Se is an essential mineral nutrient required to support the expression of some twenty-five proteins, each of which contains selenocysteine residues as essential constituents(Reference Lobanov, Hatfield and Gladyshev1). These selenoproteins have diverse functions including antioxidant protection(Reference Toppo, Flohé and Ursini2), thyroid hormone metabolism(Reference Duntas3) and Se transport(Reference Burk and Hill4). Selenoprotein expression can be reduced by deprivation of Se. SNP can also affect selenoprotein function, as in the case of cytosolic glutathione peroxidase (GPX1)(Reference Shuvalova, Kaminnyi and Meshkov5). In fact, GPX1 genotype has been associated with cancer risk(Reference Hu and Diamond6).
High Se status has been associated with reduced cancer risk, and perhaps increased risk of type 2 diabetes. The Nutritional Prevention of Cancer (NPC) Trial showed that increasing Se intake could reduce colon and prostate cancer risk(Reference Clark, Combs and Turnbull7), at least for individuals with plasma Se levels < 106 ng/ml(Reference Duffield-Lillico, Dalkin and Reid8). However, the same study indicated that Se-supplemented subjects whose plasma Se levels had increased to 180–200 ng/ml may have had increased risk of type 2 diabetes(Reference Stranges, Marshall and Natarajan9). Elevated diabetes risk was observed for subjects in the upper quintile of plasma Se ( ≥ 137·7 ng/ml) in the third National Health and Examination Survey (NHANES)(Reference Bleys, Navas-Acien and Gualler10), and among subjects in the upper quartile of plasma Se ( ≥ 147 ng/ml) in NHANES 2003–4(Reference Laclaustra, Navas-Acien and Stranges11). While increased diabetes risk was not found in response to Se-supplementation in the Se and Vitamin E Cancer Prevention Trial(Reference Lipmann, Klein and Goodman12), neither was protection against cancer detected in that cohort of men of relatively high baseline Se status (mean plasma Se 136·5 ng/ml). Chiang et al. (Reference Chiang, Shen and Kengeri13) rationalised these apparently discrepant results by suggesting that health risk may be associated with Se status according to a ‘U’-shaped dose–response curve. They demonstrated that at high dietary exposures Se can cause DNA damage in cells and induce apoptosis. This may be a mechanism whereby Se exerts its anti-cancer effect; when manifest in non-tumorigenic cells, it may also indicate Se-toxicity.
Such a bi-modal dose–response relationship would suggest that Se may be beneficial for only some individuals, such as those of relatively low Se status and that doses lower than those used previously (200 μg/d(Reference Duffield-Lillico, Dalkin and Reid8, Reference Lipmann, Klein and Goodman12)) may be effective. Should this be the case, then food-based approaches using low Se doses would be facilitated by understanding the quantitative relationship of Se intake, biomarkers of Se status and genetic or sex-based modifying factors.
Projecting the Se intake required to support an Se status target requires understanding the relationship of biomarkers of Se status and level of Se intake. Only a few multi-dose studies have been conducted from which to make such projections. Those studies have used various forms of Se supplements, including selenite which produces only minimal increases ( < 20 %) in plasma Se(Reference Duffield, Thomson and Hill14–Reference Burk, Norsworthy and Hill16), unless subjects are of low Se status, i.e. plasma Se < 55 ng/ml(Reference Xia, Hill and Byrne17). The only plasma proteins into which selenite-Se can be incorporated, selenoprotein P (SEPP1) and GPX3, are maximally expressed in non-deficient subjects. In contrast, the other major supplemental forms, selenomethionine (SeMet) or SeMet-containing supplements (Se-enriched yeast), increase plasma Se in subjects of both low(Reference Xia, Hill and Byrne17) and higher Se status(Reference Duffield, Thomson and Hill14–Reference Burk, Norsworthy and Hill16, Reference Ravn-Haren, Krath and Overvad18–Reference Combs, Midthune and Patterson21), due to the non-specific incorporation of SeMet into proteins in lieu of methionine (Met). Steady-state plasma Se concentrations are not reached for at least 9 months of SeMet supplementation(Reference Combs, Midthune and Patterson21).
