1. Introduction
Flowering plants demonstrate a wealth of phenotypic diversity, but the structure of their body pattern with well-defined body axes, organs and tissue organisation is highly similar across different species (Jürgens et al., Reference Jürgens, Ruiz, Laux, Mayer and Berleth1994). How such a stereotypical body pattern is achieved remains one of the most fascinating questions of developmental biology. Regulation of cell shape and identity is crucial in this process. Since plant cells are encased in cellulosic walls and are unable to move, proper morphology relies substantially on oriented cell division through adjustment of division plane positioning, so that daughter cells are placed in their specific locations (Smith, Reference Smith2001). Apart from controlling cell topology, positioning of division plane also determines whether a cell divides symmetrically and produces daughter cells of similar sizes, or asymmetrically with daughter cells of different sizes (Scheres & Benfey, Reference Scheres and Benfey1999). Asymmetric cell divisions, often considered formative divisions, are key to tissue formation, and require that resulting daughter cells not only differ in morphology, but most importantly possess distinct identities (Scheres & Benfey, Reference Scheres and Benfey1999). Acquisition of different identities is preceded by establishment of cellular polarity reflected in segregation of subcellular components such as hormones, mRNA, proteins and organelles between two daughter cells, and can be influenced by both intrinsic and extrinsic mechanisms (Dong et al., Reference Dong, MacAlister and Bergmann2009; Kimata et al., Reference Kimata, Higaki, Kawashima, Kurihara, Sato, Yamada, Hasezawa, Berger, Higashiyama and Ueda2016; Kimata et al., Reference Kimata, Kato, Higaki, Kurihara, Yamada, Segami, Morita, Maeshima, Hasezawa and Higashiyama2019; Kimata et al., Reference Kimata, Higaki, Kurihara, Ando, Matsumoto, Higashiyama and Ueda2020). Intrinsic regulation relies on activation of cell type-specific transcriptional networks, while extrinsic control is determined by cellular environment and involves cell-to-cell communication through different mobile molecular signals such as short miRNA, short peptides, hormones, as well as mechanical cues (Heisler et al., Reference Heisler, Hamant, Krupinski, Uyttewaal, Ohno, Jönsson, Traas and Meyerowitz2010; Schlereth et al., Reference Schlereth, Möller, Liu, Kientz, Flipse, Rademacher, Schmid, Jürgens and Weijers2010).
Plant morphogenesis and organogenesis are incredibly plastic (Dingkuhn et al., Reference Dingkuhn, Luquet, Quilot and De Reffye2005; Ramage & Williams, Reference Ramage and Williams2002). Unlike in animals, those processes continue in plants in post-embryonic development and are actively modulated by environmental cues. In fact, the vast majority of plant morphological features is formed post-embryonically. However, the basic tissue pattern with main tissue types such as epidermis, vascular and ground tissue, is formed during embryogenesis and is only reiterated in post-embryonic development (ten Hove et al., Reference ten Hove, Lu and Weijers2015). Early plant embryo development is not accompanied by the cell differentiation events that are widespread in post-embryonic development, such as root hair formation at the basal side of root epidermal cells, or suberin and lignin deposition in the root endodermis. This makes early embryogenesis a simple model allowing for dissection of pattern formation without the complication of cell differentiation pathways. Embryogenesis of most flowering plants is characterised by rather variable cell divisions – a trait highly undesirable for a model system intended for pattern formation studies (Johri et al., Reference Johri, Ambegaokar and Srivastava2013; Johri & Ambegaokar, Reference Johri and Ambegaokar1984; Pollock & Jensen, Reference Pollock and Jensen1964). Only few species, including the best characterised plant model object Arabidopsis thaliana, show a highly regular division pattern (Jürgens et al., Reference Jürgens, Ruiz, Laux, Mayer and Berleth1994; Mansfield & Briarty, Reference Mansfield and Briarty1991). Already at early stages of Arabidopsis embryogenesis comprising only 32 cells, the body axis and all major tissue types are established (ten Hove et al., Reference ten Hove, Lu and Weijers2015). The sufficiency of such a modest cell number together with the high predictability of its division pattern makes Arabidopsis embryogenesis an excellent model for studying plant body pattern formation. The versatility of this model was further cemented by development of embryo-optimised reporters of various subcellular structures, compartments and hormone response and establishment of cell fate-specific markers (Liao et al., Reference Liao, Smet, Brunoud, Yoshida, Vernoux and Weijers2015; Liao & Weijers, Reference Liao and Weijers2018).
While conventional approaches of mutant screening for defects in cell division and cell identity establishment revealed many important regulators of pattern formation (Benfey et al., Reference Benfey, Linstead, Roberts, Schiefelbein, Hauser and Aeschbacher1993; Laux et al., Reference Laux, Mayer, Berger and Jurgens1996; Scheres et al., Reference Scheres, Di Laurenzio, Willemsen, Hauser, Janmaat, Weisbeek and Benfey1995), its efficiency and sensitivity is inherently very low, particularly in species that have large gene families. Moreover, perturbations of cell division during pattern formation might lead to embryo lethality that renders mechanistic studies rather challenging. Relatively recent application of high-throughput transcriptomic and proteomic approaches has significantly advanced our understanding of pattern formation in different developmental contexts (Brady et al., Reference Brady, Orlando, Lee, Wang, Koch, Dinneny, Mace, Ohler and Benfey2007; Levesque et al., Reference Levesque, Vernoux, Busch, Cui, Wang, Blilou, Hassan, Nakajima, Matsumoto and Lohmann2006; Sozzani et al., Reference Sozzani, Cui, Moreno-Risueno, Busch, Van Norman, Vernoux, Brady, Dewitte, Murray and Benfey2010). The output of these studies was used to create publicly available databases and user-friendly tools that further facilitated elucidation of developmental processes (Le et al., Reference Le, Cheng, Bui, Wagmaister, Henry, Pelletier, Kwong, Belmonte, Kirkbride and Horvath2010; Palovaara et al., Reference Palovaara, Saiga, Wendrich, van‘t Wout Hofland, van Schayck, Hater, Mutte, Sjollema, Boekschoten and Hooiveld2017; Waese et al., Reference Waese, Fan, Pasha, Yu, Fucile, Shi, Cumming, Kelley, Sternberg and Krishnakumar2017). Apart from enabling deeper elucidation of developmental pathways, this advancement underscored the importance of quantitative approaches and incentivised generation of sensitive tools allowing for accurate quantitative analysis of dynamics of subcellular components as well as cell morphology and mechanics (de et al., Reference de Reuille, Routier-Kierzkowska, Kierzkowski, Bassel, Schüpbach, Tauriello, Bajpai, Strauss, Weber and Kiss2015; Liao et al., Reference Liao, Smet, Brunoud, Yoshida, Vernoux and Weijers2015; Liao & Weijers, Reference Liao and Weijers2018).
This manuscript aims to provide a current state of the art of the high-throughput and quantitative approaches that are presently used to shed light onto processes of plant body pattern formation using the early Arabidopsis embryo as a model. Furthermore, it outlines the current trends in the field and proposes directions for future research.
2. Quantification of cell morphology during pattern formation
The question of how a cell positions its division plane has been puzzling researchers for centuries. In the 19th century, scientists studying cell division in multicellular algae suggested that cells behave as soap bubbles and tend to minimise the interface between two daughter cells, such that cells divide along the shortest path (Errera, Reference Errera1888). However, for a given cell geometry, several short routes exist. Therefore, a cell divides along one of the shortest paths (Besson & Dumais, Reference Besson and Dumais2011). This hypothesis was successfully applied to explain cell divisions in the Arabidopsis shoot apex, however, it could not account for divisions in other contexts, particularly, where cells divide asymmetrically, for instance, during stomata development (Dong et al., Reference Dong, MacAlister and Bergmann2009). The question of how cells ‘break’ the symmetry became one of the central questions of developmental biology.
