (1) culture for M. tuberculosis and (2) culture plus clinical criteria. A positive pulmonary TB was considered if patients had (1) a culture positive for M. tuberculosis and (2) a culture positive for M. tuberculosis or high probability clinical criteria, respectively.
RESULTS
Four hundred and forty-seven clinical specimens were obtained from the respiratory tracts of patients with suspected pulmonary TB. Forty-two samples presented culture-positive for M. tuberculosis and 84 culture-negative samples were randomly selected from all negative samples recovered during the study period; two patients were further excluded owing to lack of complete clinical data in medical records. Of the 124 samples analysed, 57 were obtained from bronchoalveolar lavage (BAL), 27 from induced sputum and 40 from expectorated sputum specimens. Forty-eight samples were recovered from HIV-positive patients. Of 124 samples analysed we found seven NTM in the mycobacterial culture (the demographic data of patients are shown in Table 2).
Table 2. Demographic data of patients
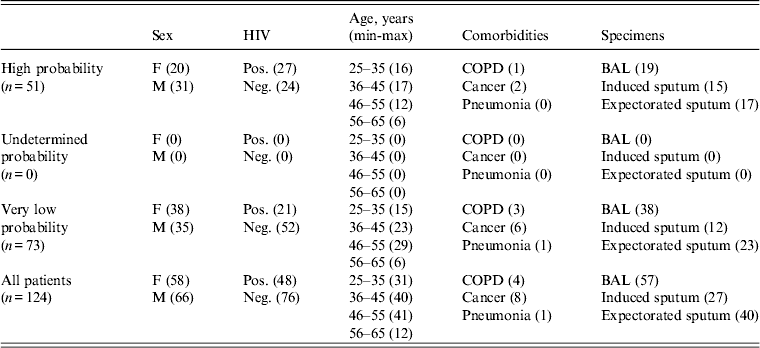
Values within parentheses indicate number of patients.
F, Female; M, male; HIV, human immunodeficiency virus; COPD, chronic obstructive pulmonary disease, BAL, bronchoalveolar lavage.
The limit of detection (LOD) of the nPCR and the rtPCR assays were 25 copies/μl and 1 copy/μl, respectively.
Seventeen AFB, 36 nPCR and 41 rtPCR assays were positive in the 42 culture-positive samples and 77 AFB, 76 nPCR and 75 rtPCR assays were negative in the 82 culture-negative samples, resulting in a Se and Sp of 97·6% [95% confidence interval (CI) 87·7–99·6] and 91·5% (95% CI 83·4–95·8) for rtPCR, and 85·7% (95% CI 72·2–93·3) and 92·7% (95% CI 84·9–96·6) for nPCR, respectively. Both NAATs presented high (>90%) Sp but the rtPCR presented higher Se than the nPCR (Table 3).
Table 3. Sensitivity, specificity and 95% confidence intervals using NAATs and AFB vs. culture

NAAT, nucleic acid amplification test; AFB, acid-fast bacilli; nPCR, nested polymerase chain reaction; rtPCR, real-time PCR.
Considering the results of the NAATs vs. culture plus clinical criteria, 22 AFB, 41 nPCR and 46 rtPCR assays were positive in the 51 patients considered as positive for pulmonary TB and 73 AFB, 72 nPCR and 71 rtPCR assays were negative in the 73 patients considered as negative for pulmonary TB, resulting in a Se and Sp of 90·2% (95% CI 79·0–95·7) and 97·3% (95% CI 90·5–99·2) for rtPCR and 80·4% (95% CI, 67·5–89·0) and 98·6% (95% CI 92·6–99·8) for nPCR, respectively. The Sp was considerably higher (>97%) for both NATTs but Se decreased (Table 4).
Table 4. Sensitivity, specificity and 95% confidence intervals using NAATs and AFB vs. culture plus clinical criteria

NAAT, Nucleic acid amplification test; AFB, acid-fast bacilli; nPCR, nested polymerase chain reaction; rtPCR, real-time PCR.
Six culture-negative samples were positive for M. tuberculosis by nPCR and seven by rtPCR. Of these, five (the same isolates for both assays) were classified as ‘High probability of TB’ by clinical criteria. One sample positive by nPCR and two by rtPCR (the same nPCR plus another sample) were classified as ‘Very low probability of TB’. There were six samples of positive culture with negative results by nPCR. One of these six culture-positive/nPCR-negative samples was also negative by rtPCR. The accuracy of nPCR and rtPCR was 90% and 94%, respectively.
