INTRODUCTION
Group A rotaviruses are recognized as the single most common cause of non-bacterial acute gastroenteritis. These viruses infect children throughout the world in the first few years of life irrespective of geographic location and socioeconomic status [Reference Cook1]. It has been estimated that rotavirus infections are associated with about 600 000 deaths of children aged <5 years annually in developing countries [Reference Parashar2].
Rotaviruses belong to the Reoviridae family and possess a genome of 11 segments of double-stranded RNA. A dual classification system has been established for rotaviruses based on two outer layer proteins, VP7 and VP4, respectively. The major neutralization epitopes on the VP7 protein have been designated as antigenic region A (amino acids 87–99), region B (amino acids 145–150) and region C (amino acids 211–223) [Reference Dyall-Smith3]. Earlier, typing of this protein involved monoclonal antibody (mAb)-based serotyping ELISA. However, a lack of readily available mAbs hampered VP4 serotyping [Reference Estes, Fields, Knipe, Howley, Chanock, Monath and Melnick4]. In recent years, application of molecular methods has allowed identification of 19 G types and 27 P types in human and animal infections [Reference Matthijnssens5]. Although clinically and epidemiologically, G1–G4 serotypes and P[8] and P[4] genotypes have acquired global importance [Reference Gentsch6, Reference Palombo7], geographical variation in the distribution of G and P types along with emergence of unusual G/P combinations have also been reported from different countries [Reference Santos and Hoshino8, Reference Gentsch9].
Data on rotavirus disease and strain surveillance have been reported from various parts of India [Reference Kang10]. In a study conducted on faecal specimens collected from patients hospitalized with acute diarrhoea from western India during 1990–1994, serotyping of rotavirus strains was carried out using mAbs to the G1–G4, G6 and G10 serotypes [Reference Kelkar and Ayachit11]. Only 25·6% of the strains were typed whereas the majority of the strains remained untypable due to non-reactivity or multireactivity. The aim of the present study was to characterize VP7 and VP4 genes of rotavirus strains representing both the categories.
METHODS
Specimens
The faecal specimens investigated in the study were collected from patients with acute diarrhoea admitted at local hospitals in Pune in 1990–1994. The patients included 85 children aged 3 months to 7 years, and five adults aged 20–38 years. Twenty-one typable (G1/G2/G3/G4) and 56 untypable strains as monitored by mAb-based serotyping ELISA [Reference Kelkar and Ayachit11] were selected for the study. Thirteen specimens collected during 2000–2002 and categorized as untypable were also investigated. Informed consent was obtained from the parents/guardians of children with diarrhoea and from adult patients prior to the sample collection.
RNA extraction and G and P typing
The faecal samples were processed to prepare 20% suspensions in 0·01 m PBS (pH 7·2), centrifuged at 10 000 rpm for 10 min at 4°C and stored at −70°C until tested. The viral RNA was extracted using Trizol LS (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The VP7 and VP4 genes were amplified by one-step RT–PCR and genotyping was carried out by multiplex PCR using primer sets and protocols described previously [Reference Gouvea12–Reference Berois15]. The PCR products were electrophoresed in 2% agarose gels containing ethidium bromide and visualized under a UV transilluminator.
Nucleotide sequencing
The strains that could not be typed by multiplex PCR were subjected to sequencing of first PCR product using Beg9 and End9 primers for the VP7 gene and con2 and con3 primers for the VP4 gene [Reference Gouvea12, Reference Gentsch14]. Briefly, the amplicons of VP7 and VP4 genes were purified on minicolumns using the QIAquick Gel Extraction kit (Qiagen, Hilden, Germany). Sequencing was carried out using ABI-Prism Big Dye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) and an ABI-Prism 310 Genetic Analyzer (Applied Biosystems). The nucleotide sequences of VP7 and VP4 genes of rotavirus strains reported in this study were deposited in the GenBank under the accession numbers DQ 886942–887001 and DQ 887002–DQ 887060, respectively.
