Introduction
Wolbachia, a bacterial genus in the Rickettsiales, are Gram-negative endosymbiotic bacteria that are primarily found in invertebrates, particularly arthropods and nematodes (Werren et al. Reference Werren, Baldo and Clark2008). Wolbachia form part of the complex invertebrate microbiome, enhancing the efficiency of a range of biological systems in host organisms, including the immune system (Pimentel et al. Reference Pimentel, Cesar, Martins and Cogni2021), nutrient acquisition (Newton & Rice Reference Newton and Rice2020, Deconninck et al. Reference Deconninck, Larges, Henri, Beaugeard, Foray and Pincebourde2024) and temperature tolerance (Hague et al. Reference Hague, Caldwell and Cooper2020), thereby impacting the host phenotype through parasitic and/or mutualistic relationships (Iturbe-Ormaetxe & O’Neill Reference Iturbe-Ormaetxe and O’Neill2007, Manoj et al. Reference Manoj, Latrofa, Epis and Otranto2021). No free-living members of the genus are known (Vancaester & Blaxter Reference Vancaester and Blaxter2023). Perhaps one of the most well-known impacts of Wolbachia is its ability to manipulate the reproduction of the host (Hyder et al. Reference Hyder, Lodhi, Wang, Bukero, Gao and Mao2024), which can, for example, initiate parthenogenesis (Werren et al. Reference Werren, Baldo and Clark2008), as well as feminization (Asgharian et al. Reference Asgharian, Chang, Mazzoglio and Negri2014) and cytoplasmic incompatibility (Chen et al. Reference Chen, Zhang and Hochstrasser2020). Other known insect endosymbionts in the same order that can induce parthenogenesis include the genera Rickettsia and Cardinium (Ma & Schwander Reference Ma and Schwander2017). Most Rickettsia species that have been observed in insects are facultative symbionts, and some can cause the death of male embryos (Giorgini et al. Reference Giorgini, Bernardo, Monti, Nappo and Gebiola2010) in, for example, certain ladybirds (Werren et al. Reference Werren, Hurst, Zhang, Breeuwer, Stouthamer and Majerus1994, von der Schulenburg et al. Reference der Schulenburg, Habig, Sloggett, Webberley, Bertrand, Hurst and Majerus2001) and beetles (Lawson Reference Lawson, Mousseau, Klaper, Hunter and Werren2001). Of these reproductive manipulations, parthenogenicity can be advantageous in the short term as it guarantees reproduction when access to mates is limited, although there is often a loss of heterozygosity over time (Jaron et al. Reference Jaron, Parker, Anselmetti, Van, Bast and Dumas2022). In extreme and isolated environments such as remote oceanic islands and in the polar regions, environmental constraints on individual movement have been suggested to select for parthenogenesis (Chown & Convey Reference Chown and Convey2016, Leihy & Chown Reference Leihy and Chown2020).
Although Wolbachia has a global distribution (Werren & Windsor Reference Werren and Windsor2000), infecting ~40% of arthropods (Nugapola et al. Reference Nugapola, de Silva and Karunaratne2017) and up to 60% of terrestrial insect species (Sazama et al. Reference Sazama, Bosch, Shouldis, Ouellette and Wesner2017), to date it has not been detected in Continental Antarctic, Maritime Antarctic or sub-Antarctic invertebrates (e.g. Charlesworth et al. Reference Charlesworth, Weinert, Araujo and Welch2019, McQueen et al. Reference McQueen, Gattoni, Gendron, Schmidt, Sommers and Porazinska2023, Serga et al. Reference Serga, Kovalenko, Maistrenko, Deconninck, Shevchenko and Iakovenko2024), including in the only insect from the Antarctic continent studied to date: the non-biting flightless midge Belgica antarctica (Diptera: Chironomidae; Maistrenko et al. Reference Maistrenko, Serga, Kovalenko and Kozeretska2023). Eretmoptera murphyi (Fig. 1) is another non-biting flightless midge that molecular analysis shows to be very closely related to B. antarctica (Allegrucci et al. Reference Allegrucci, Carchini, Convey and Sbordoni2012). It is native and endemic to the sub-Antarctic island of South Georgia (Chown & Convey Reference Chown and Convey2016), but it is now also well established and considered an invasive species on Signy Island in the Maritime Antarctic (South Orkney Islands; Hughes & Worland Reference Hughes and Worland2010, Bartlett et al. Reference Bartlett, Convey, Pertierra and Hayward2020). Unlike B. antarctica, which is a sexual species, E. murphyi reproduces parthenogenetically, and no male specimens have ever been reported (Cranston Reference Cranston1985, Bartlett et al. Reference Bartlett, Convey and Hayward2018, Kozeretska et al. Reference Kozeretska, Serga, Kovalenko, Gorobchyshyn and Convey2021). The hypothesis that Wolbachia could be responsible for the different reproductive strategies of the two species has not been investigated to date. However, recently, Serga et al. (Reference Serga, Kovalenko, Maistrenko, Deconninck, Shevchenko and Iakovenko2024) provided some new data and highlighted the absence of Wolbachia amongst all Antarctic terrestrial invertebrates whose genomes and/or microbiomes have been sequenced and are publicly available. Serga et al. (Reference Serga, Kovalenko, Maistrenko, Deconninck, Shevchenko and Iakovenko2024) specifically emphasized that, to date, no study has investigated the presence of Wolbachia in E. murphyi. Therefore, the aim of this short publication is to directly address this knowledge gap so as to contribute further insights into Wolbachia in Antarctica and to consider the possibility of other candidate bacteria that may be involved in E. murphyi’s parthenogenetic life history.

