Highlights
-
• In July 2020, a Salmonella Newport outbreak was identified in Canada. Over the course of the investigation, 515 cases were reported across seven provinces; at the time of publication, this was the largest Salmonella outbreak in Canada in over 20 years.
-
• Symptom onset dates ranged from 15 June 2020 to 29 August 2020. Ages ranged from 1 to 100 years; 54% (279/514) of the cases were female; 19% (79/421) were hospitalized; and there were three deaths reported.
-
• A novel vehicle of infection – red onions from Grower A in California, USA – was identified as the likely source of the outbreak.
-
• This outbreak emphasized the benefits of focusing on location-specific case sub-clusters and systematic linkages of cases prior to availability of WGS results as an investigative strategy, to help manage a large outbreak and improve the timeliness of case investigation.
Introduction
Salmonellosis is a nationally notifiable disease in all provinces and territories in Canada [1], and Salmonella is one of the most frequently reported enteric pathogens [2], resulting in an estimated 88,000 illnesses, 925 hospitalizations, and 17 deaths each year [Reference Thomas3]. Salmonella Newport (S. Newport) is amongst the top ten most frequently reported Salmonella serotypes in Canada, with an average of 285 isolates reported per year between 2016 and 2021. During the outbreak investigation year in 2020, 14.09% of all reported Salmonella isolates were S. Newport. For comparison, the most commonly reported serotype of Salmonella in 2020 was S. Enteritidis representing 28.91% of isolates [2]. Outbreaks of Salmonella in Canada have often been associated with eggs and poultry [Reference Bélanger4]. A Canadian Salmonella source attribution study found that chicken meat was associated with the highest average exposure to Salmonella compared to fourteen other exposure groups [Reference Christidis5]. Salmonella outbreaks have also been associated with a variety of fresh fruits and vegetables, which are increasingly recognized as a source of foodborne illness outbreaks in many parts of the world [Reference Kozak6, Reference Lynch7]. However, outbreaks involving bulb-style root vegetables, specifically onions, are rare [Reference Carstens8] and have not previously been documented in Canada.
On 15 July 2020, the United States Centers for Disease Control and Prevention (US-CDC) notified the Public Health Agency of Canada (PHAC) of a rapidly growing cluster of S. Newport cases in the USA. The same day, Alberta (AB) public health officials notified PHAC of multiple cases of Salmonella species (later serotyped as Newport) associated with congregate living facilities and restaurants. Two days later, PHAC was informed that British Columbia (BC) public health officials had also observed a large number of S. Newport cases associated with restaurant exposures. Shortly thereafter, the National Microbiology Laboratory (NML) confirmed that the cases of S. Newport identified in AB and BC were considered genetically related by WGS to each other and to the cases identified in the USA. At this time, it was determined that an international investigation into the source of these illnesses was warranted. The objective of this paper is to describe the investigative approaches used in Canada to identify the source of the S. Newport outbreak linked to a novel vehicle of foodborne illness – red onions – and to summarize the collaborative efforts undertaken to control the outbreak through public health actions.
Methods
Case definition
A confirmed case was defined as a resident of or visitor to Canada with laboratory confirmation of S. Newport, an isolate matching PulseNet Canada cluster 2007NewWGS-1MP by WGS within 0–10 wgMLST, and a symptom onset, specimen collection, or Salmonella isolation date between 1 June 2020 and 28 September 2020 inclusive.
A probable case was defined as a resident of or visitor to Canada with laboratory confirmation of S. Newport pending WGS, and symptom onset, specimen collection, or Salmonella isolation date between 1 June 2020 and 28 September 2020 inclusive.
Epidemiological investigation
Food and risk factor exposure information for cases was collected through initial interview by local public health officials using jurisdiction-specific Salmonella questionnaires. Cases were asked about exposures in the week prior to illness onset. Data from the 2015 Foodbook population-based telephone survey [9] were used to establish the expected proportion of Canadians that reported eating various food items in the past seven days. Foodbook values were restricted to the provinces and territories where cases resided and to the months in which illness onsets were reported. Comparisons between case exposure frequencies and Foodbook values were made using binomial probabilities.
After analyses of initial interview data identified food items of interest, re-interviews were conducted using standardized focused questionnaires to ask more specific questions on these suspect sources, including product type, purchase location, purchase date, brand, and method of preparation and consumption. Re-interviews were conducted by both provincial and federal epidemiologists. In the province of BC, a supplementary focused questionnaire for identified suspect sources was used at initial interview for Salmonella cases of all serotypes during the time period of the investigation.