Phylogenetic analysis
Nucleotide sequences of both the genes were aligned in Clustal W and phylogenetic trees were generated using a Kimura two-parameter distance model and Neighbour-Joining (NJ) algorithm [Reference Thompson16, Reference Kumar, Tamura and Nei17]. The reliability of the phylogenetic tree was tested by applying a bootstrap test with 1000 bootstrap replications.
RESULTS
G typing
The VP7 genes of all 90 strains in the study were typed. Of these, 35 were typed by multiplex PCR and the remaining 55 by sequencing the first PCR product. Of the 21 strains typed by ELISA, 20 showed the presence of the same G type in multiplex PCR/sequencing (G1–6, G2–5, G3–5 and G4–4) while one strain showed discordance (G4 vs. G2). Among the 69 rotavirus strains untypable by ELISA, G1–G4 and G9 specificities were detected in 35, 27, 2, 2 and 2 strains, respectively. Overall the G-type distribution in 1990–1994 showed the presence of G1 in 50·6%, G2 in 31·2%, G3 in 9·1%, G4 in 7·8% and G9 in 1·3% of the strains while in 2000–2002 it was 30·8% for G1, 61·5% for G2 and 7·7% for G9 (Table 1).
Table 1. Distribution of G and P genotypes in Pune strains

Unusual P/G combinations are highlighted in bold.
Numbers not in parentheses indicate strains from 1990 to 1994. Numbers within parentheses indicate strains from 2000–2002.
P typing
Thirty-one of the 90 rotavirus strains were typed by multiplex PCR whereas in 59 strains the VP4 gene was amplified by the primers and types were determined on the basis of sequences. P-type distribution showed P[8], P[6], P[4] and P[19] specificities, respectively, in 50%, 6·7%, 42·2% and 1·3% of the strains collected in 1990–1994 whereas in 2000–2002 P[8] was detected in 38·5% along with P[4] in 61·5% of the strains (Table 1).
Combinations of G and P types
The G- and P-typing data obtained on the 77 strains from 1990 to 1994 showed the presence of common rotavirus types in 79·2% of the cases (G1P[8], 29; G2P[4], 23; G3P[8], 4; G4P[8], 5) along with 1·3% G9P[8] (Table 1). G- and P-typing data on 13 strains from 2000–2002 indicated four G1P[8] (30·7%), eight G2P[4] (61·5%) and one G9P[8] (7·6%) (Table 1). The remaining strains (19·5%) showed unusual combinations of G and P types. These strains included, five G1P[6] (5·6%), four G1P[4] (4·4%), one G2P[8] (1·1%), three G3P[4] (3·3%), one G4P[6] (1·1%) and one G1P[19] (1·1%) (Table 1).
Nucleotide sequencing and phylogenetic analysis
Nucleotide sequences of 34 (61·8%) strains genotyped as G1 showed 90–94% identity with reference strain Wa. However, their identities were higher (99·2–100%) compared to the MVD strain isolated in Montevideo, Uruguay as confirmed phylogenetically (Fig. 1). Thirteen strains (23·6%) genotyped as G2 showed 91·9–93·7% identity with reference strain DS-1 and 96·2–99·8% identity with Asian strains (253 from Bangaluru, SC27 from Kolkata, TW93107 from Taiwan and 23G2 from Thailand), as can also be seen in Figure 1. One of the G2 strains (NIV-935070) with P[8] specificity, showed 3·3–5·9% variation from the DS-1 strain and other Pune strains with G2P[4] specificity from the 1990s. It is noteworthy that the specimens from the 1990s and 2000s revealed two major genetic lineages with a 3·5% difference in the nucleotide sequence (Fig. 1). The four G3 and three available G4 sequences showed 95·7–97·8% and 97·9–98·4% nucleotide identity with reference strains YO and Hochi from Japan, respectively. The only detected G9 strain (NIV-006835) appeared to be most closely related to strain GH3574, isolated in Ghana, both genetically (99·9%) and phylogenetically (Fig. 1).
