Impact statement
This review summarises current available knowledge on microplastic degradation by microbes and explores the novel and emerging techniques for microplastic remediation using microbes. The paper incorporates in depth discussion on microbial (bacteria, fungi and enzymatic degradation) action on microplastics and discusses why the ‘plastisphere’ is considered to be the new delicacy for the microbial communities. The paper further explores how microplastics can develop antibiotic resistance in bacteria. The effect of environmental factors and biofilms on microbial degradation of microplastics is discussed. The key knowledge gaps and future research directions were identified regarding the use of microbes for microplastic bioremediation.
Introduction
Plastics are durable, lightweight, cost-effective, and have become the most widely and frequently used synthetic materials. Plastics are almost indispensable in many aspects of human life in modern society (Chae and An, Reference Chae and An2018). Various items like shopping bags, plastic bottles, food containers, disposable cups, plumbing pipes, microwavable containers, plastic films, automotive parts, hookup wire, coaxial cables, and so on are made using plastic additives like polyethylene (PE), polystyrene (PS), polyvinyl chloride (PVC), polypropylene (PP), polyethylene terephthalate (PET), nylon, polycarbonate, polytetrafluoroethylene (PTFE) (Amobonye et al., Reference Amobonye, Bhagwat, Singh and Pillai2021). Between 1950 and 2019, the annual global production of plastics had increased dramatically from 2 million tons to 368 million tons, of this around 40% were for single-use plastics (Plastic Europe, 2020) which resulted in the rapid accumulation of plastics in the environment (Nielsen et al., Reference Nielsen, Hasselbalch, Holmberg and Stripple2019). Around 2050, the global volume of plastic wastes will reach 26 billion tonnes, of which around 50% will eventually enter various compartments of terrestrial as well as aquatic ecosystems, causing serious environmental impacts (Jambeck et al., Reference Jambeck, Geyer, Wilcox, Siegler, Perryman, Andrady, Narayan and Law2015).
Microplastics (MPs) are defined as plastic particles having size less than 5 mm and can be categorised based on primary and secondary types, sizes, shapes and polymer compositions. They are widespread in the biosphere and are potential contaminants of grave concern (Klein et al., Reference Klein, Dimzon, Eubeler and Knepper2018a; Blair Espinoza, Reference Blair Espinoza2019) and are ubiquitously found in sediments (Zamprogno et al., Reference Zamprogno, Caniçali, dos Reis Cozer, Otegui, Graceli and da Costa2021), water, sea salt (Kosuth et al., Reference Kosuth, Mason and Wattenberg2018), food (Barboza et al., Reference Barboza, Vethaak, Lavorante, Lundebye and Guilhermino2018), the atmosphere (Brahney et al., Reference Brahney, Mahowald, Prank, Cornwell, Klimont, Matsui and Prather2021) and sewage sludge. The total amount of microplastics in the aquatic ecosystems is expected to reach 12,000 metric tons (Mt) by 2050 (Geyer et al., Reference Geyer, Jambeck and Law2017). MPs are persistent (Lambert and Wagner, Reference Lambert and Wagner2018) and can bioaccumulate in the food chain and subsequently can affect plants and animals, including humans. MPs accumulation in the environment has adverse effects on the organisms including molecular stress, reduced growth rate and reproductive complications (Batel et al., Reference Batel, Linti, Scherer, Erdinger and Braunbeck2016). MPs also affect plant growth and fish reproduction (Guo and Wang, Reference Guo and Wang2019). Due to the wide range of adverse effects of MPs reducing the levels of MPs pollution is the need of the hour (Vince and Hardesty, Reference Vince and Hardesty2017). MPs contain various toxic substances added during manufacturing or collected from the environment (Andrady, Reference Andrady2011). Plastic pollution has long been regarded to be an irreversible problem (because of poor degradation and the inability to collect all plastic particles) that will affect Earth system processes. As a result, certain techniques to limit micro (plastics) loss to the environment during manufacture, use, and disposal have been proposed (Lambert and Wagner, Reference Lambert and Wagner2018). However, isolating MPs in environmental matrices might be difficult (Lambert and Wagner, Reference Lambert and Wagner2018). In recent times, bio-plastics have been developed but their biodegradation also produces MPs (Wei et al., Reference Wei, Hedenqvist, Zhao, Barth and Yin2022). Physical treatment methods like sedimentation, filtration, and so on do separate MPs but retain them in the sludge. Current practices for handling plastic wastes (e.g., recycling, landfilling, incineration) have drawbacks that potentially exacerbate existing environmental problems. For example, incinerating synthetic plastics releases volatile and hazardous waste products, such as dioxins, heavy metals, sulphides, nitrogen oxides, and furans, all of which are thought to have the potential to cause cancer (Verma et al., Reference Verma, Shankarappa, Papireddy and Gowda2016). Furthermore, downcycling (Rahimi and Garcia, Reference Rahimi and Garcia2017) and cost-ineffectiveness (Gradus et al., Reference Gradus, Nillesen, Dijkgraaf and Koppen2017) are also associated with synthetic plastic recycling. Additionally, landfilling is also not a good option since it takes a lot of space and has chances for leakage into the environment. Due to these drawbacks efforts are directed towards finding more environmental friendly approaches for managing plastic wastes. In this context, microorganisms have emerged as a likely alternative with numerous researches highlighting the capability of various microbial species for degrading MPs including PET (Taniguchi et al., Reference Taniguchi, Yoshida, Hiraga, Miyamoto, Kimura and Oda2019), PE (Restrepo-Flórez et al., Reference Restrepo-Flórez, Bassi and Thompson2014), PU (Magnin et al., Reference Magnin, Pollet, Phalip and Avérous2020), PS (Ho et al., Reference Ho, Roberts and Lucas2018) and PP (Arutchelvi et al., Reference Arutchelvi, Muniyasamy, Arkatkar, Doble, Bhaduri and Uppara2008). In this context, bioremediation of MPs has emerged as an attractive and alternate method to remove MPs from the environment. The use of microbes for MPs degradation can help in the remediation of MPs without any environmental damage (Restrepo-Flórez et al., Reference Restrepo-Flórez, Bassi and Thompson2014; Kumar Sen and Raut, Reference Kumar Sen and Raut2015; Qi et al., Reference Qi, Ren and Wang2017), thus making it a promising and safe avenue for cleaning our natural ecosystems (Shah et al., Reference Shah, Hasan, Hameed and Ahmed2008). Various efforts have also been made to screen potential plastic-degrading microbes (Hidalgo-Ruz et al., Reference Hidalgo-Ruz, Gutow, Thompson and Thiel2012) and develop biodegradable polymers. However, presently only a few microorganisms are known to be able to degrade MPs and there is a dearth of knowledge in the context of microbial degradation of MPs (Gu, Reference Gu2003).
MP are mainly considered as persistent compounds offering a large ecological niche for the colonisation of microbial communities (Kooi et al., Reference Kooi, Van Nes, Scheffer and Koelmans2017; Rummel et al., Reference Rummel, Jahnke, Gorokhova, Kühnel and Schmitt-Jansen2017). Numerous investigations have been carried out to examine the interaction between MPs and the MP colonising microorganisms, which could likely impact the behaviour of MP in the environment by altering their chemical or physical properties (Rummel et al., Reference Rummel, Jahnke, Gorokhova, Kühnel and Schmitt-Jansen2017; Roager and Sonnenschein, Reference Roager and Sonnenschein2019; Oberbeckmann and Labrenz, Reference Oberbeckmann and Labrenz2020). However, there was no consensus on whether these microorganisms specifically chose plastics to colonise. According to some research, the microbial communities on plastics do not seem to be substrate-specific (Oberbeckmann et al., Reference Oberbeckmann, Osborn and Duhaime2016). For instance, the lifetime and relative mobility of microplastics in the water column were shown to account for the differences in the microbial community compositions between floating microplastics and other substrates, such as stones and sediments (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013; Fazey and Ryan, Reference Fazey and Ryan2016). However, multiple investigations showed differences in microbial community between MP surface and the surrounding, possibly due to the limitation of nutrients or the type of MPs (Kirstein et al., Reference Kirstein, Wichels, Gullans, Krohne and Gerdts2019). Research in other areas revealed that the types of carbon sources largely drive the turnovers of microbial community compositions (Goldford et al., Reference Goldford, Lu, Bajić, Estrela, Tikhonov, Sanchez-Gorostiaga, Segrè, Mehta and Sanchez2018). While microplastics are not commonly used by microbes, some specific bacteria that break down plastic may be able to obtain carbon from them (Aravinthan et al., Reference Aravinthan, Arkatkar, Juwarkar and Doble2016; Green et al., Reference Green, Boots, Sigwart, Jiang and Rocha2016). This suggests that some microorganisms have a selection advantage when it comes to breaking down microplastics. Furthermore, certain microbes also favour adhering to the hydrophobic surface of microplastics and interacting with them (Krasowska and Sigler, Reference Krasowska and Sigler2014).
Therefore, a deeper understanding of the properties of the microbial communities colonising microplastics – such as their structure, stability, and mechanism of community assembly – based on stochastic and deterministic models was required in order to gain insight into the relationship between microplastics and the attached microorganism (Stegen et al., Reference Stegen, Lin, Konopka and Fredrickson2012; De Vries et al., Reference De Vries, Griffiths, Bailey, Craig, Girlanda, Gweon, Hallin, Kaisermann, Keith, Kretzschmar, Lemanceau, Lumini, Mason, Oliver, Ostle, Prosser, Thion, Thomson and Bardgett2018). Nevertheless, there is still a dearth of pertinent data regarding microbial communities that colonise microplastics in different environments. MP degradation in nature mainly occurs via physical, chemical and biological means (Shah et al., Reference Shah, Hasan, Hameed and Ahmed2008; Ter Halle et al., Reference Ter Halle, Ladirat, Martignac, Mingotaud, Boyron and Perez2017; Ariza-Tarazona et al., Reference Ariza-Tarazona, Villarreal-Chiu, Barbieri, Siligardi and Cedillo-González2018). Light and oxygen can cause the degradation of MPs through abiotically and enhance the microbial availability through photodegradation, thermooxidative degradation and hydrolysis (Andrady, Reference Andrady2011). In deep sediments, light and oxygen are limited and redox conditions are the vital environmental factor for MP degradation, which requires further study (Rogers et al., Reference Rogers, Carreres-Calabuig, Gorokhova and Posth2020). Under anaerobic conditions, microorganisms found on MPs utilise either plastics or other organic matters surrounding MPs as electron donors (Rogers et al., Reference Rogers, Carreres-Calabuig, Gorokhova and Posth2020). To date, various anaerobic and facultative anaerobic plastic-degrading bacterial strains have been successfully isolated from plastics (Kathiresan, Reference Kathiresan2003; Auta et al., Reference Auta, Emenike and Fauziah2017), suggesting possible biodegradation of microplastics in anoxic sediments.