Figure 1. Adult (left) and larval (right) Eretmoptera murphyi. Photographs by British Antarctic Survey.
Methods
As part of a wider insect microbiome study, E. murphyi larvae were collected from the ‘Backslope’ (unofficial name) adjacent to the British Antarctic Survey’s Signy Island research station in February 2022 during the summer. They were stored under field conditions on Signy (~2–4°C; Convey et al. Reference Convey, Newsham, Geissler, Jobson and Fox2025) before being placed in 96% ethanol and kept at -80°C on the return ship to the UK. They were maintained at this temperature at the University of Birmingham until analysis. At the University of Warwick, DNA was extracted from five individuals using a DNeasy PowerSoil Pro Kit (QIAGEN) following the manufacturer’s protocol. Universal primers were used to target and amplify the bacterial V4 region of the 16S rRNA; 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Polymerase chain reaction (PCR) involved cycles being carried out to amplify the extracted DNA (95°C for 30 s, 53°C for 30 s, 72°C for 30 s). The target region of ~300 bp was obtained by gel electrophoresis, followed by extraction and purification using a QIAquick Gel Extraction Kit (QIAGEN) following the manufacturer’s protocol. Library preparation and sequencing were carried out on an Illumina PE250 platform by Novogene, UK. A total of 333 809 bacterial sequences were obtained. The DADA2 pipeline (V1.32.0; Callahan et al. Reference Callahan, Mcmurdie, Rosen, Han, Johnson and Holmes2016) was used for quality trimming, error rate estimation, merging, chimera removal and amplicon sequence variant (ASV) feature table construction. Primer sequences were removed from the 5′ region of forward and reverse reads (19 and 20 bp, respectively), and reads were truncated at the first instance of a quality score of ≤ 10. Following dereplication, merging and chimera removal, 69.7–73.1% of bacterial reads were retained for further analysis. ASVs classified as chloroplast, mitochondria or archaea were removed from the samples. To remove sequences potentially arising from the host species (which could result in contamination and possibly reduce the microbial output, especially if the volume of DNA is low; Heravi et al. Reference Heravi, Zakrzewski, Vickery and Hu2020), a local BLAST search was performed against the closely related Antarctic chironomid Belgica antarctica (Kelley et al. Reference Kelley, Peyton, Fiston-Lavier, Teets, Yee and Johnston2014). ASVs with > 90% sequence similarity across 90% of the query length were removed to ensure host sequences were removed (Lagunas et al. Reference Lagunas, Richards, Sergaki, Burgess, Pardal and Hussain2023). Bacterial samples were rarefied to depths of 42 723 and 31 182, respectively, using the rrarefy function from the R package vegan v 2.6-6.1, with default settings (Oksanen et al. Reference Oksanen, Blanchet, Friendly, Kindt, Legendre, McGlinn and Minchin2022). Taxonomy was assigned using SILVA 138.1 prokaryotic small subunit (SSU) taxonomic training data formatted for DADA2, Zenodo (McLaren & Callahan Reference McLaren and Callahan2021). We searched for Wolbachia sequences in a database of 16S amplicon sequences amplified from whole larvae of E. murphyi.