DISCUSSION
The rapid and precise identification of M. tuberculosis plays an important role in the clinical decision in the management of patients with suspected pulmonary TB. In this study we compared the results of two NAATs with mycobacterial culture as well as with culture plus clinical data of patients with suspected TB.
As previously shown by many other studies, Se results of both NAATs evaluated in our study were significantly higher than direct detection of AFB when culture for M. tuberculosis solely was considered as gold standard. Indeed, Se of commercial NAATs have ranged from 66% to 96% [Reference Greco15, Reference Antonenka16] and from 84% to 97%, when in-house NAATs were evaluated [Reference Greco10, Reference Inoue14, Reference Gomez17–Reference Marlowe20]. When Se results were compared between both NAATs, rtPCR presented higher Se values (97·6%) than nPCR (85·7%), although there was a small overlap between the 95% CI of the Se results of these tests. Despite the relatively high Se, some samples with culture-positive results could still not be detected by NAATs. This was more notable for nPCR, which presented six false-negative results, than for rtPCR which presented only one false-negative result. This different Se values obtained in NAATs can be explained by the LOD, which was higher for nPCR than for rtPCR (25 and 1 copies/μl, respectively).
A slight decrease of Se in NAATs was observed in comparison with culture plus clinical criteria (80·4 and 90·2% for nPCR and rtPCR, respectively) in relation to culture solely (85·7% and 97·6%, respectively). In fact, there were an additional four samples, which were considered TB only by clinical criteria, which were negative for M. tuberculosis by both NAATs. One of these samples was recovered from an HIV patient with proven pleural TB with low and medium lobes consolidation of the right lung. The other three samples were from patients with positive direct examination of AFB, but negative culture. One was an HIV patient with low and medium lobes consolidation of the right lung, the other was a non-HIV patient with a previously treated TB whom data from radiological findings could not be recovered and the third sample was a non-HIV patient with previous TB and a cavitary lung lesion. In fact, in clinical practice, clinicians are recommended to treat for TB when there is a positive AFB smear in the presence of clinical or radiological findings suggesting TB [21, 22]. It might be considered that the AFB-positive samples could be due to other organisms, e.g. NTM.
Both nPCR and rtPCR in our study presented high Sp (>90%) rates for the diagnosis of M. tuberculosis, similar to the Sp observed in previous studies, which ranged from 85% to 98% for commercial tests, and 74–94% for in-house tests [Reference Greco10, Reference Inoue14, Reference Greco15].
As could be expected the Sp value increased when NAATs were compared with the culture plus clinical data in relation to culture alone, since many cases of TB cannot be confirmed by culture. The nPCR and rtPCR presented six and seven positive results, respectively, which were negative for M. tuberculosis in the microbiological culture. However, five of them had a ‘High probability’ of TB according to clinical criteria. Nevertheless, two cases were classified as ‘Very low probability’ of TB, one positive by both NAATs and the other positive only by rtPCR.
These latter occurrences could explain the reason why the Sp rates were not 100% for NAATs and why Sp was slightly higher for nPCR compared to rtPCR. It should be noted that these two patients presented a history of previous TB and this could account for the positivity of NAATs, since M. tuberculosis DNA has also been detected in patients with previous TB with no active disease [Reference Dinnes23].
The Se and Sp results of the NAATs were very similar after analysis according to the HIV status of patients (data not shown). It is of note that we found seven NTM in the mycobacterial culture and these were all negative for nPCR and rtPCR assays.
Our study confirms that NAATs may be useful for rapid and precise diagnosis of respiratory TB, with high Sp rates. We did not find a significant difference in the performance of either NAATs, regardless the comparison, although rtPCR presented Se results slightly better than nPCR, while the latter showed better results for Sp. Issues such as cost of each test in specific settings and turnaround times must be considered when choosing which test should be employed. Usually, rtPCR has been used as an alternative to conventional PCR, since it presents elevated accuracy, short turnaround times and minimal hands-on time required for the assays [Reference Espy9, Reference Halse24].
In summary, NAATs targeting the IS6110 of M. tuberculosis improve the accuracy of the diagnosis of pulmonary TB and may lead to potential positive effects for clinical management and control of the disease. There was no significant difference in the performance of either test, and the decision as to which test to use may rest with issues such as costs vs. turnaround times.
ACKNOWLEDGEMENTS
This work was supported by Fundo de Incentivo à Pesquisa e Eventos do Hospital de Clínicas de Porto Alegre (FIPE, project no. 11-0076), National Council for Scientific and Technological Development (CNPq), Ministry of Science and Technology, Brazil.
DECLARATION OF INTEREST
None.