To address this question, one should be able to recognise whether a cell divides symmetrically or asymmetrically. This task is not as trivial as it might appear, in large part, due to complex polyhedral shapes that plant cells can adopt. The early two-dimensional approaches employing classical histology could not always resolve this complexity and the question whether pattern formation during embryogenesis is accompanied by morphologically asymmetric divisions remained open (Bougourd et al., Reference Bougourd, Marrison and Haseloff2000). The recent advancement in three-dimensional (3D) imaging allowed to address this question and accurately characterise morphology of patterning during Arabidopsis embryogenesis (Figure 1) (Moukhtar et al., Reference Moukhtar, Trubuil, Belcram, Legland, Khadir, Urbain, Palauqui and Andrey2019; Truernit et al., Reference Truernit, Bauby, Dubreucq, Grandjean, Runions, Barthélémy and Palauqui2008; Yoshida et al., Reference Yoshida, de Reuille, Lane, Bassel, Prusinkiewicz, Smith and Weijers2014). This advance became an important milestone in pattern formation research and consolidated the Arabidopsis embryo as an excellent model for its elucidation. The comprehensive 3D analysis of Arabidopsis embryogenesis demonstrated that formative divisions that are accompanied by expression of fate-specific markers are morphologically asymmetric, that is generating daughter cells with different volumes (Yoshida et al., Reference Yoshida, de Reuille, Lane, Bassel, Prusinkiewicz, Smith and Weijers2014). Examples where fate acquisition correlates with asymmetry in cell division are well known in post-embryonic development (De et al., Reference De Smet, Vassileva, De Rybel, Levesque, Grunewald, Van Damme, Van Noorden, Naudts, Van Isterdael and De Clercq2008; Robinson et al., Reference Robinson, de Reuille, Chan, Bergmann, Prusinkiewicz and Coen2011). Interestingly, the analysis almost invariably identified embryos of discrete 2-, 4-, 8- and 16-cell stages, suggesting that cell divisions might be synchronised during embryogenesis.
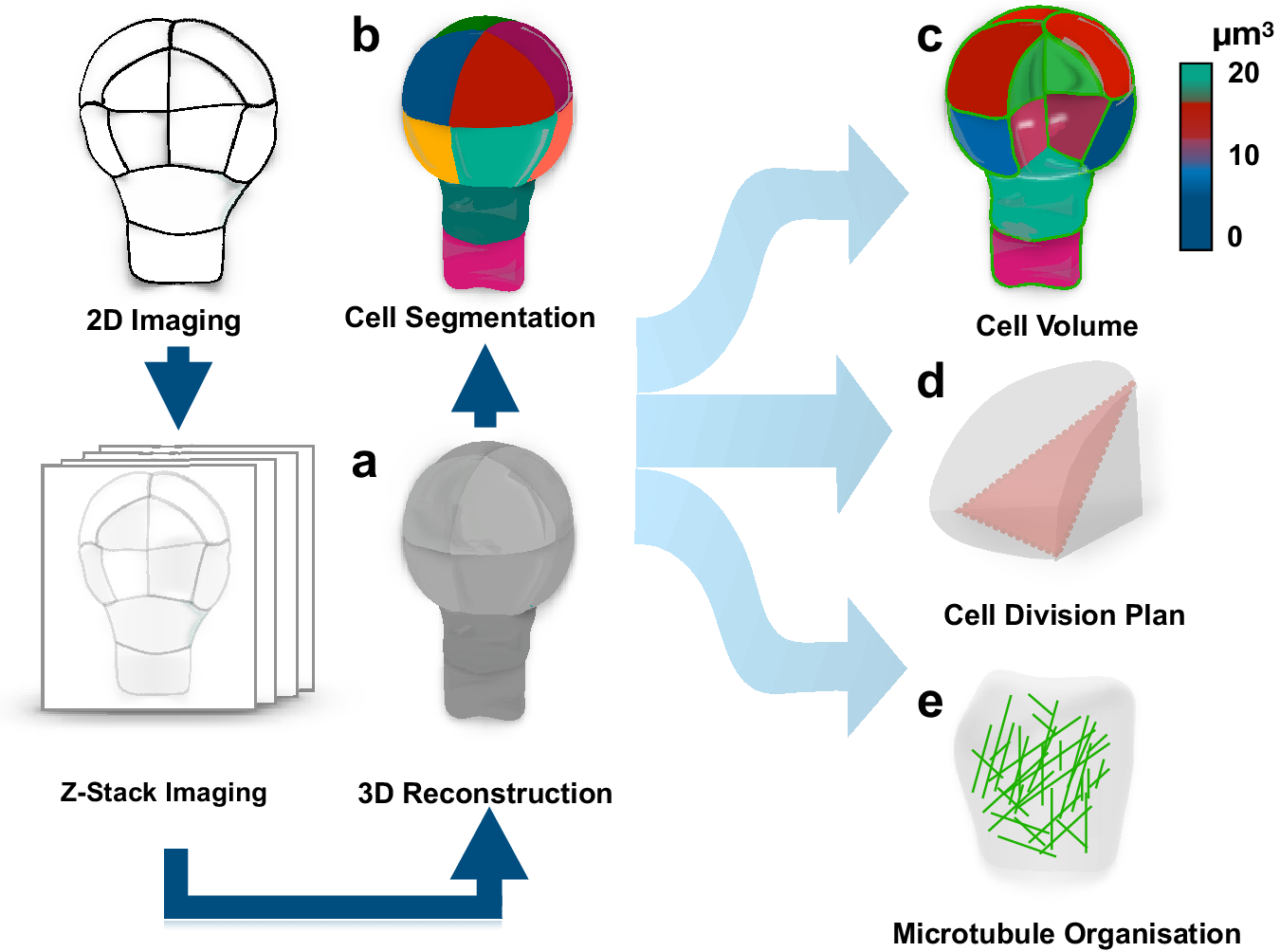
Fig. 1. Quantification of cell morphology during pattern formation. Data extracted from 3D reconstructed Arabidopsis embryogenesis (a) transferred into computational modelling environment using MorphoGraphX or PlanSeg (b) to accurately assess morphologies of individual cells (c), determine positioning of division planes (d) and trace fluorescently tagged subcellular structures (e). The data can be used in simulation modelling, allowing to trace interactions between shapes and cellular responses.
Most importantly, the study provided the first insight into how cells might disobey the shortest path rule and ‘break’ the symmetry of division. Specifically, it demonstrated that mutant embryos, constitutively expressing an inhibitor of auxin response, indole-3-acetic acid inducible 12/bodenlos (iaa12/bdl), switch from morphologically asymmetric to symmetric divisions, suggesting that breaking the symmetry is an auxin-dependent process (Yoshida et al., Reference Yoshida, de Reuille, Lane, Bassel, Prusinkiewicz, Smith and Weijers2014). A later study suggested that also the first asymmetric divisions can conform to a shortest path rule passing through the nuclear centroid, provided that newly inserted walls are curved (Moukhtar et al., Reference Moukhtar, Trubuil, Belcram, Legland, Khadir, Urbain, Palauqui and Andrey2019). Further, mechanistic studies will be required to determine the regulation of these divisions, and to address whether the same principle applies to post-embryonic development.
The accurate characterisation of cell morphologies during pattern formation was enabled by a powerful 3D image analysis tool, MorphoGraphX, that transfers recorded cell geometries into computational modelling environments (Figure 1) (de et al., Reference de Reuille, Routier-Kierzkowska, Kierzkowski, Bassel, Schüpbach, Tauriello, Bajpai, Strauss, Weber and Kiss2015). This novel computational approach instantly found application in addressing diverse questions in the field of developmental biology (Ma et al., Reference Ma, Miotk, Šutiković, Ermakova, Wenzl, Medzihradszky, Gaillochet, Forner, Utan and Brackmann2019; Sapala et al., Reference Sapala, Runions, Routier-Kierzkowska, Gupta, Hong, Hofhuis, Verger, Mosca, Li and Hay2018; Stanislas et al., Reference Stanislas, Platre, Liu, Rambaud-Lavigne, Jaillais and Hamant2018). Recently, a counterpart of this powerful tool, PlantSeg, employing principles of neural networks and allowing for high-throughput analysis of 3D images of more complex multicellular specimens, became available (Wolny et al., Reference Wolny, Cerrone, Vijayan, Tofanelli, Barro, Louveaux, Wenzl, Steigleder, Pape and Bailoni2020).
These tools provide an opportunity to explore interactions between genes and morphology during pattern formation. Their compatibility with analysis of fluorescent reporters opened possibilities for dissecting the dynamics of subcellular organisation during polarity establishment and cell division. Assessment of cell geometries with MorphoGraphX combined with modelling approaches has been successfully applied to demonstrate a link between arrangement of cortical microtubules and division plane positioning (Figure 1) (Chakrabortty et al., Reference Chakrabortty, Willemsen, de Zeeuw, Liao, Weijers, Mulder and Scheres2018).