Multi-disciplinary approaches are required in order to address complex issues regarding MP bioremediation such as screening of efficient microbes and their characterisation, evaluation of in situ toxicity, and so on. MP assessment is necessary for the preparation of appropriate feedstock for the biotransformation of MPs which are recalcitrant.
Microplastics in the environment: Distribution, accumulation, toxicity and health effects
MPs are known to occur ubiquitously in marine environment, in surface water and sediments. The presence of MP has been extensively reported in deep sea water, sea surface, on shorelines, and in aquatic organisms in several countries around the world (Gray et al., Reference Gray, Wertz, Leads and Weinstein2018; Khalik et al., Reference Khalik, Ibrahim, Anuar, Govindasamy and Baharuddin2018). The existence of MPs has been reported in diverse types of ecosystems, including permafrost, both in Arctic (Zhang et al., Reference Zhang, Gao, Kang, Allen, Wang, Luo, Yang, Chen, Hu, Chen and Du2023), Antarctica (Aves et al., Reference Aves, Revell, Gaw, Ruffell, Schuddeboom, Wotherspoon, LaRue and McDonald2022); Tibetan plateau (Wang et al., Reference Wang, Qu, Luo, Ji, Ma, Wang, Dahlgren, Zhang and Shang2023) and European alpine regions (Materić et al., Reference Materić, Kasper-Giebl, Kau, Anten, Greilinger, Ludewig, van Sebille, Röckmann and Holzinger2020, Reference Materić, Ludewig, Brunner, Röckmann and Holzinger2021). The occurrence and accumulation of MPs were also evident in several mangrove ecosystems and coral reefs (John et al., Reference John, Nandhini, Velayudhaperumal Chellam and Sillanpää2022). Occurrence of MP is also reported in different fresh water systems, including Ottawa River (Vermaire et al., Reference Vermaire, Pomeroy, Herczegh, Haggart and Murphy2017), Antuã River (Rodrigues et al., Reference Rodrigues, Abrantes, Gonçalves, Nogueira, Marques and Gonçalves2018), Yangtze River (Hu et al., Reference Hu, Chernick, Hinton and Shi2018), Vembanand Lake (Sruthy and Ramasamy, Reference Sruthy and Ramasamy2017) in North America, Portugal, China and India. These MP particles have negative effects on diverse marine organisms (Wright et al., Reference Wright, Thompson and Galloway2013) and mainly act as potential vectors for Persistent organic pollutants (POP) and other toxic pollutants (Hermabessiere et al., Reference Hermabessiere, Dehaut, Paul-Pont, Lacroix, Jezequel, Soudant and Duflos2017). The presence of MP was also reported in air using different types of sampling methods such as atmospheric sampling (Abbasi et al., Reference Abbasi, Keshavarzi, Moore, Turner, Kelly, Dominguez and Jaafarzadeh2019), dust collection (Zhang et al., Reference Zhang, Wang and Kannan2019) and wet and dry deposition (Klein and Fischer, Reference Klein and Fischer2019). The size of the MP fibres ranges from 100 to 5000 μm (Cai et al., Reference Cai, Wang, Peng, Tan, Zhan, Tan and Chen2017). In most cases, population density and proximity to urban areas are the main factors which influence the abundance of MP. The presence of MP was mostly reported in various touristic centres of China (Jiang et al., Reference Jiang, Yin, Li, Wen, Luo, Hu, Yang, Long, Deng, Huang and Liu2019), Rhine and Main Rivers, China’s Qinghai Lake (Xiong et al., Reference Xiong, Zhang, Chen, Shi, Luo and Wu2018), the Lagoon of Venice (Klein et al., 2015), Jakarta Bay (Manalu et al., Reference Manalu, Hariyadi and Wardiatno2017) and the Ottawa river (Vermaire et al., Reference Vermaire, Pomeroy, Herczegh, Haggart and Murphy2017). Both domestic and industrial sewage spillage are significant sources of MPs. According to study made by Carr et al. (Reference Carr, Liu and Tesoro2016), no MP was found in effluents of a tertiary waste water treatment plant, whereas, one plastic particle per 1.14 litre of effluent was reported from secondary waste water treatment plant. The density of the MP particles effects the vertical distribution and buoyancy of the particles. Usually, low-density MP occur more in surface zone whereas, while high-density MPs accumulate in deep seas and benthic organisms (Eerkes-Medranoet al., Reference Eerkes-Medrano, Thompson and Aldridge2015).
Most MP are toxic and known to disrupts endocrine activity. On exposure to MPs, the catalytic activity of the acetylcholinesterase (AChE) (which is essential for neurotransmission in neuromuscular junctions and brain synapses) in zebrafish larva can led to death (Chen et al., Reference Chen, Gundlach, Yang, Jiang, Velki, Yin and Hollert2017). Similar AChE activity inhibition was also reported in Dicentrarchus labrax, Artemia franciscana and Oreochromis niloticus. Moreover, body size, age influences the impact of MP on AChE activity. The presence of MP can also increase antioxidant defence response, cellular oxidative stress and lead to peroxidation of lipid which can induce disruption of membranes of presynaptic vesicles and damages the gill, muscles and liver in several fish species (Wen et al., Reference Wen, Jin, Chen, Gao, Liu, Liu and Feng2018). According to Lei et al. (Reference Lei, Liu, Song, Lu, Hu, Cao, Xie, Shi and He2018a, Reference Lei, Wu, Lu, Liu, Song, Fu, Shi, Raley-Susman and He2018b) MP caused neurodegeneration on Caenorhabditis elegans. The MP can accumulate in the gastrointestinal tract of fishes which effect the health and growth of fishes (Yin et al., Reference Yin, Chen, Xia, Shi and Qu2018). However, there is a lack of adequate data related to the impact of MP in the GI tract of marine organisms. It was also reported that both chemical composition and shape may influence the ecotoxicology of MP particles. MP present in sediments are mostly fibres followed by fragments, beads, films and foam (Kooi and Koelmans, Reference Kooi and Koelmans2019); fibres are found to induce more toxic effects compared to fragments and beads. Polysterene (PS) MP was reported to upregulate nfa, il1b and ifng1–2 gene expressions, resulting in inflammatory responses on livers, guts and gills of zebrafish, induce ROS production and decrease GSH and SOD levels (Lu et al., Reference Lu, Qiao, An and Zhang2018). MPs are considered to be potentially toxic to human health as it may enter the gastrointestinal (GI) tract by endocytosis of M cells and translocate to different parts of the body (Cox et al., Reference Cox, Covernton, Davies, Dower, Juanes and Dudas2019). According to study made by Forte et al. (Reference Forte, Iachetta, Tussellino, Carotenuto, Prisco, De Falco, Laforgia and Valiante2016), it was reported that PS nanoplastics can up-regulate IL-6 and IL-8 gene expression, resulting in inflammatory responses and morphological alterations. Moreover, high concentration of PS nanoparticles can lead to cell death through apoptosis by activating Caspase 3, 7 and 9 (Bexiga et al., Reference Bexiga, Varela, Wang, Fenaroli, Salvati, Lynch, Simpson and Dawson2011). The cytotoxicity of MP particles can also lead to DNA double-strand breaks and/or the high depletion of GSH (Paget et al., Reference Paget, Dekali, Kortulewski, Grall, Gamez, Blazy, Aguerre-Chariol, Chevillard, Braun, Rat and Lacroix2015). The study made by Xu et al. (Reference Xu, Halimu, Zhang, Song, Fu, Li, Li and Zhang2019) reported PS can affect the human lung cells and inhibit cell viability. The presence of excess MP particles can cause abnormal behaviour, produce ROS and impacts the immune system of fish. Further research is needed to understand the underlying mechanisms of MP toxicity.
Plastisphere: A new delicacy for the microbes?
Plastisphere describes a novel microbial community attached to plastics and distinct from the surroundings. Marine plastic debris provides a selective hydrophobic environment that stimulates the growth of early colonisers accelerating biofilm formation and further microbial succession (ZoBell and Anderson, Reference ZoBell and Anderson1936). Stimulation of microbial growth and respiration by inert surfaces is a well-documented phenomenon, which creates a favourable environment for microbial colonisation through micronutrient concentration (ZoBell, Reference ZoBell1943). This could play an important role for increasing microbial activity in the upper layer of ocean gyres owing to the abundant plastic debris in oligotropic areas of the oceans (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013).