Cases who reported consuming more than one type of onion were categorized to determine their ‘most likely’ onion exposure. Considerations of ‘most likely’ onion exposure included whether onions were specifically reported by the case or included in a menu description of a food item, the type of onions reported, whether the onions were consumed raw or cooked, whether the onion exposure reported was within a reasonable timeframe of illness onset, and whether the case specified a restaurant, store, or facility, that had an onion supply linking back to Grower A.
Cases were assigned to a sub-cluster if two or more cases were identified eating food prepared at the same restaurant location; shopped at the same grocery store location in the week prior to their illness onset; or were residents, visitors, or staff of the same congregate living facility (e.g. facilities that provided assisted care such as a retirement residence, assisted living facility, or nursing home). In some instances, probable cases were investigated before laboratory subtyping results were available if they were identified as having a link to an existing sub-cluster. All epidemiological data were entered into a Microsoft Access database and descriptive analysis and visualizations were performed in Stata 15 (StataCorp, College Station, Texas, TX, USA) [10] and R statistical software, v3.5.1 [11].
Clinical laboratory analysis
In Canada, all human Salmonella isolates are forwarded to provincial public health laboratories or the NML for WGS-based subtyping. WGS is completed using the standardized PulseNet Canada protocol [Reference Rumore12]. WGS data are analysed locally and then uploaded to a centralized BioNumerics v7.6 (BioMérieux) database where it is analysed by the PulseNet Canada national database team using wgMLST. Multi-jurisdictional S. Newport clusters are identified using a threshold of two or more isolates with isolation dates within the past 60 days that are related within 0–10 wgMLST allele differences. To visualize isolate relatedness, unweighted pair group method with arithmetic mean (UPGMA) dendrograms are constructed within BioNumerics v7.6 using a categorical similarity coefficient.
WGS data were exchanged with PulseNet USA to facilitate the query of the PulseNet USA databases for potential matches in the USA as well as on the National Center for Biotechnology Information (NCBI) pathogen detection pipeline. The data for isolates included in the Canadian investigation were deposited retrospectively on NCBI under BioProject PRJNA543337.
Traceback and environmental investigation
Based on findings from the epidemiologic investigation, the Canadian Food Inspection Agency (CFIA) performed traceback investigations on suspect sources with the aim of identifying commonalities. Records reviewed included purchase receipts, import documentation, invoices and/or menus from restaurants, congregate living facilities, and grocery stores. Import documentation, including invoices, were shared with the US Food and Drug Administration (US-FDA) to assess potential links of Canadian distribution to USA suppliers. A traceback visualization was created using Draw.io. A trace-forward investigation, using product distribution lists, was also performed. Using these distribution records, investigators were able to link probable cases to a common supplier based on their reported onion exposures before WGS results were available to confirm cases’ association with the outbreak. This trace-forward method was also used to determine a link between the implicated product and location-specific case sub-clusters in situations where other records were incomplete or unavailable to perform a more traditional traceback investigation.
Food and environmental samples were collected from implicated congregate living facilities, restaurants, distributors, and case homes. Samples were collected and tested using polymerase chain reactions for Salmonella by CFIA, AB Precision Labs, AB Agriculture and Forestry Agri-Food Laboratories Branch, and the BC Centre for Disease Control Public Health Laboratory. Validation studies on the onion samples were also conducted by Health Canada to determine the potential for inhibition of growth/survival of Salmonella in onions. All method validation studies were conducted following the method validation guidance published in the Health Canada Compendium of Analytical methods [13].
Results
Case characteristics
A total of 515 cases were identified in seven provinces, with AB (293) and BC (121) comprising 80% of cases (Figure 1). Symptom-onset dates ranged from 15 June 2020 to 29 August 2020. The median age of cases was 40 years (range 1–100), and 54% were female (279/514). Nineteen per cent (79/421) of cases were hospitalized, and three deaths were reported. Of the cases who visited, worked, or resided in a congregate living facility, the vast majority, 84% (72/86), occurred in AB.