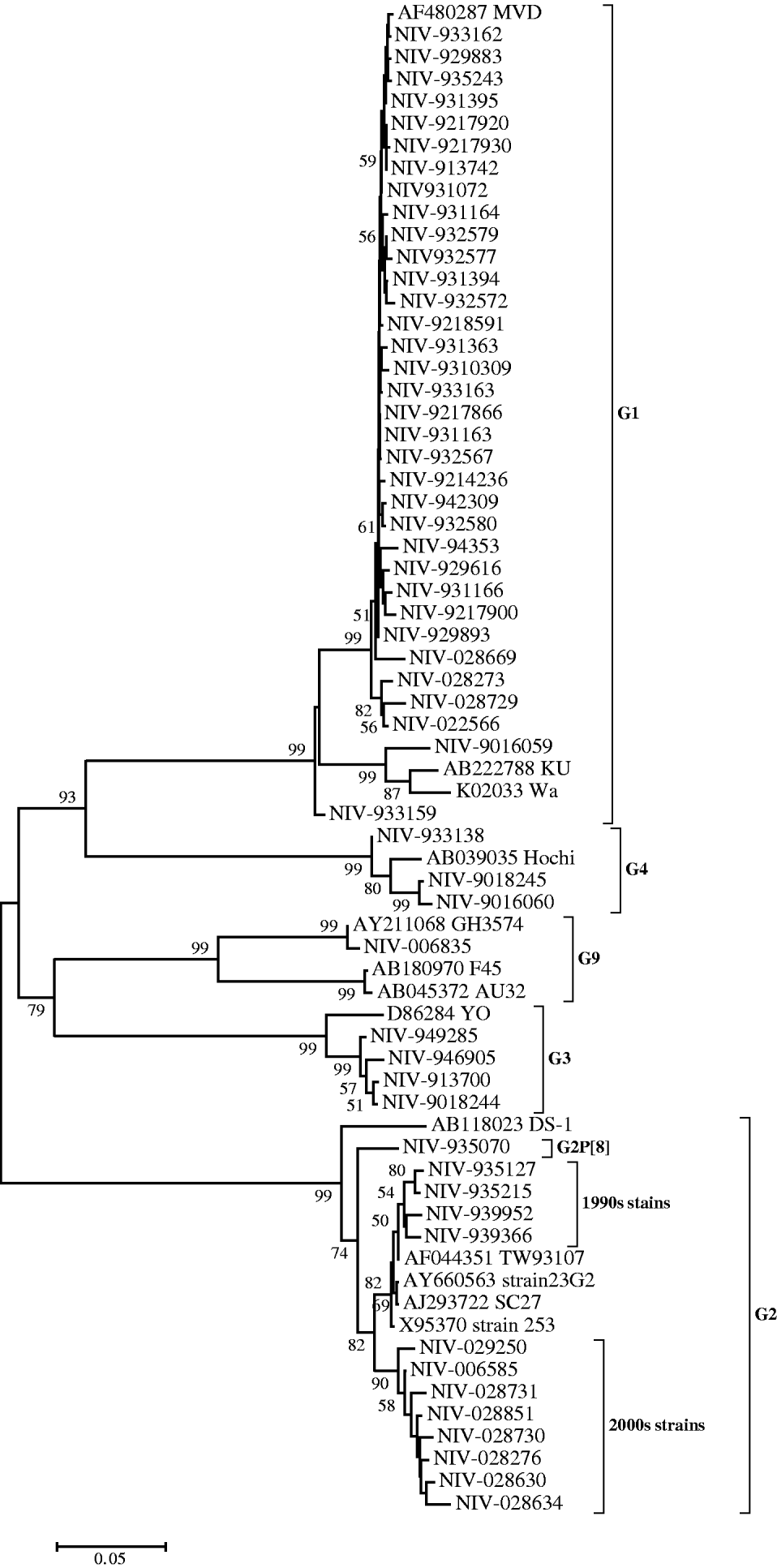
Fig. 1. Phylogenetic tree depicting the genetic relatedness of partial VP7 gene sequences of Pune rotavirus strains with other rotavirus strains. The strains are identified by the abbreviation NIV (National Institute of Virology) followed by repository number. Scale indicates genetic distance.