However, thoroughly and comprehensively, the work by Yoshida et al. (Reference Yoshida, de Reuille, Lane, Bassel, Prusinkiewicz, Smith and Weijers2014) and Moukhtar et al. (Reference Moukhtar, Trubuil, Belcram, Legland, Khadir, Urbain, Palauqui and Andrey2019) characterised embryonic cell divisions, it used fixed samples, and therefore lacked the bona fide time dimension. Hence, the real-time patterning process remains to be explored. A genuine 4D technique could be a powerful addition to the current approach and would allow, at the very least, clear up the presumptive synchronisation of cell division during embryogenesis. Addition of the real-time resolution would allow to reconstitute the sequence of events leading to asymmetric divisions and accurately track processes contributing to pattern formation. Achievement of these goals became feasible with application of an in vitro ovule cultivation system allowing tracking of early embryogenesis events (Gooh et al., Reference Gooh, Ueda, Aruga, Park, Arata, Higashiyama and Kurihara2015). Live-cell imaging was also successfully used to study mitochondria, vacuole and cytoskeleton organisation of in vitro cultivated Arabidopsis zygotes (Kimata et al., Reference Kimata, Higaki, Kawashima, Kurihara, Sato, Yamada, Hasezawa, Berger, Higashiyama and Ueda2016; Kimata et al., Reference Kimata, Kato, Higaki, Kurihara, Yamada, Segami, Morita, Maeshima, Hasezawa and Higashiyama2019; Kimata et al., Reference Kimata, Higaki, Kurihara, Ando, Matsumoto, Higashiyama and Ueda2020). Although these live-imaging techniques provide a unique opportunity to visualise processes occurring in the embryo enclosed into developing seed, they are not without their own challenges. These approaches utilise highly expressed markers that might potentially interfere with normal developmental processes. Further improvement of the live-imaging approaches will increase value of the Arabidopsis embryo as a model system.
3. Quantification of biochemical aspects of pattern formation
To understand the regulation of cell division and to address the workings of the machinery that adjusts division plane positioning during pattern formation, one needs the capacity to determine which subcellular (molecular and morphological) events precede cell division. This will require a rather comprehensive approach that will allow to interrogate the cell for the earliest changes in its transcriptome, establishment of polarity and finally, the assembly of the new cell wall. A key to tackling this problem is the development of highly sensitive methods that can track changes in gene expression at a single cell level and generation of robust quantitative reporters that could be monitored overtime. This part of the manuscript will discuss the state of the art, highlight current approaches, available reporters and tools for monitoring pattern formation.
4. The hunt for regulators and first events in cell specification
Identity of a cell can be inferred from its biochemical characteristics and morphology. Since modulation of transcription is one of the first cellular biochemical responses, one can view the emergence of specific expression profiles as the first step on the path to cell fate establishment. In order to elucidate how cell fate is established, researchers set out to find genes acting as regulators of pathways behind cell specification.
Initially, identification of genes acting as regulators of cell identity has been achieved using conventional methods, such as mutagenesis, promoter/enhancer trapping and genetic screening. Expression domains of the potential regulators have been determined using transcript localisation techniques including in situ hybridisation and the use of fluorescent reporters. This approach led to characterisation of many cell fate regulators (reviewed in ten Hove et al., Reference ten Hove, Lu and Weijers2015). Among them was A. thaliana meristem L1 layer (ATML1), a master regulator of shoot epidermal identity (Lu et al., Reference Lu, Porat, Nadeau and O'Neill1996), MONOPTEROS (MP/ARF5), a determinant of hypophysis specification (Schlereth et al., Reference Schlereth, Möller, Liu, Kientz, Flipse, Rademacher, Schmid, Jürgens and Weijers2010), HD-ZIP III genes involved in specification of the shoot apical meristem (SAM) (Prigge et al., Reference Prigge, Otsuga, Alonso, Ecker, Drews and Clark2005), PLETHORA (PLT) genes specifying root apical meristem (RAM) (Aida et al., Reference Aida, Beis, Heidstra, Willemsen, Blilou, Galinha, Nussaume, Noh, Amasino and Scheres2004; Galinha et al., Reference Galinha, Hofhuis, Luijten, Willemsen, Blilou, Heidstra and Scheres2007), WUSCHEL (WUS), regulator of SAM specification (Laux et al., Reference Laux, Mayer, Berger and Jurgens1996). WUS-related homeobox (WOX) gene family was demonstrated to mark cell fate decisions during embryogenesis (Haecker et al., Reference Haecker, Groß-Hardt, Geiges, Sarkar, Breuninger, Herrmann and Laux2004). GRAS family transcription factor SHORTROOT (SHR) and its target SCARECROW (SCR), which is also a GRAS family member, were found to control ground tissue patterning. Specifically, SHR was shown to be required for establishment of endodermal identity and both SHR and SCR control asymmetric periclinal division of cortex/endodermis initial (Benfey et al., Reference Benfey, Linstead, Roberts, Schiefelbein, Hauser and Aeschbacher1993; Levesque et al., Reference Levesque, Vernoux, Busch, Cui, Wang, Blilou, Hassan, Nakajima, Matsumoto and Lohmann2006; Scheres et al., Reference Scheres, Di Laurenzio, Willemsen, Hauser, Janmaat, Weisbeek and Benfey1995). Although these conventional approaches helped to reveal a number of important regulators of cell identity, their depth and efficiency was rather limited. Genetic screening is not an ideal approach when it comes to the hunt for regulators of cell identity establishment, since mutant for their genes might be embryo lethal. At the same time, the search for master regulators might also be hampered by genetic redundancy inherent to plant genomes.
A deeper understanding of embryonic cell specification could be attained by generation of cell type-specific whole-genome expression profiles using high-throughput techniques, such as microarray and RNA-seq technologies (Figure 2a). Such high-throughput approaches have been used in the past to obtain cell type-specific transcript profiles of post-embryonic roots (Birnbaum et al., Reference Birnbaum, Shasha, Wang, Jung, Lambert, Galbraith and Benfey2003; Brady et al., Reference Brady, Orlando, Lee, Wang, Koch, Dinneny, Mace, Ohler and Benfey2007). Owing to the facile breakdown of the root tissues to individual cells, these attempts proved very successful. Application of a similar tissue dissociation technique to embryos that are encased in developing seeds, however, turned out to be a challenging enterprise. First attempts involved expression profiling of whole seeds at different developmental stages (Girke et al., Reference Girke, Todd, Ruuska, White, Benning and Ohlrogge2000; Le et al., Reference Le, Cheng, Bui, Wagmaister, Henry, Pelletier, Kwong, Belmonte, Kirkbride and Horvath2010).

Fig. 2. Gaining insight into biochemical and mechanical responses during pattern formation in plants. (a) Study of cell identity establishment through generation of cell type-specific gene expression profiles. Following disintegration of embryos to single cells using lysis solution, cells are sorted according to the presence of fate-specific markers. The collected cells are further used in either droplet-based or plate-based single-cell RNA Seq assays that allow to interrogate cells for changes in transcriptome (b) Exploring cell-to-cell signaling during cell specification and pattern formation. A cell (pink) divides producing daughters with different fates (lilac and yellow). The molecular basis of fate specification is inferred using high-throughput approaches, such as Chip-seq and Dap-seq. (c) Visualisation of mechanical patterns in plants using mechano-probes. Cellular turgor pressure (gray shapes) causes tensile stress (red arrows) in the cell wall. The resulting tensile stress can be visualised and quantified using rotor dyes that change their fluorescence lifetime depending on the mechanical properties of the cell wall.
Generation of expression profiles of whole isolated embryos, liberated from the endosperm and the seed coat, at different developmental stages involved labour intensive excision of plant tissue using laser capture microdissection (Belmonte et al., Reference Belmonte, Kirkbride, Stone, Pelletier, Bui, Yeung, Hashimoto, Fei, Harada and Munoz2013). This technique has been recently used to compare transcript profiles of apical and basal cell lineages of the early proembryo (Zhou et al., Reference Zhou, Liu, Shen, Zhao and Sun2020). Fluorescence-activated nuclear sorting was used to separate fluorescently labelled nuclei from crude embryo preparations to generate transcript profiles of the early proembryo and the suspensor (Slane et al., Reference Slane, Kong, Berendzen, Kilian, Henschen, Kolb, Schmid, Harter, Mayer and De Smet2014). However, this approach allows to analyse only nuclear transcripts and disregards those stored in the cytosol. To capture the cytosolic transcripts, that is actively translated or stalled on ribosomes, translating ribosome affinity purification has been used to generate expression profiles of plant tissues (Mustroph et al., Reference Mustroph, Zanetti, Jang, Holtan, Repetti, Galbraith, Girke and Bailey-Serres2009; Zanetti et al., Reference Zanetti, Chang, Gong, Galbraith and Bailey-Serres2005). This technique employing bead-based affinity purification of FLAG-tagged ribosomal protein L18 driven by cell-type specific promoters allowed to assess expression profiles of Arabidopsis seedlings with high spatial resolution (Mustroph et al., Reference Mustroph, Zanetti, Jang, Holtan, Repetti, Galbraith, Girke and Bailey-Serres2009).