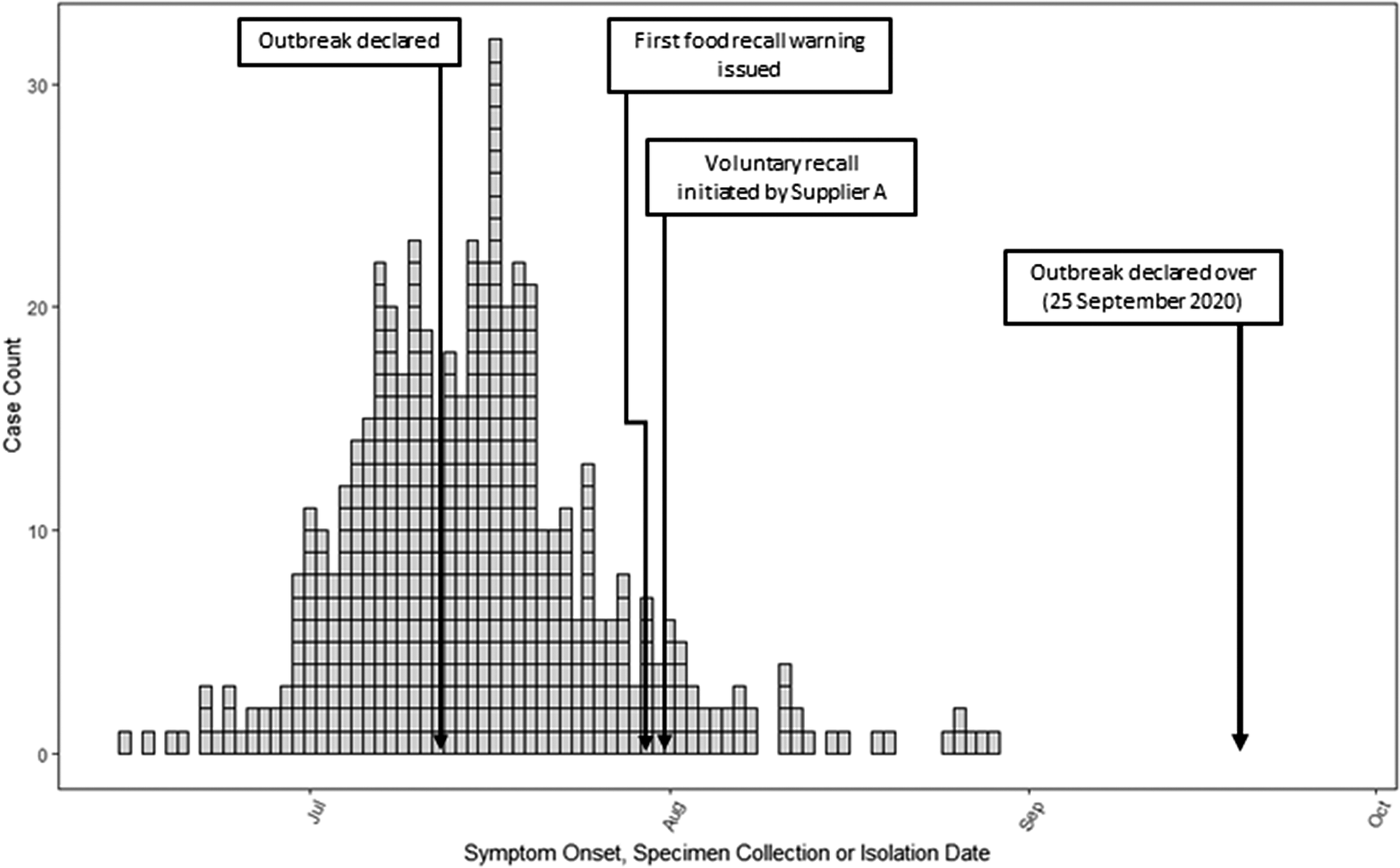
Figure 1. Distribution of Canadian Salmonella Newport outbreak cases (n = 515) by symptom onset, specimen collection or Salmonella isolation date from 15 June to 29 August 2020. Each box denotes a case.
Laboratory analysis
The 515 clinical cases were considered highly related to each other based on WGS and grouped together within 0–6 wgMLST allele differences. The WGS profile was unique in the Canadian database, and no other genetically related isolates were identified. A query of NCBI did not identify any genetically related isolates; however, a query of the PulseNet USA databases identified that the Canadian isolates were genetically related within 0–10 wgMLST allele differences to a concurrent USA investigation [Reference McCormic14].
Initial stage of the epidemiological and traceback investigation
Of 55 initial cases with exposure information available, 96% (26/27) of cases reported exposure to any onions which was significantly higher than the Foodbook reference value (83.0%; p = 0.043). Based on onion type, 85% (17/20) specifically reported consuming red onions in the week prior to becoming ill, which was significantly higher than the Foodbook reference value (38.5%, p = 0.000); 74% (14/19) reported yellow/white onions; and 52% (12/23) reported green onions, both of which were not significantly higher than the Foodbook reference values (75.6% and 48.7% respectively). Lastly, 19% (5/26) did not specify a type of onion.
Of the 17 cases who initially reported red onions, ten cases reported the exposure at a restaurant (six of which were known to be supplied by Supplier A), and seven reported the exposure at a grocery store. Of the remaining 38 initial cases who did not specifically report exposure to red onions at initial interview, seventeen resided in congregate living facilities which all received red onions from Supplier A, seven dined at restaurants known to be supplied by Supplier A, four had red onions on their purchase records with supplier information not yet known, and ten were not able to be linked to Supplier A at this time.