Early scanning electron micrographs of plastic surface biofilms (Sieburth, Reference Sieburth1975) provided clues about microbial diversity within the plastisphere. The bacterial community found on the MP surface is found to be significantly different from that in surrounding middle and upper waters or other particle types (Oberbeckmann et al., Reference Oberbeckmann, Kreikemeyer and Labrenz2018). Various factors like season, temperature, humidity, surrounding environment, polymer type, surface morphology and size of MPs influence the abundance and diversity of colonising microbial groups (Reisser et al., Reference Reisser, Shaw, Hallegraeff, Proietti, Barnes, Thums, Wilcox, Hardesty and Pattiaratchi2014; De Tender et al., Reference De Tender, Devriese, Haegeman, Maes, Ruttink and Dawyndt2015; Dussud et al., Reference Dussud, Hudec, George, Fabre, Higgs, Bruzaud, Delort, Eyheraguibel, Meistertzheim, Jacquin, Cheng, Callac, Odobel, Rabouille and Ghiglione2018a, Reference Dussud, Meistertzheim, Conan, Pujo-Pay, George, Fabre, Coudane, Higgs, Elineau, Pedrotti and Gorsky2018b). For example, studies have shown different microbial communities attached to MPs from two different oceans, and the diversity of bacteria living in water columns and bacteria attached to microplastic debris (Amaral-Zettler et al., Reference Amaral-Zettler, Zettler, Slikas, Boyd, Melvin, Morrall, Proskurowski and Mincer2015). Heterotrophic bacteria are capable of rapidly colonising plastic surfaces, which can survive longer than in the surrounding aquatic environments (Webb et al., Reference Webb, Crawford, Sawabe and Ivanova2009). The biodegradability of plastic is influenced not only by the capacity of microorganisms but also by surface texture, hydrophobicity, electrostatic interactions and free energy of the material (Falahudin et al., Reference Falahudin, Cordova, Sun, Yogaswara, Wulandari, Hindarti and Arifin2020). MPs provide a novel ecological niche for microbial growth and colonisation and serves as a carbon source. Recent studies on the basis of molecular data (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013; Bryant et al., Reference Bryant, Clemente, Viviani, Fong, Thomas, Kemp, Karl, White and DeLong2016; Dussud et al., Reference Dussud, Hudec, George, Fabre, Higgs, Bruzaud, Delort, Eyheraguibel, Meistertzheim, Jacquin, Cheng, Callac, Odobel, Rabouille and Ghiglione2018a, Reference Dussud, Meistertzheim, Conan, Pujo-Pay, George, Fabre, Coudane, Higgs, Elineau, Pedrotti and Gorsky2018b; Kirstein et al., Reference Kirstein, Wichels, Krohne and Gerdts2018) also confirmed that plastispheres are comprised of various organisms including primary producers (e.g., phototrophs), heterotrophs, predators, symbionts and decomposers. Members of genus Vibrio are reported to be enriched on microplastic surface (Frere et al., Reference Frère, Maignien, Chalopin, Huvet, Rinnert, Morrison, Kerninon, Cassone, Lambert, Reveillaud and Paul-Pont2018; Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013), however, other researchers have contradicted the claim (Schmidt et al., Reference Schmidt, Reveillaud, Zettler, Mincer, Murphy and Amaral-Zettler2014; Bryant et al., Reference Bryant, Clemente, Viviani, Fong, Thomas, Kemp, Karl, White and DeLong2016; Oberbeckmann et al., Reference Oberbeckmann, Kreikemeyer and Labrenz2018). SEM photomicrographs confirmed the presence of varied eukaryotic and bacterial microbiota on both Polypropelene (PP) and Polyethelene (PE) samples. DNA analyses validated that the communities on plastics and the surrounding water differed consistently. For example, photosynthetic filamentous cyanobacteria including Phormidium and Rivularia OTUs occurred on plastics but were absent from seawater samples where unicellular Prochlorococcus dominated the bacterial phototroph community (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013). Marine suctorian ciliates of the genus Ephelota was also present known to harbour ectosymbiotic rod-shaped bacteria (Chen et al., Reference Chen, Miao, Song, Warren, Al-Rasheid, Al-Farraj and Al-Quraishy2008). Also, sulphide-oxidising Gammaproteobacteria of the genus Thiobios was present in the samples and on the surface of stalked ciliates (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013). Moreover, polycystine colonial radiolaria were found on MP surface. The identification of the Vibrio sequence recovered in high abundance and showed similarity (100%) with various Vibrio species from the gene bank. From the Mediterranean Sea, harmful dinoflagellate species under genus Alexandrium were reported to be present on plastic marine debris (Maso et al., Reference Maso, Garce ́s, Page ́s and Camp2007) and also a few from Atlantic Ocean.
Phototrophs
Diatoms are common and omnipresent residents of plastisphere and are one of the early and dominant colonisers (Oberbeckmann et al., Reference Oberbeckmann, Loeder, Gerdts and Osborn2014; Eich et al., Reference Eich, Mildenberger, Laforsch and Weber2015; Masó et al., Reference Masó, Fortuño, De Juan and Demestre2016; Michels et al., Reference Michels, Stippkugel, Lenz, Wirtz and Engel2018; Kettner et al., Reference Kettner, Oberbeckmann, Labrenz and Grossart2019). Diatoms belonging to number of bacillariophyte genera including Navicula, Nitzschia, Sellaphora, Stauroneis and Chaetoceros (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013). Studies from Sargasso Sea reported diatoms including Mastogloia angulata, Mastogloia pusilla, Mastogloia hulburti, Cyclotella meneghiniana and Pleurosigma sp. (Carpenter and Smith, Reference Carpenter and Smith1972), and amplicon reads belonging to genera Sellaphora, Amphora and Nitzschia (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013). Furthermore, the taxa Mastogloia, Nitzschia, and Amphora have been reported from the Arabian Gulf only on the basis of morphological traits (Muthukrishnan et al., Reference Muthukrishnan, Al Khaburi and Abed2019). Metagenomic surveys (Bryant et al., Reference Bryant, Clemente, Viviani, Fong, Thomas, Kemp, Karl, White and DeLong2016) have placed diatom clades at less than 1% of the eukaryotic community suggesting their replacement as the community matures. Cyanobacteria are also present along with diatoms (Bryant et al., Reference Bryant, Clemente, Viviani, Fong, Thomas, Kemp, Karl, White and DeLong2016). Various filamentous bacteria like Phormidium, Rivularia and Leptolyngbya are also continuously reported on microplastics (Amaral-Zettler et al., Reference Amaral-Zettler, Zettler and Mincer2020). Additionally, other known photosynthetic representatives included prasinophytes, rhodophytes, cryptophytes, haptophytes, dinoflagellates, chlorarachniophytes, chrysophytes, pelagophytes and phaeophytes.
Fungi are able to form chemical bonds like carbonyl, carboxyl and ester bonds which decreased the hydrophobicity of the MP. Fungi also uses MPs as a carbon source and as a result are able to degrade them. Yamada-Onodera et al. (Reference Yamada-Onodera, Mukumoto, Katsuyaya, Saiganji and Tani2001) demonstrated Penicillium simplicissimum YK to successfully grow on solid medium supplemented with 0.5% PE after UV irradiation for 500 h. Aspergillus niger and Penicillium pinophilum was able to degrade thermo-oxidised (80°C, 15 days) low-density polyethylene (TO-LDPE) by 0.57 and 0.37% after 31 months. Additionally, the TO-LDPE weight decreased by three crystallinity and crystalline lamellar thickness units (0.4–1.8 Å), and increased small-crystal content (up to 3.2%) and mean crystallite size (8.4–14 Å) were observed. Dantzler et al. showed that serine hydrolase secreted from Pestalotiopsis microspora isolates were responsible for bio-degrading polyurethane (PUR) MP. Aspergillus tubingensis VRKPT1 and Aspergillus flavus VRKPT2 are able to degrade high-density polyethylene (HDPE) efficiently (Sangeetha Devi et al., Reference Sangeetha Devi, Rajesh Kannan, Nivas, Kannan, Chandru and Robert Antony2015) having weight loss of HDPE around 6.02 ± 0.2 and 8.51 ± 0.1%, respectively. White-rot fungi IZU154, Trametes versicolor and Phanerochaete chrysosporium have also shown excellent capability to degrade MPs (Deguchiet al.).
Photoheterotrophs and heterotrophs
In addition to phototrophs, potential photoheterotrophic bacteria of the genera Erythrobacter, Roseobacter and ‘Candidatus Pelagibacter’ (REF) are also common residents of plastisphere. Heterotrophic bacteria in seawater samples were dominated by Pelagibacter along with other free-living picoplanktonic bacterial groups with different levels of abundance in PP and PE (Giovannoni et al., Reference Giovannoni, Britschgi, Moyer and Field1990). Experiments to culture bacteria with plastic as only carbon source have given various assortments, including members of Gammaproteobacteria (Nakamiya et al., Reference Nakamiya, Sakasita, Ooi and Kinoshita1997; Yoon et al., Reference Yoon, Jeon and Kim2012) and Firmicutes (Harshvardhan and Jha, Reference Harshvardhan and Jha2013), as well as Actinobacteria (Gilan and Sivan, Reference Gilan and Sivan2013). Fungal sequences from plastic debris have also been reported by various studies (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013; Debroas et al., Reference Debroas, Mone and Ter Halle2017; Kettner et al., Reference Kettner, Rojas-Jimenez, Oberbeckmann, Labrenz and Grossart2017, Reference Kettner, Oberbeckmann, Labrenz and Grossart2019). Fungal diversity in the plastisphere is somewhat less known, however, recent studies highlighted that in brackish and freshwaters fungal assemblages are dominated by members of Chytridiomycota, Cryptomycota and Ascomycota (Kettner et al., Reference Kettner, Oberbeckmann, Labrenz and Grossart2019). From visual analysis of microbial population, members of fungal genus Malassezia were also reported (Amend et al., Reference Amend, Burgaud, Cunliffe, Edgcomb, Ettinger, Gutiérrez, Heitman, Hom, Ianiri, Jones and Kagami2019).
Studies have shown that bacterial groups belonging to phyla Bacteroidetes, Proteobacteria, Cyanobacteria and Firmicutes are frequent colonisers of MPs (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013; Dussud et al., Reference Dussud, Hudec, George, Fabre, Higgs, Bruzaud, Delort, Eyheraguibel, Meistertzheim, Jacquin, Cheng, Callac, Odobel, Rabouille and Ghiglione2018a, Reference Dussud, Meistertzheim, Conan, Pujo-Pay, George, Fabre, Coudane, Higgs, Elineau, Pedrotti and Gorsky2018b).
Bacteria belonging to genus Corynebacterium, Arthrobacter, Pseudomonas, Micrococcus, Streptomyces and Rhodococcus are capable of biodegrading plastics and have demonstrated that they can use plastics as their carbon source under laboratory conditions (Shah et al., Reference Shah, Hasan, Hameed and Ahmed2008). Interestingly, it was discovered that significant differences exist in the diversity, abundance and activity of bacterial and physiochemical characters of plastics between biodegradable and non-biodegradable plastics, indicating the presence of plastic-degrading microbes (Negoro, Reference Negoro2000).
Auta et al. (Reference Auta, Emenike, Jayanthi and Fauziah2018) described two pure bacterial cultures, Rhodococcus sp. strain 36 and Bacillus sp. strain 27 from mangrove sediments having capability for PP MP degradation. Bacillus cereus and Bacillus gottheilii were found to degrade PE (weight loss: 1.6%), polyethylene terephthalate (PET; 6.6%), and PS (7.4%) while, for B. gottheilii MP weight loss was 6.2%, 3.0%, 3.6% and 5.8% for PE, PET, PP and PS, respectively (Auta et al., Reference Auta, Emenike, Jayanthi and Fauziah2018). Stenotrophomonas maltophilia LB 2-3 was found to decrease molecular weight and tensile properties of polylactic acid (PLA) (Jeon and Kim, Reference Jeon and Kim2013a). E. coli was able to degrade polyurethanes (1–2% after 72 hours) (Uscategui et al., Reference Uscategui, Arevalo, Diaz, Cobo and Valero2016). Polypropylene films (PP) and Bioriented Polypropylene (BOPP) polymers were reported to be degraded by microorganisms to some extent (Longo et al., Reference Longo, Savaris, Zeni, Brandalise and Grisa2011). Pseudomonas aeruginosa was found to degrade PS-PLA nanocomposites (Shimpi et al., Reference Shimpi, Borane, Mishra and Kadam2012). Mohan et al. (Reference Mohan, Sekhar, Bhaskar and Nampoothiri2016) found that newly isolated Bacillus strains were able to degrade brominated high-impact polystyrene (HIPS). Bacterial strains (Enterobacter asburiae YT1 and Bacillus sp. YP1) residing in gut of waxworms were able to degrade PE (Yang et al., Reference Yang, Yang, Wu, Zhao and Jiang2014).