The nucleotide sequences of 33 (55·9%) strains from Pune, typed as P[8], appeared closely related to the RMC100 (India), OP351 (UK) and 3084376 (Australia) strains (98·8–99·8%), whereas the identity with strain KU from Japan, ranged from 92·9 to 94·4%. These relationships were also seen phylogenetically (Fig. 2). The P[8] strain (NIV-935070) with G2 specificity showed 11·7–13·3% variation with these reference strains and other Pune strains with G1P[8] specificity from the 1990s. Four out of 59 strains (6·7%) showed the P[6] specificity with 94·9–96·4% identity with prototype strain ST3 from Mexico and 98·9–99·1% identity with strains 429-np and US1205 from the United States. Twenty-one (35·5%) strains typed as P[4] showed 93·4–95·1% nucleotide identity with RV5 and DS-1 strains of Australia and the United States, respectively, and 97–99·8% identity with Indian strain RMC/G66. Interestingly, one strain with the rare genotype P[19] showed 96·7% identity and close phylogenetic clustering with the Mc323 strain from Thailand (Fig. 2).
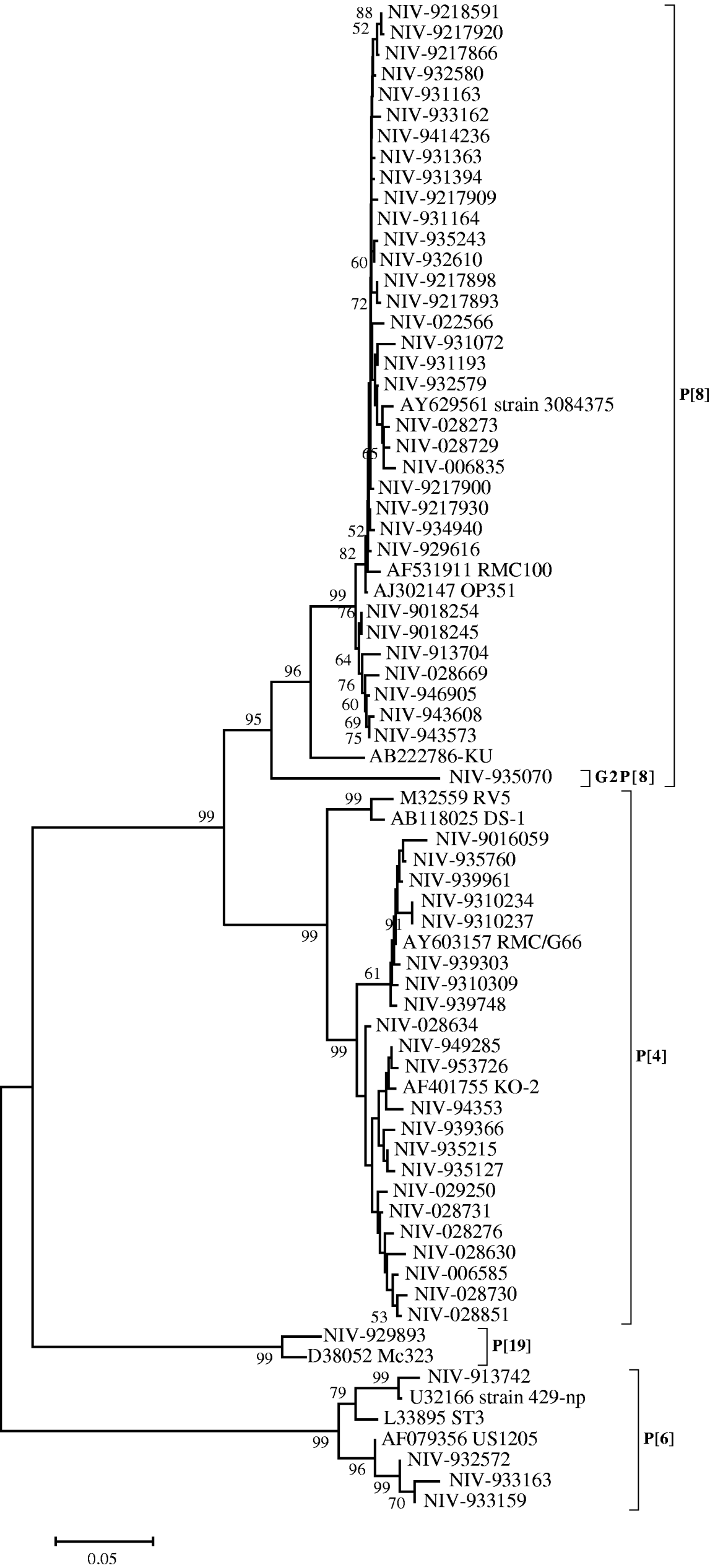
Fig. 2. Phylogenetic tree depicting the genetic relatedness of partial VP4 gene sequences of Pune rotavirus strains with other rotavirus strains. The strains are identified by the abbreviation NIV (National Institute of Virology) followed by repository number. Scale indicates genetic distance.
Analysis of the sequences of Pune strains that could not be typed by multiplex PCR showed mismatches in the primers specific to G1 (A-G at position 329), G3 (G-T, G-A, T-C, T-C at positions 486, 489, 490, 496, respectively), G9 (A-T, A-G, C-A at positions 759, 765, 773, respectively), P[4] (G-A at two positions, 478 and 489) and P[8] (GATA-ATCG at positions 348–345 and T-G at position 350) (Table 2).
Table 2. Comparative analysis of the nucleotide sequences of Pune strains and the primers used in the study

* For each genotype the original primer sequence is given first followed by sequences of the Pune strains.
The highlighted nucleotides indicate mismatches in the sequences of primers and Pune strains.
Amino-acid substitutions in VP7