Early in the investigation, restaurant and congregate living facility menus and supplier invoices were obtained and thirteen location-specific sub-cluster facilities were identified (five restaurants and eight congregate living facilities) that all received the same brand of red onions. The red onions were in two different size packages (box and carton) from importing branches of Canadian Supplier A located in three provinces. Red onion import documents identified the USA as the exporting country, and documents were shared with relevant authorities in the USA. Traceback investigation in the USA established a connection to a grower, Grower A, with growing fields in Bakersfield, California, USA [Reference McCormic14]. A traceback diagram showing the link between the thirteen initial location-specific case sub-clusters, and this grower is shown in Figure 2. This initial phase of the investigation confirmed the hypothesis that Grower A red onions were the source of the outbreak.
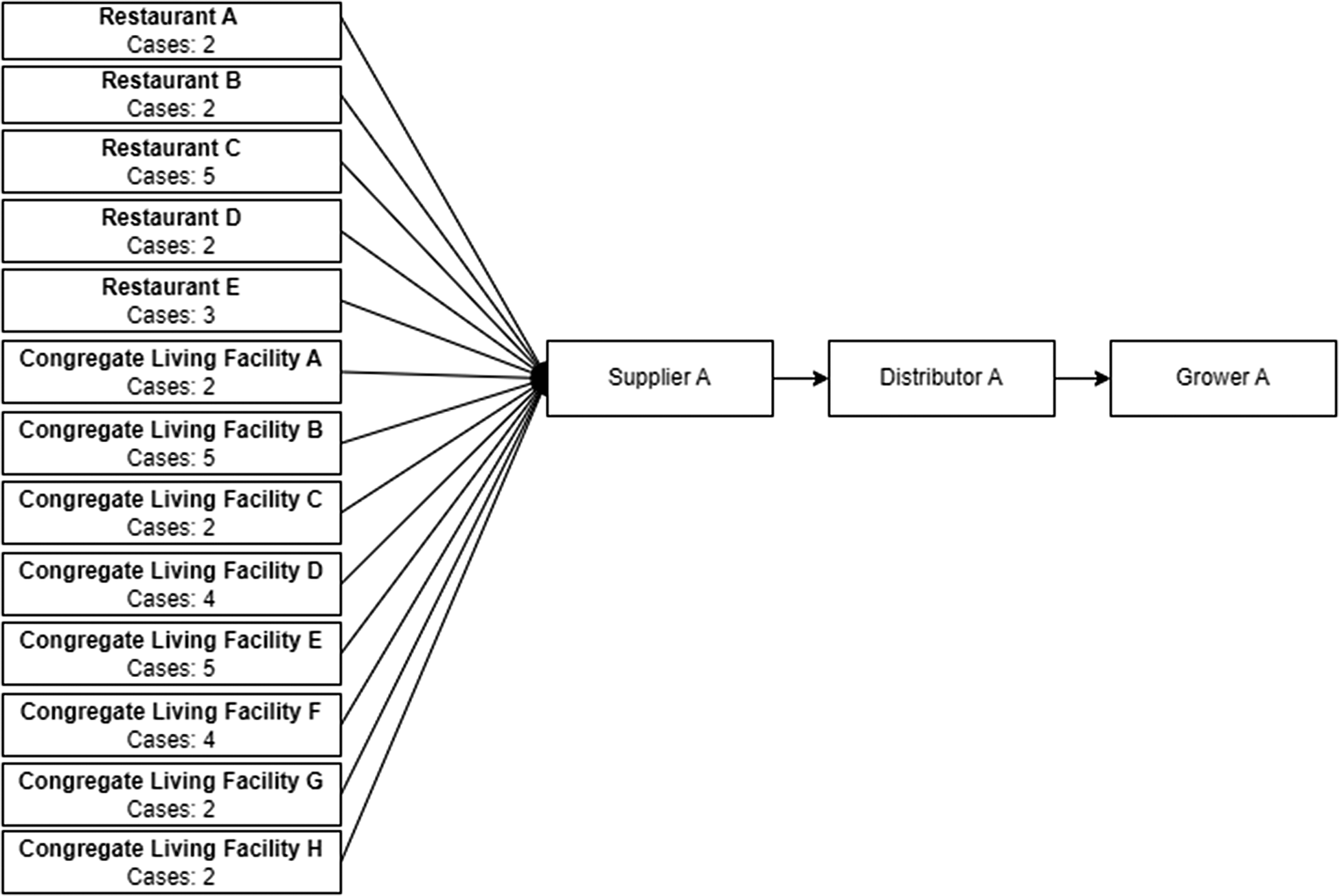
Figure 2. Red onion traceback distribution diagram for thirteen Canadian location specific case sub-clusters (restaurant or congregate living facility) with red onion exposures initially identified at the beginning of the investigation. The diagram depicts the number of cases associated with each identified sub-cluster and how they are linked to a single supplier, distributor and grower.