Park and Kim (Reference Park and Kim2019) reported a bacterial consortium consisting mainly of Bacillus sp. and Paenibacillus sp. was able to reduce the dry weight of MP particles by 14.7% and the mean diameter of MP particles by 22.8% after 60 days. Tsiota et al. (Reference Tsiota, Karkanorachaki, Syranidou, Franchini and Kalogerakis2018) reported Agios and Souda bacterial community to decrease the weight of HDPE MPs by 8 and 18% after 2 months, respectively. The gut microbes of earthworm (Lumbricus terrestris) containing members of Actinobacteria and Firmicutes genera were able to degrade LDPE MPs upon ingestion by the earthworm (Huerta Lwanga et al., Reference Huerta Lwanga, Thapa, Yang, Gertsen, Salanki, Geissen and Garbeva2018). Syranidou et al., (Reference Syranidou, Karkanorachaki, Amorotti, Repouskou, Kroll, Kolvenbach, Corvini, Fava and Kalogerakis2017) demonstrated that bacterial consortium was capable of developing dense biofilm on the weathered surface of PE and induced alterations in the surface topography and rheological properties. Exiguobacterium sp. strain YT2 isolated from gut of mealworm showed potential signs of degrading and mineralising PS MP over a time of 28 days (Yang et al., Reference Yang, Yang, Wu, Zhao, Song, Gao, Yang and Jiang2015a; Brandon et al., Reference Brandon, Gao, Tian, Ning, Yang, Zhou, Wu and Criddle2018).
Predators
Predatory ciliate Ephelota and sulphide-oxidising ectosymbiotic bacteria were reported to exist in a symbiotic relationship on the plastisphere (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013; Kettner et al., Reference Kettner, Oberbeckmann, Labrenz and Grossart2019). Positive associations between Amoebophrya and Suessiaceae on polyethylene were reported as well (Kettner et al., Reference Kettner, Oberbeckmann, Labrenz and Grossart2019). SEM and molecular data have also confirmed that choanoflagellates, radiolaria and small flagellates such as Micromonas also constitute the predatory guild in the plastisphere that devours bacteria and other organisms (Amaral-Zettler et al., Reference Amaral-Zettler, Zettler and Mincer2020).
Pathogens
Various potentially pathogenic microorganisms are reported to be attached with plastic debris (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013) including the members of the Campylobacteraceae, Aeromonas salmonicida or Arcobacter spp. from all over the world (McCormick et al., Reference McCormick, Hoellein, Mason, Schluep and Kelly2014; Kirstein et al., Reference Kirstein, Kirmizi, Wichels, Garin-Fernandez, Erler, Löder and Gerdts2016; Oberbeckmann et al., Reference Oberbeckmann, Osborn and Duhaime2016; Jiang et al., Reference Jiang, Zhao, Zhu and Li2018; Curren and Leong, Reference Curren and Leong2019). Various phototrophic species able to cause harmful algal blooms are also reported from plastic debris (Masó et al., Reference Masó, Fortuño, De Juan and Demestre2016; Casabianca et al., Reference Casabianca, Capellacci, Giacobbe, Dell’Aversano, Tartaglione, Varriale, Narizzano, Risso, Moretto, Dagnino and Bertolotto2019). Plastisphere communities can be dominated by genus Vibrio during the summer months. Moreover, they can transport potential protistan coral pathogens (Goldstein et al., Reference Goldstein, Carson and Eriksen2014) and a known fish pathogen (Virsek et al., Reference Virsek, Lovsin, Koren, Krzan and Peterlin2017). A recent study has reported the presence of Campylobacteraceae in microplastic particles which can cause gastrointestinal infections in humans (McCormick et al., Reference McCormick, Hoellein, Mason, Schluep and Kelly2014).
The knowledge regarding the microbial composition of MP-associated biofilm has revolutionised in the last half of the decade with the advent of metagenomics (Ivar do Sul et al., Reference Ivar do Sul, Tagg and Labrenz2018). The biofilm communities differ significantly from their surrounding environment (Zettler et al. Reference Zettler, Mincer and Amaral-Zettler2013; Oberbeckmann et al. Reference Oberbeckmann, Loeder, Gerdts and Osborn2014; Amaral-Zettler et al. Reference Amaral-Zettler, Zettler, Slikas, Boyd, Melvin, Morrall, Proskurowski and Mincer2015; Bryant et al. Reference Bryant, Clemente, Viviani, Fong, Thomas, Kemp, Karl, White and DeLong2016; Debroas et al. Reference Debroas, Mone and Ter Halle2017; Frere et al. Reference Frère, Maignien, Chalopin, Huvet, Rinnert, Morrison, Kerninon, Cassone, Lambert, Reveillaud and Paul-Pont2018), which is obvious since microbial community compositions on natural particles usually differ from free-living microorganisms (Crespo et al. Reference Crespo, Pommier, Fernandez-Gomez and Pedros-Alio2013; Rieck et al., Reference Rieck, Herlemann, Jurgens and Grossart2015).
The outline of plastisphere is shown in Figure 1. Different strains of bacteria and fungi capable of degrading plastics, along with the types of microplastics degraded are summarised in Table 1.

Figure 1. Plastisphere, the novel microbial community colonising and thriving on the plastic debris.
Table 1. Different strains of bacteria and fungi capable of degrading plastics, along with the types of microplastics degraded

Action of microorganisms on microplastics
Microbial degradation of MPs occurs through various steps: (1) Initial degradation of polymers to small-size particles from large polymeric structures, (2) Degradation of polymers to their oligomer, dimer and monomers and, (3) Mineralisation of MPs by microbial biomass (Blair Espinoza, Reference Blair Espinoza2019). Upon complete mineralisation, microplastics breakdown forming carbon dioxide by various enzymes, and the transformation of produced intermediates to use as a source of energy and biomass production.
The majority of synthetic polymers such as polyethylene (PE), polypropylene (PP) and polystyrene (PS) are degraded very slowly (Weinstein et al., Reference Weinstein, Crocker and Gray2016). Even the novel consortia like Enterobacter and Pseudomonas developed from cow dung showed faster biodegradation of PE and PP, demonstrated up to 15% weight loss after 120 days (Skariyachan et al., Reference Skariyachan, Taskeen, Kishore, Krishna and Naidu2021). No information is available till date about the actions of depolymerases enzymes and the microbial degradation mechanisms of these microplastics and plastic debris (Ru et al., Reference Ru, Huo and Yang2020). Bacteria, fungi and enzymes are identified based on their bioremediation capacity for PP, PE and PVC has been demonstrated based on their capacity of biofilm formation on plastic films, surface deterioration, thermal and mechanical properties alteration of plastics (Yang et al., 2015b; Auta et al., Reference Auta, Emenike, Jayanthi and Fauziah2018; Giacomucci et al., Reference Giacomucci, Raddadi, Soccio, Lotti and Fava2019; Ru et al., Reference Ru, Huo and Yang2020). Though most of the studies are based on macro-plastic films, however the same can be applied to MPs as well.
Bacterial metabolism and degradation of microplastics
Biodegradation of plastics requires the action of microbial enzymes (both bacterial and fungal) for converting them into easily metabolizable fractions (dimers, oligomers and monomers) for example lipase hydroxylase, depolymerase and protease (Haider et al., Reference Haider, Volker, Kramm, Landfester and Wurm2019). After disintegration, microorganisms feed on those products as carbon source for their growth and reproduction (Haider et al., Reference Haider, Volker, Kramm, Landfester and Wurm2019; Zuo et al., Reference Zuo, Li, Lin, Sun, Diao, Liu, Zhang and Xu2019a). Apart from that, the potential microbial degrading organisms must possess appropriate enzymatic and metabolic pathways. Physical characteristics of microplastics should facilitate attachment of microorganisms to the surface and biological reactions should not be affected by the branching pattern.
The complex interaction between the available surfaces for the colonisation of microorganisms forming biofilm was studied by Fleming et al. (Reference Fleming, Chahin and Rumbaugh2017). The attachment processes that act on MP biofilm include (1) biofouling, (2) plasticiser breakdown, (3) assault on the polymer backbone, (4) hydration and (5) organism penetration into the polymer structure.
Various bacterial taxa are renowned as plastic colonisers, for example, Arcobacter spp., Vibrio spp., Colwellia spp., Escherichia spp. and Pseudomonas spp. (Oberbeckmann and Labrenz, Reference Oberbeckmann and Labrenz2020). In research conducted by Oberbeckmann et al. (Reference Oberbeckmann, Osborn and Duhaime2016), it was reported that assemblages of microbes on the surface of polymers are not substrate-specific. Curren and Leong (Reference Curren and Leong2019) found colonisation of diverse epiplastic bacteria including Erthrobacter, Vibrio and Pseudomonas species. While another research reported rapid colonisation of low-density polyethylene (LDPE) microplastic by costal marine sediments bacteria including Arcobacter and Colwellia spp. (Harrison et al., Reference Harrison, Schratzberger, Sapp and Osborn2014). Members of bacterial families like Rhodobacteraceae and Flavobacteriaceae and the genera Albirhodobacter, Methylotenera, and Hydrogenophaga have been found to be widespread on the surface of MPs (Oberbeckmann and Labrenz, Reference Oberbeckmann and Labrenz2020). Rosato et al. ( 2020) used a PCB-dechlorinating microbial culture to study microbial colonisation of various MPs (polyethylene, polyethylene terephthalate, polystyrene, polypropylene and polyvinyl chloride) and found all the MP to be colonised by microbes and also the microbes performed dechlorination proving their potential to remediate toxicity related to PCB polluted MPs (Rosato et al., Reference Rosato, Barone, Negroni, Brigidi, Fava, Xu, Candela and Zanaroli2020).
Research indicates that using bacterial consortia in MP biodegradation is more efficient rather than using single bacterial culture. Recent research conducted on MP degradation by using microbial consortia demonstrated maximum mineralisation in around 15 days (MeyerCifuentes et al., Reference Meyer-Cifuentes, Werner, Jehmlich, Will, Neumann-Schaal and Öztürk2020).