Isolation of nuclei tagged in specific cell types (INTACT) circumvents this issue by utilising a two-component transgenic labelling system where biotin ligase (BirA) biotinylates a nuclear envelope-localised green flourescent protein [nuclear targeting fusion protein (NTF)] when co-expressed in the same cells. Biotin-tagged nuclei from crude plant tissue preparations are further isolated using streptavidin-coated beads. This technique was applied to map expression dynamics with cell type resolution at different stages of embryogenesis (Palovaara et al., Reference Palovaara, Saiga, Wendrich, van‘t Wout Hofland, van Schayck, Hater, Mutte, Sjollema, Boekschoten and Hooiveld2017). But again, INTACT allows to explore nuclear transcripts and remains blind to cytosolic ones. Significant difference between nuclear and total cellular transcript profiles suggested that nucleus-specific approaches might potentially overlook important elements of cell fate specification (Palovaara & Weijers, Reference Palovaara and Weijers2019). Therefore, future efforts should seek to achieve generation of cell type-specific total cellular transcript profiles. Another important weakness of the abovementioned approaches is their reliance on cell type-specific markers that are often missing for certain cell types during various stages of embryogenesis.
Moreover, the availability of the gene expression data boosted establishment of important molecular markers allowing for tracking specification processes in the root stem cell niche and vascular tissues (Smit et al., Reference Smit, Llavata-Peris, Roosjen, van Beijnum, Novikova, Levitsky, Sevilem, Roszak, Slane and Jürgens2020). Collectively, embryonic tissue- and cell-specific gene expression datasets became an invaluable resource for studying establishment of cell identity. Development of tools providing graphical representation of the embryonic gene expression profiles such as the Arabidopsis ePlant browser (Waese et al., Reference Waese, Fan, Pasha, Yu, Fucile, Shi, Cumming, Kelley, Sternberg and Krishnakumar2017) (http://bar.utoronto.ca/eplant/), SeedGene Network (Le et al., Reference Le, Cheng, Bui, Wagmaister, Henry, Pelletier, Kwong, Belmonte, Kirkbride and Horvath2010) (http://seedgenenetwork.net/) and AlBERTO (Palovaara et al., Reference Palovaara, Saiga, Wendrich, van‘t Wout Hofland, van Schayck, Hater, Mutte, Sjollema, Boekschoten and Hooiveld2017) (http://www.albertodb.org) made these powerful data sources easily assessable for the whole community of plant researchers.
Development of technologies allowing to infer cellular identity is undoubtedly an important milestone in developmental biology. In the future, these techniques will enable us to precisely track the establishment of cellular fates and will prove valuable in addressing the mechanisms underlying acquisition of cell identities.
5. Eavesdropping on cell-to-cell communication
Since most plant cells are pluripotent, fate establishment is profoundly influenced by the cellular context (Scheres, Reference Scheres2001; Serna et al., Reference Serna, Torres‐Contreras and Fenoll2002). Therefore, cell-to-cell communication is an essential aspect of cell specification and pattern formation in plants. To date, several key molecular cues involved in transmission of signals between cells have been identified. The most prominent among these molecules is the phytohormone auxin that modulates gene expression by promoting degradation of auxin/indole-3-acetic acid (Aux/IAA), transcriptional repressor proteins, leading to release of auxin response factors (ARFs) transcription factors, which are otherwise bound by Aux/IAA repressors. Specifically, auxin mediates binding of Aux/IAA to TIR1/AFB, a part of SCF (SKP1-CUL1-F box) complex, which results in ubiquitination and subsequent degradation of Aux/IAA. Released ARFs bind to auxin response elements (AuxREs) to activate or inhibit target gene transcription (Overvoorde et al., Reference Overvoorde, Okushima, Alonso, Chan, Chang, Ecker, Hughes, Liu, Onodera and Quach2005; Tiwari et al., Reference Tiwari, Hagen and Guilfoyle2004; Ulmasov, Hagen, et al., Reference Ulmasov, Hagen and Guilfoyle1997; Ulmasov, Murfett, et al., Reference Ulmasov, Murfett, Hagen and Guilfoyle1997; Villalobos et al., Reference Villalobos, Lee, De Oliveira, Ivetac, Brandt, Armitage, Sheard, Tan, Parry and Mao2012). Auxin is a mobile signal that is actively transported from cell to cell by plasma membrane localised AUXIN1/LIKE-AUX1 (AUX/LAX) and PIN-FORMED (PIN) proteins that facilitate its influx and efflux, respectively (Bennett et al., Reference Bennett, Marchant, May and Swarup1998; Friml, Reference Friml2003; Friml et al., Reference Friml, Vieten, Sauer, Weijers, Schwarz, Hamann, Offringa and Jürgens2003; Gälweiler et al., Reference Gälweiler, Guan, Müller, Wisman, Mendgen, Yephremov and Palme1998; Marchant et al., Reference Marchant, Kargul, May, Muller, Delbarre, Perrot‐Rechenmann and Bennett1999; Petrášek et al., Reference Petrášek, Mravec, Bouchard, Blakeslee, Abas, Seifertová, Wiśniewska, Tadele, Kubeš and Čovanová2006; Swarup et al., Reference Swarup, Friml, Marchant, Ljung, Sandberg, Palme and Bennett2001). The efflux transporters are polarly localised, assuring the directionality of the auxin flux through the plant body (Petrášek et al., Reference Petrášek, Mravec, Bouchard, Blakeslee, Abas, Seifertová, Wiśniewska, Tadele, Kubeš and Čovanová2006; Petrášek & Friml, Reference Petrášek and Friml2009). This polar transport integrates cellular and tissue polarity and underlies a multitude of developmental processes in plants (Křeček et al., Reference Křeček, Skůpa, Libus, Naramoto, Tejos, Friml and Zažímalová2009; Petrášek & Friml, Reference Petrášek and Friml2009). Interference with either biosynthesis, transport or response strongly interferes with normal development (Cheng et al., Reference Cheng, Dai and Zhao2007; Robert et al., Reference Robert, Grunewald, Sauer, Cannoot, Soriano, Swarup, Weijers, Bennett, Boutilier and Friml2015; Stepanova et al., Reference Stepanova, Robertson-Hoyt, Yun, Benavente, Xie, Doležal, Schlereth, Jürgens and Alonso2008).
Auxin is profoundly involved in pattern formation and cell specification in diverse developmental contexts in Arabidopsis: root pattern formation (Sabatini et al., Reference Sabatini, Beis, Wolkenfelt, Murfett, Guilfoyle, Malamy, Benfey, Leyser, Bechtold and Weisbeek1999), asymmetric division of the cortex/endodermis initial in post-embryonic development (Cruz-Ramírez et al., Reference Cruz-Ramírez, Díaz-Triviño, Blilou, Grieneisen, Sozzani, Zamioudis, Miskolczi, Nieuwland, Benjamins, Benjamins and Dhonukshe2012), as well as formative divisions during lateral root formation (Bishopp, Help, et al., Reference Bishopp, Help, El-Showk, Weijers, Scheres, Friml, Benková, Mähönen and Helariutta2011; Bishopp, Lehesranta, et al., Reference Bishopp, Lehesranta, Vatén, Help, El-Showk, Scheres, Helariutta, Mähönen, Sakakibara and Helariutta2011; De et al., Reference De Smet, Tetsumura, De Rybel, dit Frey, Laplaze, Casimiro, Swarup, Naudts, Vanneste and Audenaert2007; Moreno-Risueno et al., Reference Moreno-Risueno, Van Norman, Moreno, Zhang, Ahnert and Benfey2010). Similarly, pattern formation during embryogenesis critically depends on auxin signaling (Hardtke & Berleth, Reference Hardtke and Berleth1998; Jürgens, Reference Jürgens2001; Schlereth et al., Reference Schlereth, Möller, Liu, Kientz, Flipse, Rademacher, Schmid, Jürgens and Weijers2010; Yoshida et al., Reference Yoshida, de Reuille, Lane, Bassel, Prusinkiewicz, Smith and Weijers2014). Thus, auxin is involved in formative division in vasculature formation during embryogenesis (De et al., Reference De Rybel, Möller, Yoshida, Grabowicz, de Reuille, Boeren, Smith, Borst and Weijers2013; Ohashi-Ito et al., Reference Ohashi-Ito, Oguchi, Kojima, Sakakibara and Fukuda2013).