We, therefore, conducted a 12-month, randomised, double-blind, multi-dose, placebo-controlled intervention trial to determine the quantitative effects of supplemental SeMet on multiple biomarkers of Se status of healthy, Se-adequate adults. Our hypothesis was that the responses of such individuals could be used to impute the amount of oral Se necessary to raise plasma Se to given target concentrations. We previously presented the baseline findings from that study, comprising a complete assessment using multiple biomarkers of Se status(Reference Combs, Watts and Jackson22). Here, we present the responses to supplementation including subgroup analyses examining the impact of sex and selenoprotein genotype on Se biomarkers.
Subjects and methods
This study involved healthy men and women living in the vicinity of Grand Forks, ND, who volunteered and met the eligibility criteria as described previously(Reference Combs, Watts and Jackson22). Volunteers resided in their homes for the duration of the year-long study; they were requested to abstain from Brazil nuts (a significant source of Se) and dietary supplements providing ≥ 50 μg Se/d for the length of the study.
Sample size was determined from simulations based on results from the NPC trial(Reference Clark, Combs and Turnbull7) which were used to estimate the relationship between the observed change in plasma Se after 12 months of supplementation and the effective Se dose (mcg Se/d per kg0·75). For each sample size modelled, 1000 sets of random numbers were generated with the following characteristics: equal numbers of men and women randomised to each of four doses of Se (0, 50, 100 and 200 μg/d); mean body weights of the subjects in the NPC trial (women: 65 (sd 12) kg; men: 80 (sd 12) kg); predicted increase in plasma Se after 1 year of supplementation across all treatment groups of 40 (sd 10) ng/ml; a correlation structure similar to that observed in the NPC trial. These simulations indicated that, while 100 subjects would give 100 % power (α = 0·05), the length of the resulting 95 % CI would exceed 20 % of the predicted doses. Therefore, we set our recruitment target at a minimum of 240 subjects, which would give a CI of approximately 15 % of predicted dose and accommodate 20 % attrition/non-compliance. While no sex comparisons were planned, effort was made to recruit similar numbers of men and women. A flow chart showing allocation to treatment and experimental design is shown in Fig. 1.

Fig. 1 Flow chart of study design and subject participation.
Subjects visited the Research Center 2 weeks before and on the day of initiation of treatment, and at monthly intervals thereafter for 1 year. At each visit anthropometry was performed, a pill count-back was made and subjects received the next calendar pack of supplements. Each volunteer was given a cash honorarium pro-rated for the duration of his/her participation in the study.
This study was conducted according to the guidelines laid down in the Declaration of Helsinki. Oversight was provided by the University of North Dakota Human Subjects Committee (Grand Forks, ND, USA), which reviewed and approved the protocol. The purposes and procedures of the study were explained to the volunteers verbally and in writing, and written informed consent was obtained from each volunteer before his/her participation. The study is registered in the Clinical Trials Registry (ClinicalTrials.gov ID no. NCT00803699).
Dietary supplement
Subjects were randomly assigned to treatments consisting of a daily oral supplement containing either 0, 50, 100 or 200 μg Se as l-SeMet and an excipient (dicalcium phosphate) in #2 gelatin capsules (Sabinsa Corporation, Princeton, NJ, USA). The analysed Se contents of these treatments were 0·2, 56·0, 101·2 and 204·1 μg/capsule, respectively. Supplement capsules were provided in numbered 31-d bubble packs at each monthly visit to the Research Center. Compliance with the study protocol was ascertained in subject interviews and by capsule count-backs. Body weight was also recorded at each of these visits so that Se dose could be adjusted for metabolic body size (kg0·75) in the statistical analysis of the data.
Dietary intake assessment
Dietary intake was assessed at the time of randomisation to treatment and quarterly by a single self-administered FFQ patterned after the Harvard Service FFQ format(Reference Suitor, Gardner and Willett23), as described(Reference Combs, Watts and Jackson22).
Anthropometry
Body weight was measured using an electronic scale. Height was measured at the beginning of the study using a wall-mounted stadiometer.