The amino-acid alignment of G1 strains revealed two substitutions, Asn94Ser and Asp97Glu in region A, two substitutions Ser147Asn and Val149Glu in region B and one substitution Met217Thr in region C compared to KU strain of serotype 1. G2 strains showed two substitutions Ala87Thr and Asp96Asn in antigenic region A with no change in regions B and C compared to S2 strain of serotype G2. The substitutions in G3 strains were Glu99Lys in region A and Asp222Glu in region C compared to YO strain of serotype 3 while amino-acid sequences of G4 strains showed substitutions Arg143Lys and Lys149Glu in region B only. One strain with G9 specificity showed substitutions Ala87Thr in region A and Thr208Ile and Ala220Thr in region C (Table 3).
Table 3. Amino-acid substitutions in the antigenic regions of VP7 protein of Pune rotavirus strains

* Utilized for mAb generation.
DISCUSSION
In our earlier studies we utilized mAb-based ELISA for the characterization of rotavirus strains [Reference Kelkar and Ayachit11]. In these studies mAbs were found to remain either non-reactive or generate non-specific reactions with specimens containing rotavirus strains. In the present study, we report multiplex PCR and/or nucleotide sequencing based characterization of these strains.
It is worth noting that the substitutions of amino acids were identified in the antigenic regions of the majority of the rotavirus strains isolated in Pune compared to the reference strains G1 (KU), G2 (S2), G3 (Yo), G4 (ST3), and G9 (AU32) that were utilized in mAb generation for serotyping (Table 1). Mutations in antigenic regions A and C have been previously reported in several variants of G1 strains, such as KU, RV4 and K8 that showed different reactivity with mAbs [Reference Taniguchi18–Reference Kobayashi, Taniguchi and Urasawa20]. The mutation in antigenic region C was noted to be responsible for a tenfold increase in resistance to neutralization by polyclonal antiviral serum [Reference Dyall-Smith3]. Similarly, the detection of antibody escape mutants and identification of variability associated with genetic drift among strains belonging to G2 genotypes have been described [Reference Iturriza-Gomara21].