Bacterial and fungal strains, including Phaeosphaeria spartinicola, P. halima, Mycosphaerella sp. assemblages can decompose complex polymers and may potentially form biofilms on the surface and ingest MP particles (Kawai et al., Reference Kawai, Kawabata and Oda2019). Various marine hydrocarbonoclastic bacteria (e.g., Alcanivorax borkumensis) demonstrated capacity to degrade alkanes, branched aliphatic, as well as isoprenoid hydrocarbons, and alkyl cycloalkanes (Davoodi et al., Reference Davoodi, Miri, Taheran, Brar, Galvez-Cloutier and Martel2020). Researchers have also found possible potential of various marine bacteria for MP biodegradation due to their ability to form biofilms (Salvador et al., Reference Salvador, Abdulmutalib, Gonzalez, Kim, Smith, Faulon, Wei, Zimmermann and Jimenez2019). Evidence of LDPE degradation by A. borkumensis was observed while investigating biofilm formation on LDPE surface (Delacuveellerie et al., Reference Delacuvellerie, Cyriaque, Gobert, Benali and Wattiez2019). Alcanivorax sp. also interacts with plastic surface by adjusting its hydrophilicity of the cell membrane (Delacuvellerie et al., Reference Delacuvellerie, Cyriaque, Gobert, Benali and Wattiez2019). Recently, putative laccase isolated from actinomycete Rhodococcus ruber was found to be involved in PE biodegradation. Some bacterial strains obtained from sewage treatment plants, mulch films and landfills waste demonstrated the ability to utilise unpretreated PE, and PP strips as a carbon source ( Jeyakumar et al., Reference Jeyakumar, Chirsteen and Doble2013). In the presence of starch, microorganisms showed accelerated hydrolytic biodegradation of PP and PE (Cacciari et al., Reference Cacciari, Quatrini, Zirletta, Mincione, Vinciguerra, Lupattelli and Giovannozzi Sermanni1993) also the enzymatic reactions make the polymers more vulnerable to biodegradation (Ru et al., Reference Ru, Huo and Yang2020).
Fungal metabolism and degradation of microplastics
Fungi are excellent candidates for potential plastic degradation owing to their variety of metabolic potential and ability to degrade complex chemical structures (Lacerda et al., Reference Lacerda, Proietti, Secchi and Taylor2020). Marine fungus Zalerion maritimumis was reported to be successfully biodegrade PE (Paço et al., Reference Paço, Duarte, da Costa, Santos, Pereira, Pereira, Freitas, Duarte and Rocha-Santos2017). A wide diversity of epiplastic fungal species was reported from various geographic locations indicating their abundance in aquatic environments (Lacerda et al., Reference Lacerda, Proietti, Secchi and Taylor2020). Different fungal genera such as Aspergillus, Cladosporium, and Wallemia as well as a wide variety of taxa such as, Chytridiomycota and Aphelidomycota were reported from various locations (Lacerda et al., Reference Lacerda, Proietti, Secchi and Taylor2020). The fungal diversity thriving in the biofilm of microplastics found dominance of members from Chytridiomycota, Cryptomycota and Ascomycota (Kettner et al., Reference Kettner, Rojas-Jimenez, Oberbeckmann, Labrenz and Grossart2017). Brunner et al. (Reference Brunner, Fischer, Rüthi, Stierli and Frey2018) reported around 100 fungal species able to degrade MP debris highlighting the potential of marine epiplastic fungal communities. The isolated fungal strains were found successful to degrade Polyurethane (PU) but not PE polymers (Brunner et al., Reference Brunner, Fischer, Rüthi, Stierli and Frey2018). Lignin-biodegrading fungi catalyses the oxidation of aromatic and non-aromatic substrates like chorophenolic or nonphenolic compounds (polymethylmethacrylate [PMMA] and polyhydroxybutyrate [PHB]) by producing laccase (Straub et al., Reference Straub, Hirsch and Burkhardt-Holm2017).
Enzymatic degradation of microplastic: Microbial gene action and metabolism
Enzymes play a vital role in cellular functions regulation; microbes tend to modify their enzyme activity in response to changing environmental conditions. Inside the biofilm, the microbial enzymes build up a microenvironment interacting with the MP causing its degradation. Extracellular enzymes of microbes (esterase, lipase, lignin peroxides, laccase and manganese peroxides) are indispensable for increasing the hydrophilicity of plastic polymers by converting to functional carbonyl or alcohol groups, which can enhance microbial attachment and promotes biodegradation (Shahnawaz et al., Reference Shahnawaz, Sangale and Ade2019; Taniguchi et al., Reference Taniguchi, Yoshida, Hiraga, Miyamoto, Kimura and Oda2019). Extracellular enzymes such as hydrolases (such as lipases, esterase, poly (3-hydroxybutyrate) depolymerases and cutinases) operate on the plastic surface to break it down into smaller molecules (Sol et al., Reference Sol, Laca, Laca and Díaz2020). Integration of monomers transported into the cytoplasm of microbes and finally their metabolism results in their assimilation (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013). There are two types of MP degrading enzymes according to their mechanisms namely, surface modification mechanisms and degradation mechanisms (Vertommen et al., Reference Vertommen, Nierstrasz, van Der and Warmoeskerken2005). The first category of enzymes consists of a group of hydrolases (lipases, carboxylesterases, cutinases and proteases), while the polymer degrading enzymes includes oxidases, amidases, laccases, hydrolases and peroxidases (Álvarez-Barragán et al., Reference Álvarez-Barragán, Domínguez-Malfavón, Vargas-Suárez, González-Hernández, Aguilar-Osorio and Loza-Tavera2016; Gómez-Méndez et al., Reference Gómez-Méndez, Moreno-Bayona, Poutou-Pinales, Salcedo-Reyes, Pedroza-Rodríguez, Vargas and Bogoya2018). Large polymers are broken down into smaller fragments with the help of enzymes, the fragment then gets ingested by the microbes and finally gets incorporated into the metabolic cell cycle. The extracellular polymeric substance (EPS) regulates the microenvironment and maintains the microbial community (Lucas et al., Reference Lucas, Bienaime, Belloy, Queneudec, Silvestre and NavaSaucedo2008) also governing the characteristics of the biofilm determining microbial penetration rate (Lucas et al., Reference Lucas, Bienaime, Belloy, Queneudec, Silvestre and NavaSaucedo2008) and ultimately control the fate of the plastic particles.
Despite ample evidence that EPS and enzymes have a significant influence on the biodeterioration of MPs, there is currently a dearth of technical options for the total destruction of these MPs (Kumar et al., Reference Kumar, Anjana, Hinduja, Sujitha and Dharani2020). Thus, it can be said that although a variety of microplastic (PE, PP and PVC) degrading microbes has been identified but microbial enzymes known to successfully degrade PE (laccase, soybean peroxidase, manganese peroxidase) and PS (hydroquinone peroxidase) are very few (Zhang et al., Reference Zhang, Wang, Gong and Gu2004; Santos et al., Reference Santos, Gangoiti, Keul, Moller, Serra and Llama2013) and none for PP degradation (Arutchelvi et al., Reference Arutchelvi, Muniyasamy, Arkatkar, Doble, Bhaduri and Uppara2008).
Microorganisms are able to produce enzymes that can breakdown complex polymer of plastics using them as carbon source for their growth and metabolism. Their ubiquity in different environments makes them the most promising natural and sustainable approach of MPs degradation. MPs degradation by microbes is influenced by various factors, including the type and structure of the plastic, environmental conditions (e.g., temperature, pH, oxygen availability) and microbial diversity and biochemical activities (Syranidou et al., Reference Syranidou, Karkanorachaki, Amorotti, Franchini, Repouskou, Kaliva, Vamvakaki, Kolvenbach, Fava, Corvini and Kalogerakis2017a). Among the reported potential MP degraders, bacteria play a significant role through various mechanisms. Bacterial strains in the genera Pseudomonas, Bacillus and Rhodococcus have the ability to breakdown and utilise different types of MPs (Yakimov et al., Reference Yakimov, Timmis and Golyshin2007; Syranidou et al., Reference Syranidou, Karkanorachaki, Amorotti, Franchini, Repouskou, Kaliva, Vamvakaki, Kolvenbach, Fava, Corvini and Kalogerakis2017a). Various types of extracellular enzymes like lipases and esterases are produced by the bacteria which help in MPs degradation (Auta et al., Reference Auta, Emenike, Jayanthi and Fauziah2018). MPs degradation potential have also been shown by fungi, especially by the species of Aspergillus, Penicillium and Fusarium (Tournier et al., Reference Tournier, Topham, Gilles, David, Folgoas, Moya-Leclair, Kamionka, Desrousseaux, Texier, Gavalda, Cot, Gu´emard, Dalibey, Nomme, Cioci, Barbe, Chateau, Andr´e, Duquesne and Marty2020; Solanki et al., Reference Solanki, Sinha and Singh2022). Enzymes like cellulases and ligninases are secreted by fungi which are able to degrade MPs polymers (Yang et al., Reference Yang, Xu, Zhang, Hu, Gao, Cui, Grossart and Luo2022). Algae, especially diatoms, can also potentially degrade MPs with the help of their extracellular enzymes like lipases and proteases, which are able to degrade and modify MPs surface properties (Sun et al., Reference Sun, Ren and Ni2020a; Zhu et al., Reference Zhu, Li, Wang and Duan2021). Algae such as Scenedesmus dimorphus (a green alga), Anabaena spiroides (a blue-green alga), Navicula pupula (a diatom), and various species of Oscillatoria, have been observed to thrive on polythene surfaces in sewage water (Chia et al., Reference Chia, Ying Tang, Khoo, Kay Lup and Chew2020). The adhesion of these colonising algae on to the MPs surface marks the start of biodegradation, and various ligninolytic and exopolysaccharide enzymes produced by them facilitates plastic degradation (Priya et al., Reference Priya, Jalil, Dutta, Rajendran, Vasseghian, Karimi-Maleh and SotoMoscoso2022). Additionally, with biofilm formation on the plastic surface, enhanced degradation of MPs takes place due to the secretion of a range of microbial enzymes, which also facilitates the establishment of a diverse microbial community, promoting efficient microplastics degradation (Oberbeckmann et al., Reference Oberbeckmann, Kreikemeyer and Labrenz2018; Ogonowski et al., Reference Ogonowski, Motiei, Ininbergs, Hell, Gerdes, Udekwu, Bacsik and Gorokhova2018). Within the biofilm, synergistic interactions among various microorganisms creates a microenvironment favourable for enzymatic activity which further enhances the degradation potential (Wei and Zimmermann, Reference Wei and Zimmermann2017; Tournier et al., Reference Tournier, Topham, Gilles, David, Folgoas, Moya-Leclair, Kamionka, Desrousseaux, Texier, Gavalda, Cot, Gu´emard, Dalibey, Nomme, Cioci, Barbe, Chateau, Andr´e, Duquesne and Marty2020).