Sample collection and preparation
Blood and 24 h urine were collected 2 weeks before and on the day of randomisation to treatment as previously described(Reference Combs, Watts and Jackson22), as well as at quarterly intervals. Blood was collected by venepuncture in duplicate 7 ml samples into heparinised, EDTA-treated or non-treated glass tubes. Aliquots of whole blood were subjected to low-speed centrifugation to prepare erythrocyte, buffy coat, plasma and serum fractions. Urine (24 h samples) was collected in sterile polycarbonate bottles. These specimens were held at 4°C pending the completion of screening analyses; excess portions were held at − 80°C. Lymphocytes were prepared from whole blood after lysis of erythrocytes, followed by washing, centrifugation and resuspension in 1 % paraformaldehyde (1 h at 2–8°C) to stabilise the cytoplasm, and final resuspension in physiologically buffered saline. Buccal cells were collected using a sterile toothbrush according to Paetau et al. (Reference Paetau, Rao and Wiley24); cells were lysed in distilled water, and lysates were held at − 80°C for analysis.
Analytical methods
Biochemical analyses of methylation status and biomarkers of Se status for the cohort at baseline, before supplementation, were performed as previously described(Reference Combs, Watts and Jackson22). Genotyping was carried out(Reference Combs, Watts and Jackson22) for selenoprotein SNP: GPX1 (rs1050450)(Reference Hu and Diamond25), GPX4 (rs713041)(Reference Villette, Kyle and Brown26), SEPP1 (rs3877899 and rs7579)(Reference Méplan, Crosley and Nicol27) and SEP15 (rs5845)(Reference Hu, Korotkov and Mehta28). Se status was ascertained on the basis of the activity of GPX and the protein level of SEPP1 in serum, as well as the amounts of Se in plasma, buccal cells and urine. The activity of GPX3 (EC 1.11.1.9) was determined in plasma by the method of Lawrence & Burk(Reference Lawrence and Burk29). The amount of SEPP1 was measured in serum by an enzyme-linked immunoassay(Reference Hollenbach, Morgenthaler and Struck30). Se was determined in plasma, buccal cells and urine by automated electrothermal atomic absorption spectrophotometry using a reduced palladium matrix modifier and an instrument equipped with L'Vov platforms(Reference Clark, Combs and Turnbull7). Certified Standards were used (Alfa Aesar, Ward Hill, MA, USA; Perkin Elmer, Waltham, MA, USA and CPI, Santa Rosa, CA, USA) to prepare a calibration set daily with each batch. Calibration validation and calibration blanks were included at the beginning and end of the daily batch and at 10 % frequency. Matrix effects for plasma and urine were evaluated using quantitative plasma and urine standards (NIST, Gaithersburg, MD, USA; Seronorm, Billingstad, Norway and Utak, Munich, Germany) to assess the percentage recovery of the analyte in these sample matrices. There is no commercially available quantitative standard for Se in buccal cells; therefore, matrix effects of buccal cell preparations were accounted for by performing spike recoveries using certified calibration standards added directly to one of the samples.
Selenium distribution in blood
The components of plasma Se were determined from: (1) total plasma Se; (2) measured plasma GPX3 activities; (3) measured serum SEPP1 levels. To determine the amount of GPX3-derived selenocysteine from the activity of the enzyme, a rate constant of 2·8 × 104 nmol/min per mg, molecular weight 92 kD and a stoichiometry of 4 g-atoms Se per mole GPX3 were assumed(Reference Broderick, Deagen and Whanger31). For selenocysteine in glycosylated SEPP1, average molecular weight 60 kD and a stoichiometry of 9·9 g-atoms Se per mole as selenocysteine(Reference Saito, Sato and Hirashima32) were assumed. Due to its affinity for heparin, SEPP1 was measured in serum; an inherent assumption was that insignificant quantities of SEPP1 protein precipitated from serum. The difference between the total measured Se and the amounts of Se corresponding to the activity of GPX3 and measured amount of SEPP1 was taken as the amount of Se incorporated non-specifically into plasma proteins. This is presumed to be predominately protein-bound SeMet, as only very low amounts of low molecular weight Se species appear to occur in plasma( < 1–2 % of total Se)(Reference Kokarnig, Kuehnelt and Stiboller33).