Since 1995, circulation of G9 serotype has been reported from many countries in the world [Reference Santos and Hoshino8, Reference Armah22, Reference Kirkwood23]. This serotype has been identified by mAb-based ELISA and was found to be prevalent in Indian children with diarrhoea [Reference Ramachandran24, Reference Awachat and Kelkar25]. The present study indicated its circulation since 1994 in India.
Detection of unusual rotavirus strains G1P[6], G1P[4], G2P[8] and G4P[6] in the present study have been reported previously from India [Reference Ramachandran26, Reference Das27]. Interestingly, the divergence shown by the VP7 and VP4 genes of NIV-935070 strain with G2P[8] specificity is noteworthy compared to the strains with G1P[8] and G2P[4] specificities. The three G3P[4] strains and the single G1P[19] strain are being reported for the first time in India in the present study. The G3P[4] combination has been identified from other geographic regions also [Reference Santos and Hoshino8]. G1 is known to be associated with P[4], P[6], P[8] and P[9] [Reference Santos and Hoshino8, Reference Ramachandran26, Reference Abdel-Haq28, Reference Leite29]. The G1 strain found in association with P[19] in the present study is a rare genotype that was first described in a porcine rotavirus strain 4F by Burke et al. [Reference Bruke, McCrae and Desselberger30]. Further, P[19] strains (Mc323 and Mc345) identified in human rotavirus infections in Thailand were identified as being closely related to porcine P[19] strain [Reference Okado31]. Thus, the strain G1P[19] reported in this study may be a human–porcine reassortant.
The P[6] genotype previously thought to be restricted to asymptomatic infections has also been identified in symptomatic infants recently [Reference Iturriza-Gomara32]. Three P[6] strains of our study clustered with strains causing symptomatic infections while one was closer to a strain associated with an asymptomatic infection (Fig. 2).
It should be noted that the molecular techniques employed in the present study provided typing of rotavirus strains that escaped reactivity or showed multireactivity with mAbs. A high correlation (95%) was found between serotyping ELISA and genotyping techniques for typable specimens. The discordance seen in a small proportion of samples was probably due to non-specific reactions in the ELISA. It is interesting to note that the uncommon combinations of G and P types caused a significant number of infections (Table 1). The segmented nature of the rotavirus genome is known to cause reassortments among the genes. This is facilitated in vivo with multiple infections. However, despite the occurrence of infections with uncommon strains mixed infections were not detected in patients in the present study by multiplex PCR and sequencing.
It should be noted that 61·1% and 65·5% of the strains of the present study remained untypable for VP7 and VP4 genes, respectively, by multiplex PCR. Sequence analysis of the strains showed mismatches in the primers specific to G1, G3, G9, P[4] and P[8] (Table 2). This is in agreement with earlier studies reporting the failure to type rotavirus strains due to mismatches in the primer sets [Reference Rahman33–Reference Iturriza-Gomara35].
To summarize, the present study documents predominance of rotavirus G1P[8] and G2P[4] types in the years 1990–1994 and 2000–2002, respectively. It also revealed significant contribution of unusual rotavirus strains in causing diarrhoea. The data are of epidemiological importance and imply that such strains are circulating in western India. Complete molecular characterization of these strains will be useful in the identification of additional components that may be needed in current development of rotavirus vaccines.
ACKNOWLEDGEMENTS
The authors are grateful to Dr A. C. Mishra, Director, NIV for his support. Financial support provided to J.K.Z. by the Indian Council of Medical Research (ICMR), New Delhi, India is gratefully acknowledged.
DECLARATION OF INTEREST
None.