The Arabidopsis genome encodes 6 TIR/AFB, 29 Aux/IAA and 23 ARF proteins (Weijers & Wagner, Reference Weijers and Wagner2016). Differential TIR/AFB–Aux/IAA and Aux/IAA–ARF interactions likely contribute to the complexity and diversity of auxin responses (Piya et al., Reference Piya, Shrestha, Binder, Stewart and Hewezi2014; Villalobos et al., Reference Villalobos, Lee, De Oliveira, Ivetac, Brandt, Armitage, Sheard, Tan, Parry and Mao2012). Moreover, differential expression of ARFs during embryogenesis, observed in the study using fluorescent reporter lines, suggested that auxin response machinery might be adjusted to elicit cell type-specific transcriptional responses (Rademacher et al., Reference Rademacher, Lokerse, Schlereth, Llavata-Peris, Bayer, Kientz, Rios, Borst, Lukowitz and Jürgens2012). Information of transcription factor and DNA binding interaction from Chromatin immunoprecipitation followed by sequencing (ChIP-seq) (Park, Reference Park2009) or DNA affinity purification sequencing (DAP-seq) (Bartlett et al., Reference Bartlett, O'Malley, Huang, Galli, Nery, Gallavotti and Ecker2017) in combination with high-resolution single cell expression profiles will help to gain a deeper insight into workings of the auxin response machinery (Figure 2b).
Auxin dependent cell-to-cell communication is mediated not only through the transport of auxin, but also through intercellular movement of its downstream effectors. Auxin response factor5/monopteros (ARF5/MP) expressed in the embryo proper drives hypophysis specification, thereby regulating non-cell autonomous cell fate establishment. Target of MP7 (TMO7), which expression is activated by ARF5/MP in the embryo, mediates this regulation by moving from the embryo proper to the hypophysis precursor (Schlereth et al., Reference Schlereth, Möller, Liu, Kientz, Flipse, Rademacher, Schmid, Jürgens and Weijers2010).
The ability to track auxin signaling through visualisation of transcriptional response was crucial for the studies exploring the role of auxin in cell-to-cell communication and pattern formation. This was first enabled by the use of a synthetic auxin response reporter DR5 consisting of 7–9 AuxRE repeats (ARF binding sites) and marking sites of transcriptional auxin response by activating fused reporter genes such as β-glucuronidase, fluorescent proteins or luciferase (Friml et al., Reference Friml, Vieten, Sauer, Weijers, Schwarz, Hamann, Offringa and Jürgens2003; Moreno-Risueno et al., Reference Moreno-Risueno, Van Norman, Moreno, Zhang, Ahnert and Benfey2010; Ulmasov et al., Reference Ulmasov, Hagen and Guilfoyle1997). However, the DR5 reporter failed to visualise computationally predicted auxin gradients and therefore was referred to as ‘auxin maxima’ reporter (Grieneisen et al., Reference Grieneisen, Xu, Marée, Hogeweg and Scheres2007). More sensitive visualisation of auxin responses was achieved by introduction of a new reporter, DR5v2, harbouring higher affinity ARF binding sites ( Liao et al., Reference Liao, Smet, Brunoud, Yoshida, Vernoux and Weijers2015). However, the most prominent advancement was achieved by development a two-component ratio-metric auxin response reporter, R2D2, that allowed for the most accurate visualisation and a semi-quantitative analysis of auxin readout in both embryonic and post-embryonic development (Liao et al., Reference Liao, Smet, Brunoud, Yoshida, Vernoux and Weijers2015). This quantitative reporter has been successfully used to visualise auxin response in specific embryonic tissues, such as vasculature, ground tissue, protoderm, hypophysis and suspensor (Möller et al., Reference Möller, Colette, Xiang, Williams, López, Yoshida, Smit, Datla and Weijers2017; Roodbarkelari et al., Reference Roodbarkelari, Du, Truernit and Laux2015; Smit et al., Reference Smit, Llavata-Peris, Roosjen, van Beijnum, Novikova, Levitsky, Sevilem, Roszak, Slane and Jürgens2020).
Besides auxin, several other phytohormones have been shown to be involved in pattern formation during embryogenesis. Thus, antagonistic interaction of auxin with cytokinin was demonstrated to control embryonic root stem-cell niche establishment. The study employed two-component-output sensor fused to luciferase to visualise cytokinin signaling (Müller & Sheen, Reference Müller and Sheen2008). Such mutually inhibitory activity of these two hormones was detected in pattern formation and cell specification in other developmental contexts outside embryogenesis (; Bishopp, Help, et al., Reference Bishopp, Help, El-Showk, Weijers, Scheres, Friml, Benková, Mähönen and Helariutta2011; Chang et al., Reference Chang, Ramireddy and Schmülling2013; Mähönen et al., Reference Mähönen, Bishopp, Higuchi, Nieminen, Kinoshita, Törmäkangas, Ikeda, Oka, Kakimoto and Helariutta2006). It is conceivable that the interaction of hormonal cues might be used to couple positional information with fate establishment. In order to address this possibility, one has to be able to track and quantify the readout of the hormonal cues, which presently remains an obstacle due to the lack of proper tools (other than auxin-specific ones). A novel set of synthetic and modular hormone activated Cas9-based repressors (HACRs) reporters allowing for visualisation of auxin, gibberellin and jasmonates, and hold great promise to overcome this obstacle (Khakhar et al., Reference Khakhar, Leydon, Lemmex, Klavins and Nemhauser2018). HACRs include deactivated Cas9 (dCas9) protein fused to a highly sensitive hormone-responsive degron domain and a part of the TOPLESS transcriptional repressor. Association of dCas9 with a guide RNA targets HACR to a promoter (such as pUBQ1), leading to transcriptional repression of a reporter (nuclear targeted Venus-Luciferase). Upon hormone accumulation, the degron sequence targets the HACR for ubiquitination and subsequent proteasomal degradation. Thus, in parallel to the natural hormone response, hormone accumulation triggers relief of repression on HACR target genes (reporter).
The system was transformed to A. thaliana that were genetically engineered for consistently producing a fluorescent protein. When there was no hormone, HACRs repress fluorescent gene which is consequent to an absent of fluorescent signal. If the hormone is present, HACRs is degraded and therefore fluorescent signal is present. With the ability of reducing fluorescent signal at different time point, HACRs system can finely detect hormonal level and track re-programming in developing plants.
Since cells need to transmit different types of information to their neighbours, for example, topology, stress, identity, differentiation state, one might expect the means of cell–cell communication to be rather sophisticated and involve different types of signals to avoid any miscommunication. In this light, small non-coding RNA molecules such as microRNA (miRNA) have emerged as an important signaling element, mediating cell–cell communication in plant development. Embryonic miRNAs were demonstrated to mediate regulation of gene expression by modifying the stability of embryonic messenger RNA, and thereby coordinating cellular transcript profiles during development. Thus, miR165/166 were shown to be important for establishment of shoot apical meristem at the globular stage of embryo development by repressing HD-ZIP lll genes (Müller & Sheen, Reference Müller and Sheen2008; Vashisht & Nodine, Reference Vashisht and Nodine2014). Furthermore, miR165/166 were also shown to be expressed in the basal-peripheral region, and were suggested to non-cell autonomously control expression of HD-ZIP III genes in specification of RAM during embryogenesis (Miyashima et al., Reference Miyashima, Honda, Hashimoto, Tatematsu, Hashimoto, Sato-Nara, Okada and Nakajima2013). Another type of miRNA, namely miR394, is required to maintain apical meristem and stem cell niche by repressing putative F-box protein (leaf curling responsiveness) (Song et al., Reference Song, Huang, Dalmay and Yang2012). Using fluorescent reporter lines, miR394 was found to first appear in the most outer layer of shoot apex (L1) and at later stages extend its expression to the inner layer (L2 and L3), suggesting a role of this miRNA in maintenance of three niches, including protoderm (in L1), ground meristem (in L2) and procambium (in L3).
Comprehensive analyses of miRNA distribution and dynamics in embryogenesis provided an opportunity to gain a deeper insight into the roles of these molecules in pattern formation (Plotnikova et al., Reference Plotnikova, Kellner, Schon, Mosiolek and Nodine2019; Seefried et al., Reference Seefried, Willmann, Clausen and Jenik2014). The study by Plotnikova and co-workers suggested that over 300 miRNAs operate during embryogenesis, and identified 59 high-confidence miRNA targets (Plotnikova et al., Reference Plotnikova, Kellner, Schon, Mosiolek and Nodine2019). Previously, similar large-scale miRNA datasets have been used in development of user-friendly tools, such as miRBase (Kozomara & Griffiths-Jones, Reference Kozomara and Griffiths-Jones2014) (http://www.mirbase.org/), psRNATarget (Dai et al., Reference Dai, Zhuang and Zhao2018) (http://plantgrn.noble.org/psRNATarget/) and plant microRNA database (Yi et al., Reference Yi, Zhang, Ling, Xu and Su2015) (http://structuralbiology.cau.edu.cn/PNRD). These resources hold a great potential to advance our perception of the roles of miRNA in pattern formation during embryogenesis.