Addressing the burning issue of plastic pollution requires detailed knowledge of the MPs degradation mechanism for implementation of effective remediation (Lopez-Pedrouso et al., Reference Lopez-Pedrouso, Varela, Franco, Fernandez and Aboal2020; Othman et al., Reference Othman, Hasan, Muhamad, Ismail and Abdullah2021). A wide range of enzymatic processes including hydrolysis, oxidation, reduction and esterification, can breakdown polymer bonds or modify polymer functional groups (Lopez-Pedrouso et al., Reference Lopez-Pedrouso, Varela, Franco, Fernandez and Aboal2020; Othman et al., Reference Othman, Hasan, Muhamad, Ismail and Abdullah2021). These enzymatic processes are crucial for breakdown of plastic polymer chains into smaller molecules that can serve as microorganisms’ source of carbon and energy. One of the most common plastics in our daily life polyethylene terephthalate (PET) can be degraded by PET hydrolases (Carniel et al., Reference Carniel, Valoni, Nicomedes, Gomes and Castro2017), which can be obtained from microorganisms like Ideonella sakaiensis and Rhodococcus sp. (Yoshida et al., Reference Yoshida, Hiraga, Takehana, Taniguchi, Yamaji, Maeda, Toyohara, Miyamoto, Kimura and Oda2016; Wei and Zimmermann, Reference Wei and Zimmermann2017) and offers a promising avenue for PET plastic waste management. Also, cutinases have shown excellent ability in PET degradation (O’Neill et al., Reference O’Neill, Araújo, Casal, Guebitz and Cavaco-Paulo2007; Furukawa et al., Reference Furukawa, Kawakami, Tomizawa and Miyamoto2019) especially, microorganisms Thermobifida alba and Thermobifida fusca can achieve this. Additionally, Microbes like Trametes versicolor and Pycnoporus cinnabarinus with the help of laccases oxidise phenolic compounds, influencing the fate of PS and PU (Bilal et al., Reference Bilal, Iqbal and Barcelo2019; Ghatge et al., Reference Ghatge, Yang, Ahn and Hur2020; Yao et al., Reference Yao, Xia, Dou, Du and Wu2022). Notably, in plastic degradation lignin peroxidases and manganese peroxidases, obtained from microorganisms such as Phanerochaete chrysosporium and Pleurotus ostreatus, have shown promise (Mukherjee and Kundu, Reference Mukherjee and Kundu2014; Ojha et al., Reference Ojha, Pradhan, Singh, Barla, Shrivastava, Khatua, Rai and Bose2017; Rovaletti et al., Reference Rovaletti, De Gioia, Fantucci, Greco, Vertemara, Zampella, Arrigoni and Bertini2023). Excellent plastic degrading potential is also shown by Cellulases, particularly against PS (Koeck et al., Reference Koeck, Pechtl, Zverlov and Schwarz2014; Pathak, Reference Pathak2017; Yan et al., Reference Yan, Wei, Cui, Bornscheuer and Liu2021c). The likes of Clostridium thermocellum and Cellulomonas fimi contribute to our understanding of this multifaceted enzymatic approach to plastic waste management.
Can microplastic develop antibiotic resistance in bacteria?
In the context of antibiotic resistance, two freshwater studies demonstrated microplastic-associated assemblages having an increased transfer frequency of a plasmid coding for trimethoprim resistance (Arias-Andres et al., Reference Arias-Andres, Klumper, Rojas-Jimenez and Grossart2018) and higher abundance of the gene int1, a proxy for anthropogenic pollution (Eckert et al., Reference Eckert, Di Cesare, Kettner, Arias-Andres, Fontaneto, Grossart and Corno2018).
MPs can selectively enrich both antibiotics and antibiotic-resistant bacteria on their surfaces in various environments (Wu et al., Reference Wu, Pan, Li, Li, Bartlam and Wang2019; Su et al., Reference Su, Zhang, Zhu, Shi, Wei, Xie and Shi2020; Sun et al., Reference Sun, Cao, Duan, Wang, Ding and Wang2020b; Wang et al., Reference Wang, Xue, Li, Zhang, Pan and Luo2020). Zhang et al. (Reference Zhang, Lu, Wu, Wang and Luo2020) isolated and characterised antibiotic-resistant marine bacteria from MP particles which showed the presence of several multidrug-resistant marine bacteria including pathogenic Vibrio species (Zhang et al., Reference Zhang, Lu, Wu, Wang and Luo2020). Other studies showed the presence of multidrug-resistant pathogens from Vibrio sp. (Laverty et al., Reference Laverty, Primpke, Lorenz, Gerdts and Dobbs2020) and E. coli (Song et al., Reference Song, Jongmans-Hochschulz, Mauder, Imirzalioglu, Wichels and Gerdts2020) on marine microplastics. A study published recently described whole genome sequencing (WGS) of antibiotic-resistant microorganisms isolated from marine plastics (Radisic et al., Reference Radisic, Nimje, Bienfait and Marathe2020).
Enrichment of various Antibiotic Resistant Genes (ARGs) like sul1, tetA, tetC, tetX and ermE was reported from plastic particles in both sea and freshwater (Wang et al., Reference Wang, Xue, Li, Zhang, Pan and Luo2020) and selective enrichment of strB, blaTEM, ermB, tetM and tetQ was seen on MP particles from landfill leachates (Shi et al., Reference Shi, Wu, Su and Xie2020). Lu et al. (Reference Lu, Lu and Liu2020) reported the presence of upto 43 different ARGs on MP surface from vegetable soil using advanced high-throughput qPCR screening.
Using Shotgun metagenomics 64 different ARG subtypes providing resistance against 13 different antibiotics on macroplastics and microplastics were collected from North Pacific Gyre (Yang et al., Reference Yang, Liu, Song, Ye, Lin, Li and Liu2019). This research and numerous others have discovered clinically relevant ARGs on MP particles, such as sul1, tetA, tetC, tetX, ermE, aac(3), macB and blaTEM, which are typically present in human infections (Alcock et al., Reference Alcock, Raphenya, Lau, Tsang, Bouchard, Edalatmand, Huynh, Nguyen, Cheng and Liu2020), suggesting that MPs serves as reservoirs of clinically important antibiotic resistance genes. The numerous antibiotics and active metabolites present in various chemicals and heavy metals (Godoy et al., Reference Godoy, Blázquez, Calero, Quesada and Martín-Lara2019; Chen et al., Reference Chen, Gu, Bao, Ma and Mu2020; Mammo et al., Reference Mammo, Amoah, Gani, Pillay, Ratha, Bux and Kumari2020; Wang et al., Reference Wang, Xue, Li, Zhang, Pan and Luo2020) adsorbed to MPs leads to multidrug resistance among different bacteria within microbial biofilm resulting in active selection of antibiotic resistance on MP surfaces.
Imran et al. (Reference Imran, Das and Naik2019) have advocated that the development and spread of multiple drug-resistant human pathogens occur by co-contamination of MPs and heavy metals through co-selection mechanisms.
Role of environmental factors in microbial degradation of microplastics
Various environmental factors like pH, moisture, salinity and temperature play vital roles in bioplastic degradation (Gong et al., Reference Gong, Duan and Zhao2012). While, factors like temperature and ionic strength facilitate the colonisation of microbes and biofilm formation (Rummel et al., Reference Rummel, Jahnke, Gorokhova, Kühnel and Schmitt-Jansen2017), the presence of nutrients and other pollutants, as well as the availability of light and pressure are known to affect the nature and extent of microbial attachment (Harrison et al., Reference Harrison, Hoellein, Sapp, Tagg, Ju-Nam and Ojeda2018). The surrounding environment is a major player for biofilm structuring and low nutrient and high salinity forms substrate-specific assemblages (Oberbeckmann et al., Reference Oberbeckmann, Kreikemeyer and Labrenz2018). An optimum temperature is required which favours enzymatic activity and microbial growth (Shruti and Kutralam-Muniasamy, Reference Shruti and Kutralam-Muniasamy2019). Moisture content and salinity are other important factors for biodegradation (Gong et al., Reference Gong, Duan and Zhao2012). Bioplastic degradation is kick-started by water uptake followed by the breakage of ester bonds. pH of the medium also affects the rate of reaction, alkaline condition favours hydrolysis of PLA (Elsawy et al., Reference Elsawy, Kim, Park and Deep2017). Comparative analysis revealed that the plastic material is a minor factor determining MP-associated biofilms while the most important factor was geographical region (Amaral-Zettler et al., Reference Amaral-Zettler, Zettler, Slikas, Boyd, Melvin, Morrall, Proskurowski and Mincer2015).
Keeping the above facts in mind, it can be inferred that finding a natural environment for complete bioplastic degradation is a difficult task. A potential solution to this situation is the use of engineered microorganisms which can degrade bioplastics with highest efficiency while withstanding extreme environmental conditions (Danso et al., Reference Danso, Schmeisser, Chow, Zimmermann, Wei, Leggewie, Li, Hazen and Streit2018b).
Role of microbial biofilms in microplastic degradation
Biofilms are diverse microbial communities consisting of bacteria, fungi, algae, and so on thriving on any surface submerged and in spatial proximity (Donal, Reference Donlan2002). The microorganisms are collectively benefited by having a stable consortia, availability of nutrients, and protection from desiccation (Zettler et al., Reference Zettler, Mincer and Amaral-Zettler2013). Biofilm can affect microplastic structure and function in various ways, with the help of various enzymes. These enzymes can transform surface properties, additive degradation and metabolic by-product release, thus can determine the fate of MPs in marine environments (Miao et al., Reference Miao, Wang, Hou, Yao, Liu, Liu and Li2019).
MP biodegradation starts off after the attachment of first microbes and development of biofilm on the plastic surface (plastisphere). The plastisphere comprises of various microbial communities developing into a biofilm (Urbanek et al., Reference Urbanek, Rymowicz and Mirończuk2018). The plastic surface encourages microbial colonisation and biofilm formation leading to a reduction in polymer buoyancy and hydrophobicity (Lobelle and Cunliffe, Reference Lobelle and Cunliffe2011). Various polymer additives being easily metabolised, encourages microorganisms’ initial attachments and biofilm formation (Ru et al., Reference Ru, Huo and Yang2020). Microbes ensure surface attachment by various mechanisms like, cell surface charge changes and hydrophobicity and modifications in EPS production, pioneer microbes then modify surface morphology for additional microbial colonisation. Various substratum property like crystallinity, melting temperature, roughness, and so on might influence microbial community assemblages during colonisation stages (Rummel et al., Reference Rummel, Jahnke, Gorokhova, Kühnel and Schmitt-Jansen2017). Various factors affecting biofilm degradation of MPs are summarised in Figure 2; biofilm formation process and microplastic degradation are shown in Figure 3.