Overall epidemiological and traceback investigation
One hundred and eighty-three cases were re-interviewed to gather more specificity on onion exposures. In total, exposure information from initial interviews and/or re-interviews was available for 80% (414/515) of all cases. Of the 414 cases, 71% (295/414) had sufficient onion exposure information and reported consuming onions in the week prior to becoming ill (Table 1). Of the 295 cases that reported onion exposure, 80% (235) of cases reported red onion, 14% (41) of cases reported white onion, and 6% (19) did not specify the type of onion consumed.
Table 1. Number of Salmonella Newport outbreak cases reporting onion exposure one week prior to illness onset, by onion type and exposure setting/location, Canada, July to September 2020
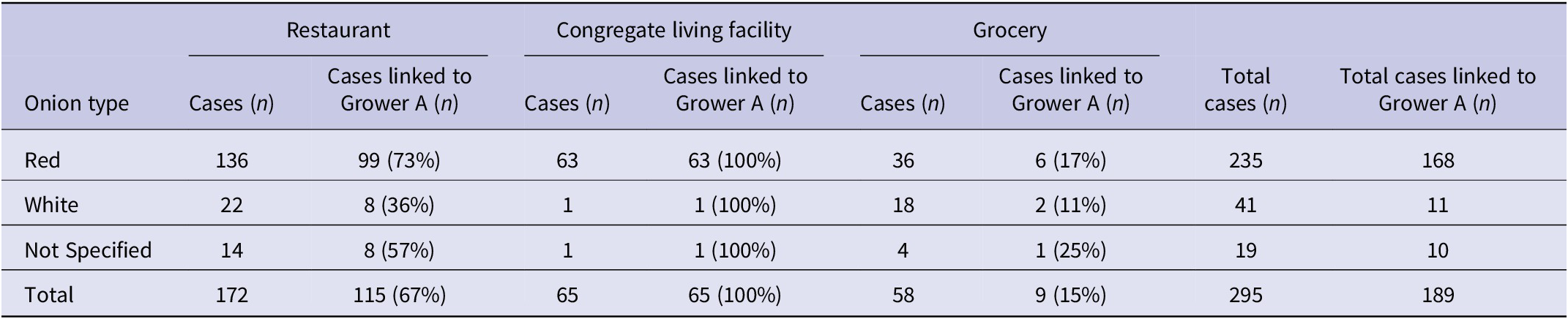
Of the 235 cases who reported red onion exposure, 58% (136) reported the exposure from a meal at a restaurant; 27% (63) reported the exposure from visiting, working at, or residing in a congregate living facility; and 15% (36) reported the exposure from a grocery store (Table 1).
In total, over fifty restaurant and congregate living facility menus and supplier invoices were obtained, as well as product distribution documents for over 65,000 shipment transactions. Based on these documents, it was further identified that Canadian Supplier A, through Distributor A, was the sole supplier of red onions to the majority of congregate living facilities that had outbreak cases. There were no commonalities observed amongst green, yellow, and other fresh onions received by the facilities during the relevant exposure periods as not all facilities received these types of onions during the period of consumption, or more than one brand was in distribution.