However, profound understanding of how plant cells are specified during pattern formation requires higher resolution that can be gained through generation of single cell expression profiles using advanced techniques, such as assays employing a droplet-based platform Drop-seq (Macosko et al., Reference Macosko, Basu, Satija, Nemesh, Shekhar, Goldman, Tirosh, Bialas, Kamitaki and Martersteck2015) or plate-based platform CEL-seq (Hashimshony et al., Reference Hashimshony, Senderovich, Avital, Klochendler, de Leeuw, Anavy, Gennert, Li, Livak and Rozenblatt-Rosen2016), Seq-well (Gierahn et al., Reference Gierahn, Wadsworth, Hughes, Bryson, Butler, Satija, Fortune, Love and Shalek2017) and SMART-seq (Picelli et al., Reference Picelli, Faridani, Björklund, Winberg, Sagasser and Sandberg2014) (Figure 2a). Specifically, SMART-seq and CEL-seq offer methodologies an efficient way to profile low number of single cells by sorting cells into microwells or plates, that host all the steps from the cell lysis to construction of cDNA libraries (Hagemann-Jensen et al., Reference Hagemann-Jensen, Ziegenhain, Chen, Ramsköld, Hendriks, Larsson, Faridani and Sandberg2020; Hashimshony et al., Reference Hashimshony, Senderovich, Avital, Klochendler, de Leeuw, Anavy, Gennert, Li, Livak and Rozenblatt-Rosen2016; Picelli et al., Reference Picelli, Faridani, Björklund, Winberg, Sagasser and Sandberg2014). Other approaches such as Drop-seq and inDrop utilise separation of cells through encapsulating individual cells into nanoliter-sized droplets, followed by collecting all samples in a single tube. With few handling steps, the droplet-based approaches are suited for profiling of large numbers of cells at once (Macosko et al., Reference Macosko, Basu, Satija, Nemesh, Shekhar, Goldman, Tirosh, Bialas, Kamitaki and Martersteck2015; Zilionis et al., Reference Zilionis, Nainys, Veres, Savova, Zemmour, Klein and Mazutis2017). In comparison with droplet-based techniques, plate-based assays allow for a greater flexibility in terms of discrimination of individual cell samples (individual wells) by quality. Moreover, they enable interruption of the assay or long-term storage of samples at ultra-low temperatures. With these advanced scRNA-seq technologies, generation of highly accurate single-cell transcript profiles of the first steps of plant development is within reach. However, the outcome and the current enigma of the cell type-specific adjustment of molecular composition is the positioning of division plane, that will ultimately contribute to pattern formation.
6. Digging into the cell interior
Positioning of division plane during asymmetric divisions is crucial for pattern formation in plants. However, the cues and the machinery that adjust division planes remain to be understood. Asymmetric divisions are preceded by the establishment of cellular polarity that is reflected by the segregation and organisation of subcellular components, such as hormones, mRNA, proteins and organelles (Dong et al., Reference Dong, MacAlister and Bergmann2009; Kimata et al., Reference Kimata, Higaki, Kawashima, Kurihara, Sato, Yamada, Hasezawa, Berger, Higashiyama and Ueda2016; Kimata et al., Reference Kimata, Kato, Higaki, Kurihara, Yamada, Segami, Morita, Maeshima, Hasezawa and Higashiyama2019).
Specific dynamics of cytoskeletal elements, namely actin and microtubules, was shown to precede the first asymmetric zygotic division in Arabidopsis (Kimata et al., Reference Kimata, Higaki, Kawashima, Kurihara, Sato, Yamada, Hasezawa, Berger, Higashiyama and Ueda2016). Additionally, actin dynamics has been suggested to be important for polar auxin transport (Dhonukshe et al., Reference Dhonukshe, Grigoriev, Fischer, Tominaga, Robinson, Hašek, Paciorek, Petrášek, Seifertová and Tejos2008). Microtubule organisation was shown to strongly correlate with PIN1 localisation in the Arabidopsis shoot apex (Heisler et al., Reference Heisler, Hamant, Krupinski, Uyttewaal, Ohno, Jönsson, Traas and Meyerowitz2010). Microtubules were also reported to guide deposition of cellulose fibers, thereby affecting the direction of cellular growth (Bringmann et al., Reference Bringmann, Landrein, Schudoma, Hamant, Hauser and Persson2012; Paradez et al., Reference Paradez, Wright and Ehrhardt2006). Both microtubules and actin are required for early events in lateral root formation (Barro et al., Reference Barro, Stöckle, Thellmann, Ruiz-Duarte, Bald, Louveaux, von Born, Denninger, Goh and Fukaki2019). Spatial organisation and dynamics of other organelles and compartments are also important to consider. For example, polar vacuole distribution is essential for accurate zygotic division, while nuclear dynamics and migration are important for asymmetric divisions during stomata formation (Kimata et al., Reference Kimata, Kato, Higaki, Kurihara, Yamada, Segami, Morita, Maeshima, Hasezawa and Higashiyama2019; Muroyama et al., Reference Muroyama, Gong and Bergmann2020) and lateral root formation (De et al., Reference De Rybel, Vassileva, Parizot, Demeulenaere, Grunewald, Audenaert, Van Campenhout, Overvoorde, Jansen and Vanneste2010; Vermeer et al., Reference Vermeer, von Wangenheim, Barberon, Lee, Stelzer, Maizel and Geldner2014). Activity of endomembrane trafficking is essential for polar localisation of auxin transporters (Doyle et al., Reference Doyle, Vain and Robert2015; Kleine-Vehn et al., Reference Kleine-Vehn, Dhonukshe, Swarup, Bennett and Friml2006). So far, the roles of dynamics and organisation of subcellular structures in asymmetric divisions and auxin transport, were studied using either Arabidopsis zygote of post-embryonic organs, and until recently were completely overlooked in embryogenesis. Tackling these questions using the model of the Arabidopsis embryo with its highly stereotypical division pattern would allow to accurately link subcellular dynamics and division plane positioning.
A recently developed collection of embryo-optimised fluorescent subcellular reporters now allows to tackle this question. These reporters, named Arabidopsis cellular markers for embryogenesis (ACE), are based on a set of available markers for various subcellular structures driven by embryo-specific promoters such as RPS5A and WOX2, allowing to track their organisation and dynamics (Liao & Weijers, Reference Liao and Weijers2018). Actin and microtubule reporters from this series, namely ACE-14 and ACE-15, allowed to reveal the distinct organisation of the cytoskeletal elements at different stages of embryogenesis. Reporters for the Golgi complex and endosomal elements, GOT1p (ACE-09) and RabC1 (ACE-07), respectively, enabled tracing early secretion events. Plasmodesmatal marker PDCB1 (ACE-13), allowed visualisation of patterns of plasmodesmata channels essential for passive transport and cell-cell communication. ACE-11 reporter marking nuclear pore complex could help to address whether nuclear dynamics is also important for formative divisions during embryogenesis.
The importance of organisation of subcellular structures in asymmetric divisions was strongly emphasised by a study employing live-time imaging to trace the cytoskeleton in zygote polarisation and division (Kimata et al., Reference Kimata, Higaki, Kawashima, Kurihara, Sato, Yamada, Hasezawa, Berger, Higashiyama and Ueda2016). This work highlighted the necessity to include the time dimension in studies elucidating establishment of polarity and pattern formation. The current knowledge of patterning during embryogenesis was gained from analysis of fixed samples and the time dimension was included by screening of a large amount of fixed samples, so that the whole duration of early embryogenesis was monitored (Yoshida et al., Reference Yoshida, de Reuille, Lane, Bassel, Prusinkiewicz, Smith and Weijers2014). Addressing how embryonic patterning occurs in real-time became a reality with development of in vitro ovule culture system that allows normal ovule growth and embryo development from zygote combined with live imaging (Gooh et al., Reference Gooh, Ueda, Aruga, Park, Arata, Higashiyama and Kurihara2015).