Apoptosis and DNA damage assessment
In order to determine whether this level of Se induced adverse apoptotic responses, that process was evaluated in lymphocytes from a random subset of sixty subjects (thirty women, thirty men) at both 0 and 12 months. Apoptosis was induced by treatment with either H2O2 (22 μm), cycloheximide (9·3 mm) or no agent (180 min, 37°C) followed by permeabilisation with 70 % ethanol. Cellular DNA was labelled using terminal deoxynucleotidyl transferase and bromolated deoxyuridine (BrdU) triphosphate nucleotides, after which cells were probed in the dark with fluorescein~BrdU mAb (APO-BrdU™; Phoenix Flow Systems, Inc., San Diego, CA, USA, catalogue no. AU1001). DNA was stained using propidium iodide (PI)/RNase A and within 2 h samples were analysed by flow cytometry using a four-colour instrument (Epics-XL, Beckman-Coulter, Miami, FL, USA) equipped with a 488 nm laser. BrdU and PI were detected by absorbance at 525 and 620 nm, respectively. BrdU incorporation was plotted v. PI incorporation with a logical gate excluding doublets. A total of 15 000 events were collected (flow rate 200–600 events/s) in the region encompassing the main population of intact single leucocytes. Data were analysed using Summit™ Offline flow cytometry analysis software (Cytomation, Inc., Fort Collins, CO, USA).
The extent of DNA damage in peripheral blood lymphocytes was measured by the alkaline comet assay as described by Waters et al. (Reference Waters, Shen and Glickman34). The extent of DNA damage was assessed under basal conditions and after ex vivo exposure of lymphocytes to 22 μm-H2O2. Damage was scored in 200 cells randomly selected from each sample by a single examiner blinded to treatment. SYBR Green 1 stained nucleoids were examined at 200 × magnification using an epifluorescence microscope. Each cell was scored visually by the method of Duthie & Collins(Reference Duthie and Collins35): no damage (type 0); mild to moderate damage (types 1 and 2) and extensive DNA damage (types 3 and 4). Extent of damage was expressed as the percentage of cells of types 3 and 4.
Statistical analyses
All statistical analyses were performed using SAS version 9.1.3 (SAS Institute, Inc., Cary, NC, USA). Data for buccal Se, urine Se, SEPP1, folate, homocysteine (Hcy), vitamin B12 and thyroid-stimulating hormone were highly skewed for which reason they were logarithmically transformed so that their distributions would more closely approximate normal. For these variables, geometric means with the ± 1 sd confidence limits are reported; other data are expressed as arithmetic means and standard deviations. Nucleic acid data were extracted using Robust Multi-Array Average (RMA) in mAdb (a Multi-Array database tool) (National Cancer Institute, National Institutes of Health) and analysed by paired t tests (P < 0·05), adjusting for multiplicity using the false discovery rate test.
Repeated-measures ANOVA was used to test for effects of supplemental Se level over time. When appropriate, Tukey's contrasts were used to compare the supplement levels at each individual time point. Regression analysis was used to model the change in Se status given the effective Se dose, calculated as the Se dose consumed (adjusted for reported compliance) per metabolic body size defined as kg0·75. Categorical variables were included in the regressions to test for effects of sex or genotype.
Results
A total of 261 subjects (106 men, 155 women) were enrolled in the study and randomised to the treatments. Of these, 243 subjects completed the 12-month study, for an attrition rate of 7 %. Of the dropouts (eight men, ten women), none complained of side-effects. Compliance with the treatment protocol, as ascertained by monthly capsule count-backs for subjects completing the study, was 97 %.
The study subjects were genotyped(Reference Combs, Watts and Jackson22) for selenoprotein SNPs that have been correlated with cancer incidence or mechanistically implicated in carcinogenesis(Reference Suitor, Gardner and Willett23–Reference Villette, Kyle and Brown26), and exhibited the following genotype frequencies. The prevalence of dominant alleles in this population was: GPX1 (Pro198Leu; rs1050450: 46 % C/C, 43 % T/C, 11 % T/T), GPX4 (3′-UTR (untranslated region); rs713041: 28 % C/C, 52 % T/C, 20 % T/T), SEPP1 (Ala234Thr; rs3877899: 58 % G/G, 38 % A/G, 4 % A/A and 3′-UTR; rs7579: 44 % G/G, 44 % A/G, 12 % A/A) and SEP15 (3′-UTR; rs5845: 65 % C/C, 31 % T/C, 4 % T/T).