Figure 2. Various factors affecting biofilm degradation of microplastics. Reprinted after Sun et al. (Reference Sun, Xiang, Xiong, Fang and Wang2023), under a creative commons licence, open access.

Figure 3. Bacterial colonisation, biofilm formation and degradation of microplastics. Reprinted after Sun et al. (Reference Sun, Xiang, Xiong, Fang and Wang2023), under a creative commons licence, open access.
Biofilm maturation into complex structures is apparently achieved through quorum sensing (QS) processes among cells. QS Signalling gene gets accumulated in the external environment and regulates neighbouring cells specific gene expression. A few bacterial species use QS to coordinate and regulate the disassembly of the biofilm (Sharma et al., Reference Sharma, Misba and Khan2019). Various researches have suggested active QS involvement in the organisation and development of multispecies biofilms in the marine environment (Hmelo, Reference Hmelo2017).
After colonisation, biodeterioration of the polymer follows leading to loss in physical integrity and loss of polymer’s mechanical properties (Kumar et al., Reference Kumar, Anjana, Hinduja, Sujitha and Dharani2020). The process is primarily achieved by exoenzymes secreted by microbes. At this point, the microbe’s EPS offers strong adherence to the polymer surface, and the enzymes’ catalytic activity commences the dissolution of the polymeric structure. EPS and enzymes are thought to have significant influences on the biodeterioration of MPs (Kumar et al., Reference Kumar, Anjana, Hinduja, Sujitha and Dharani2020). The deteriorated polymers are then converted to oligomers, dimers and monomers through biofragmentation and finally get assimilated (Amobonye et al., Reference Amobonye, Bhagwat, Singh and Pillai2020). The catalytic axctivities of microbial enzymes control the biodegradation of MPs mediated by biofilm promoting fragmentation of the polymer. The fragments formed through enzymatic depolymerisation can be assimilated by microbes resulting in increase in their biomass (Degli-Innocenti, Reference Degli-Innocenti2014). These enzymes hydrolyze polymers by a nucleophilic attack on the carbonyl carbon, resulting in either readily assimilable oligomeric or monomeric elements or compounds that require more processing before they can be digested by the microorganisms (Kumar et al., Reference Kumar, Anjana, Hinduja, Sujitha and Dharani2020).
During assimilation, the fragmented polymers gets integrated in the microbial cells (Lucas et al., Reference Lucas, Bienaime, Belloy, Queneudec, Silvestre and NavaSaucedo2008). Enzymes and carrier proteins are involved in assimilation of monomers with catabolic cycles for energy production (Hosaka et al., Reference Hosaka, Kamimura, Toribami, Mori, Kasai, Fukuda and Masai2013; Durairaj et al., Reference Durairaj, Hur and Yun2016). The assimilation step is mostly followed by mineralisation, in which the polymers get completely degraded and final products like CO2, N2, H2O and CH4 are released. Complete mineralisation of PET has been reported to produce acetic acid which gets used in the Krebs cycle or integrated into lipid synthesis (Wilkes and Aristilde, Reference Wilkes and Aristilde2017). Polymer physiochemical properties such as hydrophobicity, surface energies and functional groups influence the formation of biofilm (Bhagwat et al., Reference Bhagwat, O’Connor, Grainge and Palanisami2021). For biological sedimentation, the conditioning layer is very important, which actually depends on the roughness, hydrophobicity and chemical nature of the initial matrix surface (Tu et al., Reference Tu, Zhou, Zhang, Liu, Luo, He and Luo2020).
Different biochemical processes in microbe-mediated microplastic degradation and related biotechnological interventions are shown in Figure 4.

Figure 4. Different biochemical processes in microbe-mediated microplastic degradation and related biotechnological interventions. Reprinted after Zhou et al. (Reference Zhou, Kumar, Sarsaiya, Sirohi, Awasthi, Sindhu, Binod, Pandey, Bolan, Zhang, Singh, Kumar and Awasthi2022) (Elsevier), licence number 5647011139377.
Microbial remediation of microplastic: Novel and emerging techniques
Absorption of microplastics by green algae
Algal cells can be effectively used for the breakdown of complex polymeric materials (Manzi et al., Reference Manzi, Abou-Shanab, Jeon, Wang and Salama2022). These algal cells may interact with MP particles and alter their properties which may determine the adsorption rate and fate of these MP particles (Kershaw et al., Reference Kershaw and Rochman2015). Algal consortium can be used in degradation of MP polymers as they do not require carbon source in the growth media and can easily adapt to different environmental conditions. Microalgae are known to produce biofilms on the surface of MP particles and help in the degradation process by producing ligninolytic and exopolysaccharide enzymes, this polymers on the other hand act as a source of carbon which promotes the growth of algal cells. The crucial processes that promote the degradation of MP particles includes corrosion, hydrolysis penetration and fouling (Chia et al., Reference Chia, Ying Tang, Khoo, Kay Lup and Chew2020). Green algae such as Oscillatoria subbrevis and Phormidium lucidum have been reported to colonise the surface of LDPE and degrade it without any pretreatment (Sarmah and Rout, Reference Sarmah and Rout2018). Recent advancement in biotechnology have led to the production of genetically modified microalgal cell factories capable of producing hydrolytic enzymes having the ability to degrade plastics. Green microalgae Chlamydomonas reinhardtii was genetically modified to produce polyethylene terephthalate hydrolase, able to degrade polyethylene terephthalate films and terephthalic acid (Kim et al., Reference Kim, Park, Tran, Cho, Choi, Lee and Kim2020). These microalgae affect the vertical flux of the polymers by varying the density of the polymers which can be incorporated into hetero-aggregates promoting adsorption. However, this process is poorly understood (Tadsuwan et al., Reference Tadsuwan and Babel2021).
Application of whole cell biocatalysis and microplastic immobilisation
Biocatalysis uses whole cells of bacteria, fungi, microalgae and plants to catalyse organic reaction. This provides high enantioselectivity and is considered as low or non-toxic, ecofriendly and green alternative to treat pollutant. Recent publications on biocatalysis concentrates on the use of enzymes and overexpression of enzymes in genetically engineered microbes but they are considered to be expensive as it requires resources to renew cofactors required for enzyme activities (Monti et al., Reference Monti, Ottolina, Carrea and Riva2011; Sheldon and Pereira, Reference Sheldon and Pereira2017). Biocatalysis may result in chemical transformation which may help in preparation of chiral compounds and organic compounds. They can effectively reduce carbon–carbon double bonds using highly selective processes and mild reactions. Thus, whole cell biocatalysis can be a sustainable technique in organic synthesis (Iqbal et al., Reference Iqbal, Rudroff, Brigé, Van Beeumen and Mihovilovic2012). Biocatalytic hydrogenation of alkanes can also serve as a crucial strategy for metal-assisted hydrogenation reactions. MP contains complex structures which can be effectively degraded and immobilised using whole cell biocatalysis.
Microplastic biodegradation by hyperthermophilic composting technology
Hyperthermophilic composting (hTC) is done at high temperature above 90°C using hyperthermophilic bacteria (Yu et al., Reference Yu, Tang, Liao, Liu, Zhou, Chen, Rensing and Zhou2018). As this process is performed at a very high temperature it leads to more efficient bioconversion within short period. Moreover, hTC is also effective for the reduction of antibiotic resistance genes and mobile genetic elements in sewage sludge due to the involvement of high temperature (Liao et al., Reference Liao, Zhao, Cui, Chen, Yu, Geisen, Friman and Zhou2019). Sewage sludge is one of the major sources of MP particles and removal of these MP particles by conventional treatment procedures can be ineffective. hTC can be effectively used for in situ biodegradation of sludge-based MPs. In a study made by Chen et al. (Reference Chen, Zhao, Xing, Xie, Yang, Cui, Lu, Liao, Yu, Wang and Zhou2019) hTC reported removal of 43.7% of MP from sewage sludge. hTC was also capable of degradation of 7.3 % of the PS-MPs at 70°C in 56 days through bio-oxidation. High-throughput sequencing showed the presence of Bacillus, Geobacillus and Thermus was directly linked with hTC which accelerate MP degradation and associated −C−C− bonds cleavage at high temperature (Manzur et al., Reference Manzur, Limon-Gonzalez and Favela-Torres2004). In addition, hTC leads to the killing of pathogens associated with MP and lacks nitrification and denitrification processes, preventing the excess loss of nitrogen (Kanazawa et al., Reference Kanazawa, Ishikawa, Tomita-Yokotani, Hashimoto, Kitaya, Yamashita, Nagatomo, Oshima, Wada and Force2008).
Future research directions in microplastic bioremediation
Studies regarding accumulation, characterisation, identification of microplastics are numerous, however, studies regarding their mitigation are still lagging. Biodegradation process of microplastics can be understood following four common approaches namely, (1) the depletion of substrates, (2) accumulation of biomass, (3) reaction products and (4) changes in substrate properties. Additionally, standard processes for plastic biodegradation involves producing microbial films on the polymeric surfaces followed by thier breakdown into smaller pieces (<20 μm). However, the methods are not economical and also not widely applicable. The micro-Fourier transform infrared (FTIR) can facilitate the mapping of samples, multiple polymer characterisation and identification of irregular shaped MPs and is widely accepted (Gong et al., Reference Gong, Kong, Li, Li, Li and Zhang2018), however, the method is quite expensive and time-consuming. Other analytical methods employed for MP identification includes scanning electron microscope-energy dispersive spectroscopy (SEM-EDS), pyrolysis-gas chromatography-mass spectrometry and ESEM-EDS (Horton et al., Reference Horton, Walton, Spurgeon, Lahive and Svendsen2017). MPs are classified as an emerging pollutants due to a paucity of MPs data from soil and water, which limits knowledge of their environmental impact. It is difficult to show the ecotoxicological risks of microplastics on the surrounding ecosystems due to a lack of quantitative data (Harrison et al., Reference Harrison, Ojeda and Romero-Gonzalez2012; Goel, Reference Goel2017; Gong et al., Reference Gong, Kong, Li, Li, Li and Zhang2018). In a nutshell, microplastic research is still in its infancy and more research is needed to address links between plastics and microplastics generation.