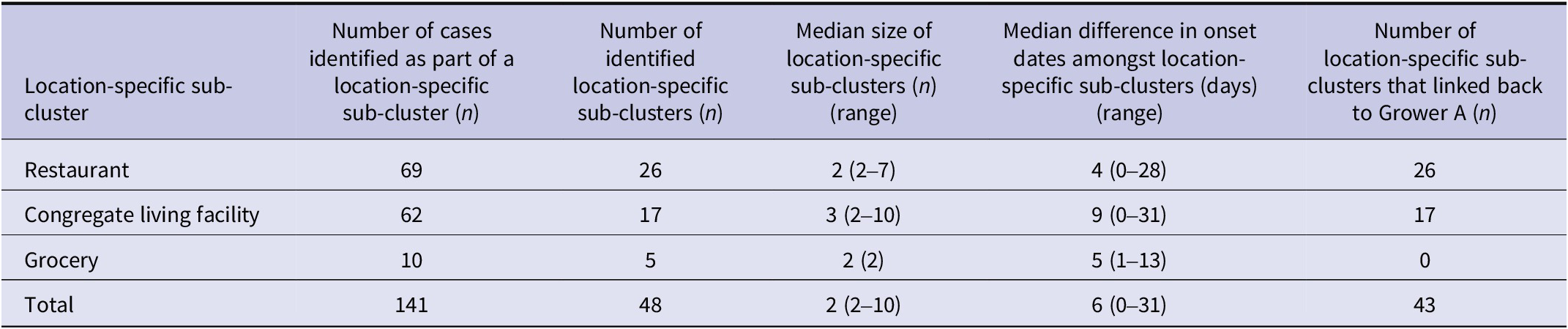
Notably, 48% (141/295) of cases with onion exposure information were identified as part of forty-eight location-specific case sub-clusters during this investigation (54.2% (26) at restaurants, 35.4% (17) at congregate living facilities, and 10.4% (5) at grocery stores) (Table 2). Of note, there were no household clusters identified during this investigation. Overall, traceback or trace-forward methods during the investigation identified that 89% (43/48) of location-specific case sub-clusters shared the common onion supplier of interest – Supplier A. Ninety-three per cent (40) of these location-specific case sub-clusters received red onions and 7% (3) only received other bulb onions from this supplier. Lastly, the geographical distribution of Grower A onions was consistent with the geographical distribution of outbreak cases.
Table 2. Number of location-specific Salmonella Newport outbreak cases identified as part of a sub-cluster by exposure setting/location, Canada, July to September 2020
Environmental investigation
Sixty-five food and environmental samples were collected and tested during this investigation, including thirty-nine red onions, six yellow onions, four unspecified onions, and sixteen environmental (onion bins/netting) samples. Salmonella was not detected in any of the tested samples. Internal validation studies performed on the samples demonstrated that Salmonella could be recovered at sufficiently low inoculation levels from the onion matrix and that onion homogenates did not appear to affect the growth of healthy Salmonella cultures. The US-FDA also conducted field investigations at the exporter of interest, where a number of water, soil, and sediment samples collected were positive for seventeen different Salmonella serotypes including S. Newport; however, the outbreak strain was not found.
Control measures in Canada
One week after the investigation began in Canada, on 30 July 2020, a Public Health Notice was issued advising Canadians in implicated provinces to avoid consuming red onions and products containing red onions imported from the USA. On the same day, CFIA posted a food recall warning for red onions imported from the USA by Supplier A and distributed in the five Canadian provinces in which cases were reported at that time. On 1 August 2020, Grower A voluntarily recalled red, yellow, white, and sweet yellow onions, distributed to the USA and Canada as these products were harvested together with potential for cross-contamination. Nine additional food recall warnings were posted between 1 August 2020 and 14 August 2020 as new information became available, such as additional distributors of onions or products made with onions from Grower A (e.g. salsa, remoulade, crab cakes, salads, pasta salads, and sandwiches). The outbreak was declared over on 28 September 2020.
Discussion
The investigation team coordinated the response to the largest multi-provincial Salmonella outbreak in Canada in over 20 years, resulting in 515 confirmed cases across seven provinces, associated with a novel vehicle of infection. The epidemiological and traceback investigations demonstrated that the likely source of this outbreak was red onions, a raw agricultural commodity that had not been previously associated with foodborne illness outbreaks in Canada.
Apart from the large outbreak size and novel suspect source, what made this investigation unique in Canada were the epidemiological methods that were used for hypothesis generation. The location-specific case sub-clusters provided confidence that Salmonella cases likely shared a common exposure due to the common exposure location and, in the case of restaurant and grocery store sub-clusters, a common exposure date. As a result, it was possible to investigate probable cases prior to availability of WGS rather than initiating investigation when the WGS results were known, which is the typical approach for the investigation of national Salmonella clusters. By focusing the investigation on location-specific case sub-clusters which has been used in previous outbreak investigations [Reference Behravesh15–Reference Laughlin17] and initiating investigation of probable cases prior to availability of laboratory subtyping, red onions were identified as the likely source of the outbreak only one week after initiation of the collaborative investigation. This is a useful strategy that can be leveraged to help manage future large, fast-paced outbreaks.