This cohort was of relatively high Se status, as indicated by the baseline values of the biomarkers of Se status, e.g. plasma Se level (142·0 (sd 23·5) ng/ml) and an estimated average Se intake of 109 (sd 44) μg/d(Reference Combs, Watts and Jackson22). We found no evidence that the background intake of Se from dietary sources, as estimated by a quarterly FFQ, differed significantly between treatment groups or changed significantly during the course of the study. In consideration of the analysed Se contents of the supplements, these treatments provided estimated total Se intakes of approximately 109, 165, 210 and 313 μg/d. Thus, all of the subjects were consuming Se at levels greater than the recommended daily allowance, which was set on the basis of maximal expression of GPX3. There is no recommendation for the concentration of Se in plasma.
Supplementation with SeMet produced significant increases in some but not all biomarkers of Se status. Dose-dependent increases in the Se contents of plasma (Fig. 2(a)) and urine (Fig. 1(b)) manifested within 3 months, with apparent plateaus being reached by 9–12 months. No significant treatment effect on GPX3 activity was detected, although that parameter showed a small ( < 2 %) but significant increase over the course of the study (Fig. 2(c)).

Fig. 2 Time-courses of changes in (a) plasma Se level (diet, P < 0·0001; month, P < 0·0001; diet × month, P < 0·0001), (b) urinary Se (diet, P < 0·0001; month, P < 0·0001; diet × month, P < 0·0001) and (c) glutathione peroxidase 3 (GPX3: diet, P < 0·75; month, P < 0·0009; diet × month, P < 0·34) activity in response to l-selenomethionine supplementation. Values are means, with their standard errors represented by vertical bars.
The 1-year changes in the levels of Se in plasma, urine and buccal cells, and in the non-specific component of plasma Se, each showed significant linear relationships with effective Se dose (μg/d per kg0·75) (Fig. 3). No changes were observed for GPX3 activity or SEPP1 level. The dose-dependent increase in urinary Se excretion of women was 74 % greater (P = 0·001) than that of men (Fig. 4), and for individuals of both sexes with the GPX1 T/T genotype was 59 % greater (P < 0·006) than those with the GPX1 C/C genotype (Fig. 5). These differences were significant within 3 months of SeMet supplementation. Individuals with GPX1 T/T genotype also had significantly lower (7 %, P < 0·05) plasma Se levels than those with the GPX1 C/C genotype at baseline(Reference Combs, Watts and Jackson22). No other differences in responses to SeMet-supplementation were associated with the selenoprotein genotypes tested.

Fig. 3 Relationships of 1-year changes in (a) plasma Se (Δplasma Se = 12·6+18·2 (effective dose); R 2 = 0·60, P = 0·0001), (b) glutathione peroxidase 3 (GPX3) activity (Δplasma GPX = 0·06 − 0·001 (effective dose); R 2 = 0·00 001, P = 0·91), (c) urinary Se (Δurine Se = 17·4+18·6 (effective dose); R 2 = 0·44, P = 0·0001), (d) selenoprotein P (SEPP1; ΔSEPP1 = 0·15+0·018 (effective dose); R 2 = 0·002, P = 0·51), (e) buccal cell Se (Δbuccal cell Se = 4·6+2·6 (effective dose); R 2 = 0·22, P = 0·0001) and (f) non-specific plasma Se (Δnon-specific plasma Se = 8·4+16·9 (effective dose); R 2 = 0·58, P = 0·0001) with effective Se dose (μg/d per kg0·75/d). For each data set, linear regressions (![]() ) and their 95 % CI (
) and their 95 % CI (![]() ) and 95 % prediction intervals (
) and 95 % prediction intervals (![]() ) are shown.
) are shown.

Fig. 4 Relationships of responses of (a, c) plasma and (b, d) urinary Se levels to selenomethionine supplementation for men (M) and women (W): (a, b) dose–responses over time and (c, d) 1-year dose–responses to effective Se dose (μg/d per kg0·75). For each data set, the linear regression is shown for each sex. (d) M: Δurine Se = 20·4+12·0 (effective dose) and W: Δurine Se = 20·4+20·9 (effective dose).