7. Probing cell mechanics
For decades regulation of cell division plane positioning in plants has been attributed to biochemical cues (Pillitteri et al., Reference Pillitteri, Guo and Dong2016). Compelling evidence suggests that mechanical stimuli are as important as biochemical cues in instructing division plane orientation and regulating key processes in plant development, such as differentiation, expansion, cell fate specification and polarity (Heisler et al., Reference Heisler, Hamant, Krupinski, Uyttewaal, Ohno, Jönsson, Traas and Meyerowitz2010; Louveaux et al., Reference Louveaux, Julien, Mirabet, Boudaoud and Hamant2016; Nakayama et al., Reference Nakayama, Smith, Mandel, Robinson, Kimura, Boudaoud and Kuhlemeier2012; Sampathkumar, Krupinski, et al., Reference Sampathkumar, Krupinski, Wightman, Milani, Berquand, Boudaoud, Hamant, Jönsson and Meyerowitz2014; Sampathkumar, Yan, et al., Reference Sampathkumar, Yan, Krupinski and Meyerowitz2014). However, what is the origin of mechanical forces in plant tissues?
Plant cell walls experience tensile stress caused by cellular turgor pressure. To resist this stress, cell walls alter their mechanical properties by depositing cellulose microfibrils and other components, such as pectins and xyloglucans (Braybrook & Jönsson, Reference Braybrook and Jönsson2016). Since a cell shares its cellulosic walls with its neighbours that might have different growth rates and direction, mechanical properties of a single cell might be highly anisotropic. Such mechanical anisotropy reflects stress patterns present in tissues and instructs positioning of division plane. This phenomenon was first demonstrated on the model of the Arabidopsis shoot apex (Nakayama et al., Reference Nakayama, Smith, Mandel, Robinson, Kimura, Boudaoud and Kuhlemeier2012). It has been noted that in its central region, where stress is isotropic, cells divide along the shortest path (symmetrically), following Errera’s rule. However, in its boundary region, where stresses are highly anisotropic, Errera’s rule is broken and cells divide along the maximal tensile stress, which is a longer path (Louveaux et al., Reference Louveaux, Julien, Mirabet, Boudaoud and Hamant2016). Corroborating the notion that cell divisions are under mechanical control, cell ablation led to re-arrangement of stress pattern and resulted in re-orientation of cell divisions in the shoot apex (Heisler et al., Reference Heisler, Hamant, Krupinski, Uyttewaal, Ohno, Jönsson, Traas and Meyerowitz2010). The mechanical stimulus in this experiment has been translated into subcellular response, resulting in re-polarisation of the auxin transporter PIN1, and re-organisation of microtubules. Such profound effect on cellular polarity in response to mechanical stimuli is expected to be accompanied and/or preceded by changes in gene expression profiles. A number of genes which expression is under mechanical control has been identified. Most prominent among those are TOUCH genes (Braam, Reference Braam2005; Hamant & Haswell, Reference Hamant and Haswell2017; Landrein et al., Reference Landrein, Kiss, Sassi, Chauvet, Das, Cortizo, Laufs, Takeda, Aida and Traas2015). To understand how mechanical control of cell division is executed, one has to address following questions: (a) how mechanical stimuli are perceived and (b) translated into biochemical responses; (c) what is the chain of subcellular events that leads to adjustment of division plane in response to mechanical cues?
Cells can perceive mechanical signals through activation of mechanosensitive ion channels. Upon activation, such channels allow for ion passage down the electrochemical gradient (Hamant & Haswell, Reference Hamant and Haswell2017; Hamilton et al., Reference Hamilton, Jensen, Maksaev, Katims, Sherp and Haswell2015; Peyronnet et al., Reference Peyronnet, Tran, Girault and Frachisse2014). Interestingly, a recent study demonstrated that blocking calcium channels prevents PIN1 re-polarisation and organogenesis in the SAM, corroborating the notion that ion fluxes are located upstream of establishment of polar protein domains in mechanotransduction (Li et al., Reference Li, Yan, Bhatia, Altinok, Afik, Durand-Smet, Tarr, Schroeder, Heisler and Meyerowitz2019).
Recently, cortical microtubules started to emerge as possible sensory elements of the cellular interior. The capacity of microtubule arrangement to respond to mechanical stimuli and to correlate with tissue stress patterns allowed to consider this structures as a proxy for mechanical stress distribution (Hamant et al., Reference Hamant, Inoue, Bouchez, Dumais and Mjolsness2019; Heisler et al., Reference Heisler, Hamant, Krupinski, Uyttewaal, Ohno, Jönsson, Traas and Meyerowitz2010; Louveaux et al., Reference Louveaux, Julien, Mirabet, Boudaoud and Hamant2016; Sampathkumar, Krupinski, et al., Reference Sampathkumar, Krupinski, Wightman, Milani, Berquand, Boudaoud, Hamant, Jönsson and Meyerowitz2014). A recent study demonstrated that organisation of microtubules determined by cellular geometry instructs cell division (Chakrabortty et al., Reference Chakrabortty, Willemsen, de Zeeuw, Liao, Weijers, Mulder and Scheres2018). This suggests that the feedback between geometry and mechanics might be mediated through these cytoskeletal elements (Chakrabortty et al., Reference Chakrabortty, Willemsen, de Zeeuw, Liao, Weijers, Mulder and Scheres2018; Hamant et al., Reference Hamant, Inoue, Bouchez, Dumais and Mjolsness2019). To gain a deeper insight into regulation of this interaction and to shed light onto translation of mechanical stimuli into cellular responses we need to be able to trace subcellular components with high spatial and temporal resolution.
To understand how mechanical cues shape plant development, one has to be able to determine tissue stress patterns. Our current understanding of mechanical stress patterns in plant tissues has been gained from studies using atomic force microscopy, measuring stress experienced by the cell wall. This approach allowed to determine stress patterns of the epidermal cell layer of SAM, leaf pavement cells and hypocotyl epidermal layer (Peaucelle et al., Reference Peaucelle, Wightman and Höfte2015; Sampathkumar, Krupinski, et al., Reference Sampathkumar, Krupinski, Wightman, Milani, Berquand, Boudaoud, Hamant, Jönsson and Meyerowitz2014). However, this technique could not be applied to probe mechanical properties of deeper tissue layers, leaving questions about the role of mechanical stimuli in pattern formation unanswered. Another question that remains to be addressed is how mechanical stimuli from the cell wall are translated into subcellular response. As the cell wall is separated from the cellular interior by the plasma membrane, it is tempting to speculate that mechanical cues are translated into properties of that barrier compartment. The recent development of sensitive cell-permeable mechano-probes that allow to visualise tension patterns of the plasma membrane and porosity of the cell wall provided an excellent opportunity to fill this knowledge gap and to visualise the stress patterns of inner tissues of plant organs (Figure 2c) (Michels et al., Reference Michels, Gorelova, Harnvanichvech, Borst, Albada, Weijers and Sprakel2020). Combined with reporters for subcellular structures and tools for 3D image analysis such mechano-probes provide an excellent opportunity to dissect the relationship between mechanical stress patterns, morphology and biochemical responses and to reveal the feedback regulation between these elements at cellular, tissue and organ levels.
8. Application of modelling approaches in elucidation of pattern formation
Experimental molecular studies, discussed in the previous sections, typically focus on individual elements encoded by a genome or small arrays of elements to derive their interaction and to explore their contribution to biological processes. Since such studies can only deal with a limited number of elements at a time, they are restricted in power. In part, this stems from the fact that molecular pathways orchestrating biological processes are multicomponent, nonlinear and involve complex feedback regulatory loops. The large number of different input signals controlling a single biological process further adds up to the complexity. This renders experimental molecular approaches unable to provide comprehensive insights into the complex machineries driving biological processes. For instance, as shown in previous sections, a process of cell division is instructed by biochemical signals involving transcription factors, miRNA, hormones, and so on, being at the same time under mechanical control and dependent on cellular environment and cell and organ geometry.
Development of mathematical modelling approaches empowered researchers to tackle such complex problems and to test contributions of individual elements and input signals and their interactions to an output trait. Theoretical approaches also enable prediction of regularities and processes that cannot be tested or visualised due to certain technical limitations. Thus, although accurate experimental visualisation of auxin distribution was not possible, computational studies enabled its modelling and pinpointed the importance of polar auxin transport in the root tip and emphasised its role in the RAM establishment and root zonation (Grieneisen et al., Reference Grieneisen, Xu, Marée, Hogeweg and Scheres2007; Mironova et al., Reference Mironova, Omelyanchuk, Yosiphon, Fadeev, Kolchanov, Mjolsness and Likhoshvai2010), predicted role of AUX/LAX influx carriers and PIN efflux transporters in control of auxin distribution in the root tip (Band et al., Reference Band, Wells, Fozard, Ghetiu, French, Pound, Wilson, Yu, Li and Hijazi2014) that was further experimentally supported on the model of Arabidopsis embryo (Robert et al., Reference Robert, Grunewald, Sauer, Cannoot, Soriano, Swarup, Weijers, Bennett, Boutilier and Friml2015). Studies coupling auxin distribution predictions with expression profiles demonstrated that auxin influx and efflux coordinate gene expression during lateral root development (Péret et al., Reference Péret, Middleton, French, Larrieu, Bishopp, Njo, Wells, Porco, Mellor and Band2013).