MPs have become a potential substrate for colonisation in the oceans, with members of the family Sphingomonadaceae in particular selectively colonising microplastic polymers. Based on our reanalysis of available literature and critical review, available information and evidences of MPs degradation by microbes are not adequate. According to Tagg and Labrenz (Reference Tagg and Labrenz2018) for ensuring environmental compatibility and sustainability, collective actions are needed in order to better understand microbial degradation of plastics focused research on understanding the microbial pathways that can potentially degrade plastic are needed.
Future studies should consider genomics and proteomics approach in order to amplify the rate of microbe-mediated degradation of MP. The use of fungi for degrading MP effectively is now receiving increased attention. Fungi that are able to withstand oxidation and corrosion will be identified and screened for investigating their degradation properties on MPs that are difficult to degrade paving the way towards new and innovative strategies focused at mitigating the environmental impacts of MPs (Paco et al., Reference Paço, Duarte, da Costa, Santos, Pereira, Pereira, Freitas, Duarte and Rocha-Santos2017). Since MP biodegradation is a complex issue testing the efficacy of the process under real-world conditions is of prime importance.
Although advanced and innovative methods have been developed for isolating strains capable of degrading MPs, still the number of bacteria selected for screening is mostly limited to taxa like Bacillus, Pseudomonas, Chelatococcus and Lysinibacillus fusiformis. However, the bacterial degradation efficiency of MPs is quite low (0–15%) and takes a long time (usually 0–3 months) showing that microbial degradation of MP is a slow and time consuming process. Thus, future studies should look into improving the bacteria-mediated degradation potential of MPs with the help of modern molecular techniques like in vitro transcription, in situ hybridization, high-throughput sequencing and PCR. Also, multiple bacterial consortia have been identified from various environments (Sangwan and Wu, Reference Sangwan and Wu2008) which have potential for MP degradation. However, these are complex processes due to the involvement of various microorganisms and multiple enzymes. Therefore, in depth studies are needed to get a clear idea about the factors influencing and mechanisms involved in the degradation process.
Also, the progression of the whole process of biofilm formation needs to be studied in detail, especially the microorganisms initiating the colonisation of a novel MP particle in different environments and the successional process leading to further colonisation by other microorganisms. This will help us in getting an idea about the whole process of MP biodegradation.
Future research should be oriented towards microbial remediation of plastic, especially understanding the microbial degradation mechanism and factors affecting it under real-world conditions so that it can be applied in real-world situations and can perform optimally and especially in developing innovative approaches towards microbial remediation through novel methods which are environmentally sound and sustainable.
Conclusions
Human activities have long continued to modify planet earth, but now it is time to access our impacts on the planet. Plastics have provided many benefits and have made our life easier. Perhaps it is time to reconsider the importance we place on contemporary conveniences. Planet Earth, with its many ecosystems and over 1030 microbial inhabitants (Flemming and Wuertz, Reference Flemming and Wuertz2019), has discovered a mechanism to biotransform and even to some extent degrade plastic materials. MPs are an emerging class of contaminants having ubiquitous presence in every environment and affecting them negatively. Currently, MPs are a serious issue not only in the aquatic ecosystem but also in terrestrial ecosystems. In this scenario, there is an urgent need to develop strategies for mitigating this problem. Very little knowledge is available on key depolymerase and the mechanisms of biodegradation of Low biodegradable MPs (e.g., PP, PE, PS, PVC) and the most common depolymerising method is pyrolysis. Thus, more research efforts are needed to identify depolymerases from the plastic-degrading strains. Surface-modifying enzymes and esterase’s (e.g., cutinases, lipase, PETase) have been tested for MPs and their catalytic efficiency towards initial PET hydrolysis varies depending on crystallinity and environment conditions. Thus, to protect enzymes within an engineered cellular environment application of whole-cell biodegradation has been recommended. Nevertheless, degradation of biodegradable MPs takes place under assisted conditions. Regarding the highly crystalline MPs sustained thermal hydrolysis has been recommended to make the polymer bioavailable to microbes. For composting biodegradation, uncertainty remains in the requirement for specific collection and composting facilities and various contaminated and toxic MPs residues. Thus, the primary focus should be to reduce MP footprint through the use of natural alternatives (e.g., jojoba beads, pumice, ground nutshells) and MPs isolation in environmental matrices in order to reduce MPs release into the environment. Future studies should aim at multi-omics approaches to decipher biochemical transformation mediated by microbes. Furthermore, biotransformation of plastic debris by microbes may potentially have an important role in the generation of micron- and sub-micron-scale polymer particles, which could impact human health and food security.
In marine environments, biofilms induce transformation of surface properties, the degradation of additives, and the release of metabolic by-products with the help of modifying/hydrolysing microbial enzymes. Thus, biofilms can be potential candidates for mitigation of microplastics. However, we have limited knowledge about microbial communities on MPs coated by biofilm, factors affecting their colonisation and their interaction with plastic substrate. There is an urgent need for studies regarding epispastic marine microbial communities and their ability to remediate MPs from aquatic environment. Bioengineered microbes and modified enzyme systems can provide exciting avenues of research in this regard.
Open peer review
To view the open peer review materials for this article, please visit http://doi.org/10.1017/plc.2024.26.
Author contribution
Conceptualization: S.B.; Submission: S.B.; Supervision: S.B.; Writing – original draft: A.T., A.B., S.D., S.B.; Writing – review and editing: S.D., S.B.
Financial support
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Competing interest
The authors declare none.


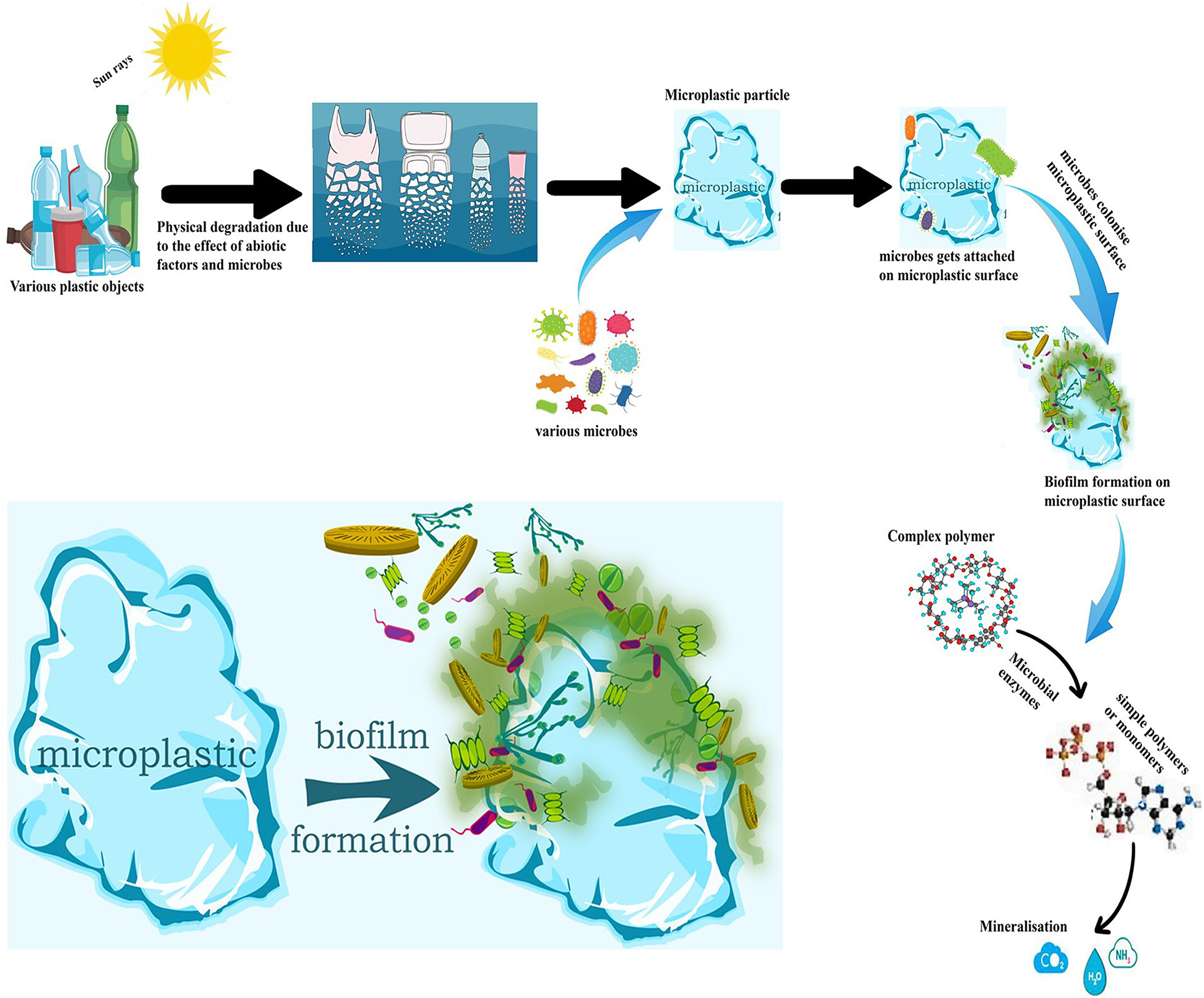






Comments
To the Editor, Date: 13.10.2023.
Cambridge Prisms: Plastics,
Cambridge University Press.
Dear Editor,
Please find enclosed the manuscript of the review article entitled “The good, the bad and the ugly: Critical insights on the applications of microbes in microplastic degradation”. We would like to submit this manuscript in “Cambridge Prisms: Plastics” Journal for possible publication.
This review summarizes current available knowledge on microplastic degradation by microbes and explores the novel and emerging techniques for microplastic remediation using microbes. It begins by highlighting the “good” aspects, where microbes exhibit the potential to break down microplastics through enzymatic activity and biofilm formation. The mechanisms underlying these degradation processes are explored, shedding light on the microbial diversity and plastic-specific enzymes involved. Conversely, the “bad” aspects of microbial involvement are examined. Microbes can facilitate the transport and bioaccumulation of microplastics in various ecosystems, potentially exacerbating their harmful effects. The “ugly” side of microplastic degradation includes the production of harmful byproducts during microbial breakdown, raising concerns about secondary pollution and toxicity. Although there are some papers highlighted microplastic degradation by microorganisms, this paper summarizes all the novel and emerging research methods and approaches, supported with comprehensive diagrams. We hope that this paper would attract many researchers working in the area of plastic pollution, bioremediation and microbial technologies.
Your early actions would be highly appreciated. I look forward to hearing from you.
Best regards,
Sayan Bhattacharya,
Assistant Professor,
School of Ecology and Environment Studies,
Nalanda University, India.