Fig. 5 Relationships of 1-year changes in urinary Se levels of subjects of different glutathione peroxidase (GPX1) genotypes and effective Se dose (μg/d per kg0·75); ■: GPX1 679 T/T (Leu198Leu); ●: GPX1 679 T/C (Leu198Pro); ◆: GPX1 679 C/C (Pro198Pro). For each data set, the linear regression is shown. (a) GPX1 679 T/T: Δplasma Se = 12·8+20·0 (effective dose); GPX1 679 T/C: Δplasma Se = 12·8+16·4 (effective dose); GPX1 679 C/C: Δplasma Se = 12·8+19·5 (effective dose). (b) GPX1 679 T/T: Δurine Se = 17·3+25·4 (effective dose); GPX1 679 T/C: Δurine Se = 17·3+19·3 (effective dose); GPX1 679 C/C: Δurine Se = 17·3+16·0 (effective dose).
The major components of plasma Se were estimated from the difference between total plasma Se and the measured activities and amounts, respectively, of GPX3 and SEPP1, as described previously(Reference Combs, Watts and Jackson22). By these estimates, the increase in plasma Se produced by SeMet-supplementation was limited to the non-specific component of that compartment (Table 1).
Table 1 Summary of responses to selenium supplementation
(Mean values and standard deviations)

GPX, glutathione peroxidase; SEPP1, selenoprotein P; OHdG, hydroxydeoxyguanosine; TSH, thyroid-stimulating hormone; CHX, cycloheximide.
* Women, n 143; men, n 97.
† See text for calculation.
‡ Women, n 30; men, n 30.
§ Damage scores 3 or 4 (see text for details).
SeMet-supplementation produced no significant effects on spontaneous or induced apoptosis of peripheral lymphocytes (Table 1). Similarly, no significant difference in spontaneous or induced lymphocyte DNA damage was detected in the alkaline comet assay (Table 1). No significant effects of SeMet-supplementation were detected on the levels of folate, vitamin B12, Hcy, thyroid hormones or buffy coat HbA1c (Table 1).
Discussion
The baseline plasma Se level of this cohort, 142·0 (sd 23·5) ng/ml, shows that this population was adequately nourished with respect to Se prior to supplementation. Nevertheless, plasma Se increased with SeMet-supplementation, reaching a new steady state within 9–12 months. That change, from the initial steady state to the other, after a year of SeMet-supplementation was described by the following relation:
where ΔSe is steady-state change in plasma Se concentration; and Sein is regular Se intake as SeMet expressed as μg/d per kg0·75.
That the y-intercept is significantly different from 0 (P < 0·003) reflects an 8·9 % increase in mean plasma Se level of the non-supplemented members of the cohort during the course of the study. Apart from this apparently secular effect, the amount of increased intake of Se (as SeMet) required to raise plasma Se concentration to a particular target level (Sepl-target) is:
where Sepl is current plasma Se concentration, ng/ml; and Sepl-target is target plasma Se concentration, ng/ml.
The increase in plasma Se in this cohort did not involve increases in the specific selenoproteins in plasma (GPX3, SEPP1), indicating that the baseline Se intake, estimated to be 109·1 (sd 43·6) μg/d(Reference Combs, Watts and Jackson22), was sufficient to support maximal expression of each. Instead, the response of plasma Se was explained by changes solely in the non-specific component of plasma Se, which comprised 46 % of plasma Se at baseline and increased, over 1 year of SeMet supplementation, to 46·6, 56·3, 61·8 and 70·5 % of plasma total Se with 0, 50, 100 and 200 μg Se/d, respectively. That increase is described by the following relation:
where ΔSenon-spec is the steady-state change in non-specific plasma Se level; and Sein is regular Se intake as SeMet expressed as μg Se/d per kg0·75 (y-intercept is not significantly different from 0 (P < 0·08)).