While investigation of some of these location-specific case sub-clusters was limited due to a combination of poor recall, use of proxy respondents, and complex clinical histories, particularly in congregate living facilities, that made it challenging to determine the relevant exposure period, the availability of menus and supplier invoices presented an opportunity for comparison and identification of commonalities between facilities. Additionally, collecting menus and supplier invoices proved advantageous for location-specific case sub-clusters linked to restaurants, where most cases reported a sandwich or burger exposure. Given the variety of sandwiches and burgers consumed by cases, and the concurrent USA investigation that was seeing an early Mexican-style foods signal, it was apparent that a raw produce ingredient or topping was likely common across cases, which served as a helpful starting point for identifying suspect sources. Review of restaurant menus and supplier invoices, in combination with available records from affected congregate living facilities, ultimately helped to identify red onions as the likely source, as all sandwiches or burgers reported by cases contained red onion and no other commonalities were identified amongst other food items of interest such as tomatoes or lettuce. Additionally, investigating location-specific case sub-clusters was essential as the suspect source was an ingredient and thus not often reported as an exposure by outbreak cases and the product brand was unknown to outbreak cases, as often happens in produce-related outbreaks. Using the above investigative strategies in tandem supported timely case investigations, which helped to reduce the potential impact of recall bias, and swift outbreak source identification.
After the identification of red onions from Grower A as the likely source of the outbreak, the product distribution lists provided by the grower allowed the investigative team to determine whether case sub-cluster locations had received the implicated product. This process enabled the team to triage and organize the large volume of outbreak cases and assisted with early prioritization of cases for re-interview. This systematic approach was also used to link case exposures back to red onions from Grower A to ensure that there were no other sources or suppliers involved in the large outbreak. Ultimately, 5 of the 48 location-specific case sub-clusters, and 36% (106) of cases were not able to be linked back to Grower A. This finding could be attributed to poor case recall, and investigators not having access to all distribution lists (including secondary and tertiary distribution) which prevented systematically linking cases and clusters to the Grower A onion supply chain. Although the majority of cases reported onion exposure, it is also possible that those that did not were infected as a result of the Salmonella bacteria contaminating other food items due to cross-contamination with cutting boards and knives used to prepare Supplier A onions. As mentioned above, some cases may not have been able to accurately recall consuming onions within their exposure period, especially for meals consumed outside the home.
Furthermore, fresh fruits and vegetables are increasingly recognized as a source of foodborne illness outbreaks in many parts of the world [Reference Kozak6, Reference Lynch7]; however, the size of this outbreak and the vehicle of infection are both unique within Canada. Excluding the current outbreak, ten Canadian multi-provincial foodborne illness outbreaks had been associated with fresh produce exposure over the past decade (2010–2020). The number of confirmed cases in these outbreaks ranged from 4 to 399, with a median of 65 cases [18–27]. Interestingly, the majority of these multi-provincial foodborne illness outbreaks associated with fresh produce exposure (n = 7, 64%) were due to Escherichia coli O157:H7 contamination of romaine lettuce and other leafy greens. The current outbreak was 8.5 times larger than the median size of other Canadian multi-provincial foodborne illness outbreaks associated with fresh produce exposure over the past decade. Additionally, given that the implicated onions in this investigation were distributed by one of the largest food service distributors in Canada, it is likely that the true magnitude of this outbreak was even greater, considering the known impact that the COVID-19 pandemic had on enteric case reporting [Reference Dougherty28]. Notably, prior to the current outbreak, onions had never been implicated as a suspect or confirmed outbreak source in Canada. Between 2016 and 2018, there were three suspected onion-related outbreaks documented in the USA caused by S. Javiana [29]; however, these were significantly smaller in size, ranging from 29 to 149 cases.
Additionally, the epidemiology of both the Canadian and USA investigations was very similar in this bi-national outbreak. The US-CDC investigation identified 1127 cases in the USA from forty-eight states, showing a similarly broad geographic distribution to the Canadian outbreak. Case onset dates ranged from 19 June to 11 September 2020. Case ages ranged from 0 to 102 years, with a median of 41. Fifty-eight per cent of cases were female. Twenty-four per cent (167/705) of cases were hospitalized, and no deaths were reported [Reference McCormic14]. Ninety-one per cent of cases interviewed (344/380) reported consuming any type of onions or foods likely containing onions in the week prior to becoming ill. Of the 208 people who were asked what types of onions they ate, 66% (137) reported consuming red onions, 63% (130) reported consuming white onions, and 53% (110) reported consuming yellow onions [Reference McCormic14]. International collaboration between USA and Canadian public health and food safety partners proved key to finding commonalities across investigations, identifying a source, and pursing public health action.
At its conclusion, over 18,000 tons of recalled onions were implicated in this investigation. Recalled onions were sold in an assortment of packages, in bulk, under a variety of brands, and were distributed to multiple grocery stores, restaurant chains, and congregate living facilities across Canada and the USA.