The use of gene expression data for the modelling of genetic networks has long been exercised to explore various aspects of plant development and physiology (De, Reference De Jong2002). Specific focus on the modelling of genetic networks was laid in studies describing pattern formation and identity specification. Thus, gene expression visualised by confocal imaging was used to develop a computational model of bisymmetric patterning of the root vascular system predicting interplay of SHORTROOT, miRNA165/6, PHABULOSA, auxin transport and cytokinin signaling (Muraro et al., Reference Muraro, Mellor, Pound, Lucas, Chopard, Byrne, Godin, Hodgman, King and Pridmore2014). Importance of auxin-cytokinin interaction in vascular tissue patterning was further supported by De et al. (Reference De Rybel, Adibi, Breda, Wendrich, Smit, Novák, Yamaguchi, Yoshida, Van Isterdael and Palovaara2014). The study identified a genetic network underlying an auxin-driven cytokinin biosynthesis in the early development of the Arabidopsis embryo and ensuring a correct vascular tissue patterning. Modelling study using live imaging data combined with gene expression profiling and chromatin immunoprecipitation revealed importance of transcriptional repression of WUS and a set of transcription factors to prevent premature differentiation of SAM proliferation zone (Yadav et al., Reference Yadav, Perales, Gruel, Ohno, Heisler, Girke, Jönsson and Reddy2013). Previously confocal imaging data were used to model WUS expression domain in SAM (Jönsson et al., Reference Jönsson, Heisler, Reddy, Agrawal, Gor, Shapiro, Mjolsness and Meyerowitz2005). Modelling approaches also predicted various aspects of epidermal cell lineage, including root hair and trichome patterning and stomata development (Ryu et al., Reference Ryu, Zheng, Huang and Schiefelbein2013). A recent study using the model of the Arabidopsis floral meristem and employing high-resolution time-lapse imaging provided an insight into the link between gene regulation and morphogenesis (Refahi et al., Reference Refahi, Zardilis, Michelin, Wightman, Leggio, Legrand, Faure, Vachez, Armezzani and Zhao2021).
Modelling studies covered various aspects of plant development at the organ and organismal levels, describing cell division patterns during lateral root formation (von et al., Reference von Wangenheim, Fangerau, Schmitz, Smith, Leitte, Stelzer and Maizel2016) and establishment of apical–basal axis during embryogenesis (Wabnik et al., Reference Wabnik, Robert, Smith and Friml2013). Theoretical studies also aimed at linking gene expression networks with organ shape. Thus, a computational model has tested the significance of the reciprocal repression between CUC2 and the PIN1 module in the determination of leaf margin shape (Bilsborough et al., Reference Bilsborough, Runions, Barkoulas, Jenkins, Hasson, Galinha, Laufs, Hay, Prusinkiewicz and Tsiantis2011). Relatively recent advance of 3D imaging techniques enabled development of models explaining the relationship between cell shape, dynamics of subcellular structures and division plane orientation. Thus, orientation of the cortical microtubule arrays was suggested to determine positioning of division planes during embryogenesis (Chakrabortty et al., Reference Chakrabortty, Willemsen, de Zeeuw, Liao, Weijers, Mulder and Scheres2018). Another modelling study on embryo development suggested the possibility that cell divisions are guided by nuclear positioning (Moukhtar et al., Reference Moukhtar, Trubuil, Belcram, Legland, Khadir, Urbain, Palauqui and Andrey2019), drawing parallel with the role of nuclear movement in asymmetric divisions during post-embryonic development (De et al., Reference De Rybel, Vassileva, Parizot, Demeulenaere, Grunewald, Audenaert, Van Campenhout, Overvoorde, Jansen and Vanneste2010; Kimata et al., Reference Kimata, Kato, Higaki, Kurihara, Yamada, Segami, Morita, Maeshima, Hasezawa and Higashiyama2019; Muroyama et al., Reference Muroyama, Gong and Bergmann2020; Vermeer et al., Reference Vermeer, von Wangenheim, Barberon, Lee, Stelzer, Maizel and Geldner2014). However, since the nucleus occupies a substantial part of the cellular volume in early embryo cells (Liao & Weijers, Reference Liao and Weijers2018), it remains to be investigated if and how regulated nuclear position can contribute to division orientation in these cells.
The dynamic interplay of geometrical and genetic inputs has attracted significant interest during the past decade. In this light, a novel modelling framework, morphodynamics, has been developed to explain complex temporal and spatial interactions of growth and signaling (Bassel et al., Reference Bassel, Stamm, Mosca, de Reuille, Gibbs, Winter, Janka, Holdsworth and Smith2014; Jönsson et al., Reference Jönsson, Gruel, Krupinski and Troein2012). The recently emerged mechanical aspect of plant development was incorporated into computational studies to explain positioning of division plane orientation, cytoskeleton behaviour and polarity of stomatal stem cells (Louveaux et al., Reference Louveaux, Julien, Mirabet, Boudaoud and Hamant2016; Sampathkumar, Yan, et al., Reference Sampathkumar, Yan, Krupinski and Meyerowitz2014).
A fundamental progress over the past two decades is the link between computational modelling, molecular biology and advanced morphology analysis. Coupled with the recently developed approach of visualising tissue mechanical patterns (Michels et al., Reference Michels, Gorelova, Harnvanichvech, Borst, Albada, Weijers and Sprakel2020), this framework holds a great potential in achieving a more comprehensive perception of pattern formation in plants (Figure 2).
9. Conclusions and future perspectives
During the past decade the Arabidopsis embryogenesis emerged as a versatile model allowing to address many aspects of plant development. Its low cell number and the stereotypical division pattern rendered the Arabidopsis embryo model to be simple enough for the use in modelling studies, while the presence of all basic tissue types and organs made this system sufficient to explore cell specification and establishment of the body pattern. The recent development of embryo-optimised reporters and the advance of imaging techniques further secured the embryo in the role of a good model system.
However, as any model, Arabidopsis embryo system has its limitations. Specifically, its location inside the developing seed made the use live imaging approaches challenging until this issue was countered by the development of in vitro ovule culture systems (Gooh et al., Reference Gooh, Ueda, Aruga, Park, Arata, Higashiyama and Kurihara2015).
Despite these challenges, the use of Arabidopsis embryo model holds potential for establishing new methodologies. A typical example is a combination of the existing reporters for subcellular structures with the recently developed mechano-probes that emerged as an excellent tool capable of addressing how mechanical cues instruct cell division and pattern formation. Future development of high sensitivity techniques that would allow to trace subcellular components at high resolution in real time and techniques enabling single molecule imaging would help to accurately reconstruct the sequence of intracellular events leading symmetry breaking during cell division.
Finally, its highly predictable division pattern makes the Arabidopsis embryo a perfect model to address the question of symmetry breaking. The study by Yoshida et al. (Reference Yoshida, de Reuille, Lane, Bassel, Prusinkiewicz, Smith and Weijers2014) demonstrated the robustness of this system and uncovered auxin response as a requirement for symmetry breaking. It still remains to be determined if the same principle operates in other developmental (post-embryonic) contexts and a general question arises: How far we can extrapolate principles applying to the embryo model? We, therefore, anticipate the future research to focus on comparing different developmental contexts of the same species as well as cognate contexts of different species as it appears to be the final obstacle for Arabidopsis embryo to enjoy the most coveted feature of any model system – universality.
Acknowledgements
The authors thank members of the Weijers and Sprakel labs for inspiring discussions.
Financial support
This work was financially supported by the Netherlands Organisation for Scientific Research (NWO; ECHO grant 711.018.002 to J.S) and the European Research Council (ERC; Advanced Grant ‘DIRNDL’; Contract number 833867 to D.W).
Conflict of interest
The authors declare no conflicts of interest.
Authorship contributions
Y.H. and V.G. prepared the first draft of the manuscript and figures. J.S. and D.W. provided input to the draft and all authors contributed to writing the final version.
Data availability statement
No new data or code are presented in this paper.





Comments
Dear Olivier,
It is my pleasure to submit our invited review paper on the plant embryo as a model for quantitative plant biology. In our review, we have attempted to provide a forward-looking view on how this developmental system can be used to address questions related to the mechanisms underlying multicellular plant development. The review is over the 5000 word limit, and we welcome any feedback on whether you would like us to shorten the text.
We look forward to your feedback..
Kind regards,
Dolf