Sexually dimorphic aspects of Se status and response to supplementation have recently been reviewed by Schomburg & Schweizer(Reference Schomburg and Schweizer36). Previously, Méplan et al. (Reference Méplan, Crosley and Nicol37) observed that response of the predominate plasma selenoprotein, SEPP1, to Se supplementation differed by sex. The same group noted that the effect of SNP on the expression of selenoprotein GPX4 depended on sex(Reference Méplan, Crosley and Nicol38). Our present study results show that SeMet-supplementation increased urinary Se output more in women than men, suggesting significant, sex-specific differences in Se metabolism. In rodents, it has been shown that selenoprotein biosynthesis is sexually dimorphic in the liver and kidney(Reference Stoedter, Renko and Hög39, Reference Riese, Michaelis and Mentrup40) even though total Se concentrations and SEPP levels in serum do not differ. Accordingly, Se supplementation in mice affected selenoprotein biosynthesis in a sex-specific manner, indicating that hepatic metabolism of dietary Se differs in males and females(Reference Schomburg, Riese and Renko41). It is possible that such an effect may underlie the sexual dimorphic urinary Se excretion that we observed in this study involving human subjects.
We also found increased urinary Se output in individuals with the GPX1 T/T relative to those of the GPX1 C/C genotype. Differential turnover of the GPX1 enzyme, which accounts for some 60 % of liver Se(Reference Cheng, Ho and Ross42), is likely to affect whole-body Se balance; this may explain the increased urinary output. It is also possible that the urinary response related to the fact that individuals with GPX1 T/T have significantly lower (7 %) baseline plasma Se levels than individuals with GPX1 C/C(Reference Combs, Watts and Jackson22). Cominetti et al. (Reference Cominetti, de Bartoli and Prgatto43) observed a similar difference for morbidly obese women of lower Se status in response to Se from Brazil nuts; however, in their relatively small (n 37) study, they were unable to detect the significant genotype-related differences in baseline plasma Se which we observed in the present cohort(Reference Combs, Watts and Jackson22). Hu & Diamond(Reference Hu and Diamond25) found that the GPX1 activity response of cultured breast cancer cells ectopically expressing the GPX1 T/T genotype was less than that of cells expressing the GPX1 C/C genotype. That this difference may be related to cancer risk is suggested by the findings of GPX1 genotype as a determinant of the risk of lung cancer among male smokers in the Finnish Alpha-Tocopherol Beta-Carotene (ATBC) Trial(Reference Ratnasinghe, Tangrea and Andersen44) and breast cancer in American women(Reference Suitor, Gardner and Willett23, Reference Hu and Diamond45), as well as the loss of heterozygosity in lung tumours(Reference Moscow, Schmidt and Ingram46).
Gonzalez et al. (Reference Gonzalez, Huerta and Alvarez-Uria47) found serum Se and Hcy concentrations to vary inversely, explaining 5·8 % of the variance of the latter (2·2 % of the variance was accounted for by serum folate concentration). Finding a 63 % reduction (P = 0·013) in the risk of elevated Hcy concentrations for subjects with serum Se levels in the highest tertile, they concluded that Se should be considered as a potential factor for lowering serum Hcy. The present study, however, detected no effects of SeMet supplementation on circulating levels of folate, vitamin B12 or Hcy.
While Se is required for each of the deiodinases involved in thyroid hormone metabolism, that Se supplementation did not affect thyroid hormone levels suggests that those selenoproteins were maximally expressed in these subjects before supplementation. These findings are consistent with our previous results with another cohort of Se-adequate adults(Reference Broome, McArdle and Kyle15), although another group(Reference Hawkes and Keim48, Reference Hawkes, Keim and Richter49) reported such changes.
These results provide no evidence of adverse effects of this level of SeMet supplementation. No effects on lymphocyte DNA damage were observed, although DNA damage was reported for morbidly obese women with the GPX1 679 T/T genotype(Reference Cominetti, de Bartoli and Prgatto43) and have been observed in animal models in response to selenite(Reference Lu, Kaeck and Jiang50). Neither did SeMet affect spontaneous or peroxide-induced apoptosis of peripheral lymphocytes.
A limitation of the present study was the use of SeMet as the intervention agent. The biological utilisation of this form of Se is known to be affected by dietary Met(Reference Sunde, Gutzke and Hoekstra51–Reference Butler, Beilstein and Whanger53), due to the competition of Met and SeMet(Reference Butler, Beilstein and Whanger53) for incorporation into general proteins. This cohort was adequately nourished with respect to total protein, and thus Met. Therefore, Se-enriched yeast, which is mostly SeMet, would be expected to have given comparable results. However, supplementation with inorganic Se, which does not contribute to the non-specific fraction of plasma Se as defined in this study, would be expected to have increased primarily urinary Se and, perhaps, its precursors in the non-protein bound fraction of plasma(Reference Kokarnig, Kuehnelt and Stiboller33).