With such a large-scale outbreak, there were several challenges faced by the investigative team. First, as is the case with many produce items, onion distribution channels from farm to fork are complex and multifaceted, involving numerous transfers throughout the transportation process and onion packaging at the consumer level is limited, presenting challenges in narrowing the scope of recalls and effectively communicating risks to the public [Reference Marshall30]. Due to this limitation, the first public health notice contained a broad message, stating, ‘do not eat any red onions imported to Canada from the USA’. This type of communication, a readily available risk management tool, has been used previously with produce related outbreaks [Reference Irvin31]. Second, large case counts and varying types of data used in this outbreak investigation also created challenges for existing data management infrastructure. These challenges can slow down the speed of an investigation and delay source identification and, as such, point to the need for flexible data management systems and a robust and skilled public health workforce to adapt. During this investigation, this challenge was overcome by prioritizing data entry and analysis, as well as alternation between staff concurrently accessing the data management system to minimize system overload. Third, the coinciding COVID-19 pandemic and the demand it placed on public health resources necessitated flexibility in the use of available resources. Extensive collaboration and communication between affected public health jurisdictions was essential to gather and analyse the large volume of data associated with the outbreak investigation and to take public health action. Lastly, despite the number of representative samples analysed, the Salmonella outbreak strain was not identified in any of the collected product samples. Possible reasons for this could be that samples were collected at a time when contaminated lots were no longer in distribution (e.g. by the time the outbreak was detected due to the usual reporting delays inherent in laboratory-based outbreak detection and when the recalls were issued any remaining products had either been used up or discarded), or contamination may have been heterogeneous amongst the affected lots [32]. While the root cause of contamination is unclear [Reference McCormic14], the US-FDA is leading work on prevention strategies related to bulb onions [33].
Despite the challenges described above, the collaboration between partners during this investigation was noteworthy. The PHAC team provided support to provincial and local public health partners who were faced with resource limitations due to simultaneously responding to the COVID-19 pandemic, and the NML provided serotyping and WGS support to provincial laboratories experiencing high testing volumes, allowing new cases to be identified as quickly as possible. International sharing of traceback and trace-forward information between US-FDA and CFIA led to the identification of the supplier of interest to both the Canadian and USA investigations, which in turn led to timely public health action that helped protect the health and safety of all Canadians. In total, 1642 Canadian and USA cases were identified between the two investigations; the successful investigation of this large-scale outbreak highlights the need for continued collaboration in future investigations.
Data availability statement
The data from this study are not publicly available due to privacy concerns and legislative requirements.
Acknowledgements
The authors thank the following organizations for their contribution to the outbreak investigation: local public health colleagues and public health laboratories in British Columbia, Alberta, Saskatchewan, Manitoba, Ontario, Quebec, and Prince Edward Island; British Columbia Centre for Disease Control; British Columbia Public Health Laboratory; Alberta Health; Alberta Health Services; Saskatchewan Ministry of Health; Manitoba Health; Public Health Ontario; Ontario Ministry of Health; Ministère de la Santé et des Services Sociaux du Québec; Laboratoire de santé publique du Québec; Prince Edward Island Department of Health and Wellness; Canadian Food Inspection Agency; the National Micorbiology Laboratory; Health Canada; the Public Health Agency of Canada; the US Centers for Disease Control and Prevention; and the US Food and Drug Administration.
Author contribution
Investigation: Z.D.M., C.R.S., L.H., L.C., A.J.M., A.H., A.K., C.N., E.B., J.J.L., J.M.C., M.T., K.P., V.M., L.D.; Writing – review & editing: Z.D.M., C.R.S., L.H., L.I., L.C., A.T., A.J.M., A.H., A.K., C.N., C.M., D.S., E.B., J.J.L., J.M.C., M.T., K.P., R.A., V.M., L.D.; Conceptualization: C.R.S., L.H., L.I., A.T., A.J.M., D.S., J.J.L., J.M.C., M.T., R.A., L.D.; Data curation: C.R.S., L.H., L.C., A.J.M., A.K., C.M., E.B., J.J.L., M.T., V.M., L.D.; Formal analysis: C.R.S., L.H., L.I., A.T., A.J.M., A.K., C.M., D.S., E.B., J.J.L., R.A., L.D.; Methodology: C.R.S., L.H., L.C., A.J.M., A.K., C.N., E.B., J.J.L., J.M.C., M.T., V.M., L.D.; Validation: C.R.S., L.H., L.I., A.T., A.J.M., D.S., J.J.L., M.T., R.A., L.D.; Writing – original draft: C.R.S., A.H., D.S., J.M.C., L.D.; Resources: L.H., L.C., C.M.; Visualization: L.H., L.I., L.C., A.T., A.J.M., C.M., D.S., E.B., J.J.L., M.T., R.A., L.D.; Project administration: L.C., A.H., J.M.C.; Supervision: A.H., A.K., C.N., D.S.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the United States Centers for Disease Control and Prevention.