Conclusions
The present study demonstrates the feasibility of tracking responses to SeMet supplementation in a cohort that is not deficient in the nutrient. In such a cohort, biomarkers with clear functional significance that are limited by Se deficiency, such as GPX3 activity and SEPP1 level, are already maximally expressed. Therefore, they are of no utility as indicators of further increases in Se status. Such tracking demands the use of biomarkers related to tissue/body Se pools. We have demonstrated the use of four such biomarkers: plasma total Se, plasma non-specific Se, buccal cell Se and urinary Se. Each responded to SeMet supplementation; those responses were linear with effective Se dose (i.e. corrected for body mass) over the period of time under investigation. The non-specific component of plasma Se, which we imputed from direct measurements of total plasma Se, SEPP1 and GPX3 activity, was responsive to SeMet supplementation, reflecting the incorporation of SeMet into plasma proteins.
Because SeMet is the dominant form of Se in foods(Reference Olson, Novachek and Whitehead54, Reference Thavarajah, Vandenberg and George55), the algorithms generated in this study can be used to predict the steady-state levels of total and non-specific Se in plasma that will be achieved with any particular regular intake of dietary Se. By this relation, an individual with a plasma Se level of 85 ng/ml would have to consume 1·2 μg Se/d per kg0·75 to support the plasma total Se level (106 ng/ml) associated with prostate cancer risk reduction in the NPC Trial(Reference Duffield-Lillico, Dalkin and Reid8), whereas an intake of 3·4 μg Se/d per kg0·75 would be required to reach the plasma total Se level (147 ng/ml) associated with increased type 2 diabetes risk in the NHANES 2003–4(Reference Laclaustra, Navas-Acien and Stranges11). This indicates that Se supplementation targeting cancer risk reduction can be safely achieved.
These results suggest risk categories in which personalised needs for Se can be considered. That women retained ingested Se less well than men suggests that they may have greater dietary Se needs. That the same phenomenon was observed for GPX1 genotype suggests that genotype may also be a determinant of Se need. While the algorithm presented here has application as a general guideline for supplementation, personalised strategies will need to consider the influences of sex and genotype.
Acknowledgements
We appreciate the contribution of the supplements used in this trial by the Sabinsa Corporation. We gratefully acknowledge the expert contributions of the following members of the Grand Forks Human Nutrition Research Center staff: Brenda Ling for subject recruiting; Wesley Canfield, for subject health oversight; Emily Nielsen, Doreen Rolshoven, Judy Schumacher, and Ruth Christianson, for coordinating subject visits and sample collection; Sandy Botnen, Teresa Numedahl, Wendy Mayer, Pat Wilson and Kay Williams for sample preparation and clinical biochemical analyses; Bonnie Hoverson, Elvira Bell, Sue Sherette, Charlene Kuntz and Doris Zidon and Mary Jo Peltier for preparation and distribution of supplement packets; Laura Idso for sample preparation and enzyme analyses; Eric Uthus for advice on several analytical methods; and Craig Lacher and Bill Martin for Se analyses. The authors' responsibilities were as follows: G. F. C., J. C. W., C. D. D. and J. A. M. designed the research; G. F. C., M. I. J., D. J. W., H. Z., J. I., L. S., A. H., C. S. H., E. C. C. and D. J. W. conducted the research; L. K. J. analysed the data; G. F. C. and M. I. J. wrote the paper and had responsibility for its final content. None of the authors had any competing interests to declare. This research was supported by the Grand Forks Human Nutrition Research Center CRIS project no. 5450-51000-036-00D and NCI-ARS Interagency Agreement 07-0A-5450-330, and Deutsche Forschungsgemeinschaft DFG (Scho849/2-2, GraKo 1208) and Deutsche Krebshilfe (10-1792 Scho2) (to L. S.).


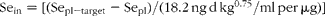 .
.