After sequencing and rarefaction revealed 13 sequences belonging to the order Rickettsiales, some of which have known impacts on insect reproduction (Hagimori et al. Reference Hagimori, Abe, Date and Miura2006, Giorgini et al. Reference Giorgini, Bernardo, Monti, Nappo and Gebiola2010, Owashi et al. Reference Owashi, Arai, Adachi-Hagimori and Kageyama2024), these were selected for phylogenetic comparison with reference sequences. These reference sequences were obtained through BLAST analysis, which was performed for each ASV using the 16S ribosomal RNA database. The top results for each ASV (with the highest percentage similarity) were used as reference sequences in generating the phylogenetic tree. To place the ASVs in a broader context, three Wolbachia strains (all insect endosymbionts) were also selected from the SILVA 138.2 SSU database. These were chosen based on sequence quality, species-level annotation and host diversity to reflect phylogenetic variation across strains. To explore potential links to reproductive manipulation, two additional sequences from taxa that are known to facilitate parthenogenesis were sourced from GenBank and included in the phylogenetic analysis. These were Rickettsia sp. (accession no. L28944.1), which induces parthenogenesis in the larval endoparasitoid Neochrysocharis formosa (Hagimori et al. Reference Hagimori, Abe, Date and Miura2006), and Rickettsia bellii strain RML369-C (accession no. NR_074484.2), which facilitates cytoplasmic incompatibility (a similar reproductive method to parthenogenesis that is shared with Wolbachia) in the green mirid Nesidiocoris tenuis (Owashi et al. Reference Owashi, Arai, Adachi-Hagimori and Kageyama2024) and parthenogenesis in the parasitoid wasp Pnigalio soemius (Giorgini et al. Reference Giorgini, Bernardo, Monti, Nappo and Gebiola2010). Our ASV subset and reference sequences were aligned using the AlignSeqs() function in the DECIPHER package. A multiple sequence alignment was converted into a phylogenetic data object using phyDat() from the phangorn package, and a pairwise genetic distance matrix was computed with dist.ml() based on a maximum likelihood model of nucleotide substitution. These values were used to infer genetic similarity between our ASVs and the reference sequences, with lower values indicating greater genetic similarity. The phylogenetic tree was constructed using NJ().
Results
A total of 2013 bacterial ASVs were generated across all samples, and 1971 were classified to at least the phylum level. The average number of unique ASVs in each sample was 677. A search of the database found no Wolbachia sequences present in the samples of E. murphyi. Thirteen ASVs were assigned to the order Rickettsiales (Table I), to which Wolbachia belongs (Serga et al. Reference Serga, Kovalenko, Maistrenko, Deconninck, Shevchenko and Iakovenko2024). Of these, five were assigned to the genus Rickettsia and eight were unassigned beyond the order level. None were assigned to species level. BLAST results indicated that 9/13 of our Rickettsiales ASVs were most closely matched to known Rickettsia species with high sequence identities (Table I). In particular, ASV_1, ASV_2, ASV_1143 and ASV_1321 showed the highest similarity (98.50–98.88%) to Rickettsia hoogstraalii strain Croatica, whereas ASV_584 and ASV_1143 were closely matched to Rickettsia conorii strain Malish 7 (98.50%). At lower similarity, ASV_1327 aligned most closely with Rickettsia rhipicephali (95.13%), ASV_1715 with Rickettsia peacockii (93.26%) and ASV_1734 with Rickettsia honei (91.85%). Phylogenetic reconstruction confirmed that the majority of ASVs clustered with a parthenogenesis-associated Rickettsia sp. (accession L28944.1), with pairwise genetic distances ranging from 0.0112 to 0.2087 (Fig. 2 & Table I), corresponding to ~79–99% sequence similarity (Yarza et al. Reference Yarza, Yilmaz, Pruesse, Glöckner, Ludwig and Schleifer2014). No ASVs showed close similarity to Wolbachia reference strains.
Table I. Taxonomic assignment of the Rickettsiales amplicon sequence variant (ASV) subset using the SILVA 138.1 prokaryotic small subunit (SSU) taxonomic training data formatted for DADA2, Zenodo. No ASVs were assigned to species level. Closest BLAST search results to each ASV and the associated percentage identities are shown. Closest reference sequences for the Rickettsiales ASV subset with the associated genetic distances (measured as estimated substitutions per nucleotide site, calculated using maximum likelihood models) were used to construct the phylogenetic tree.
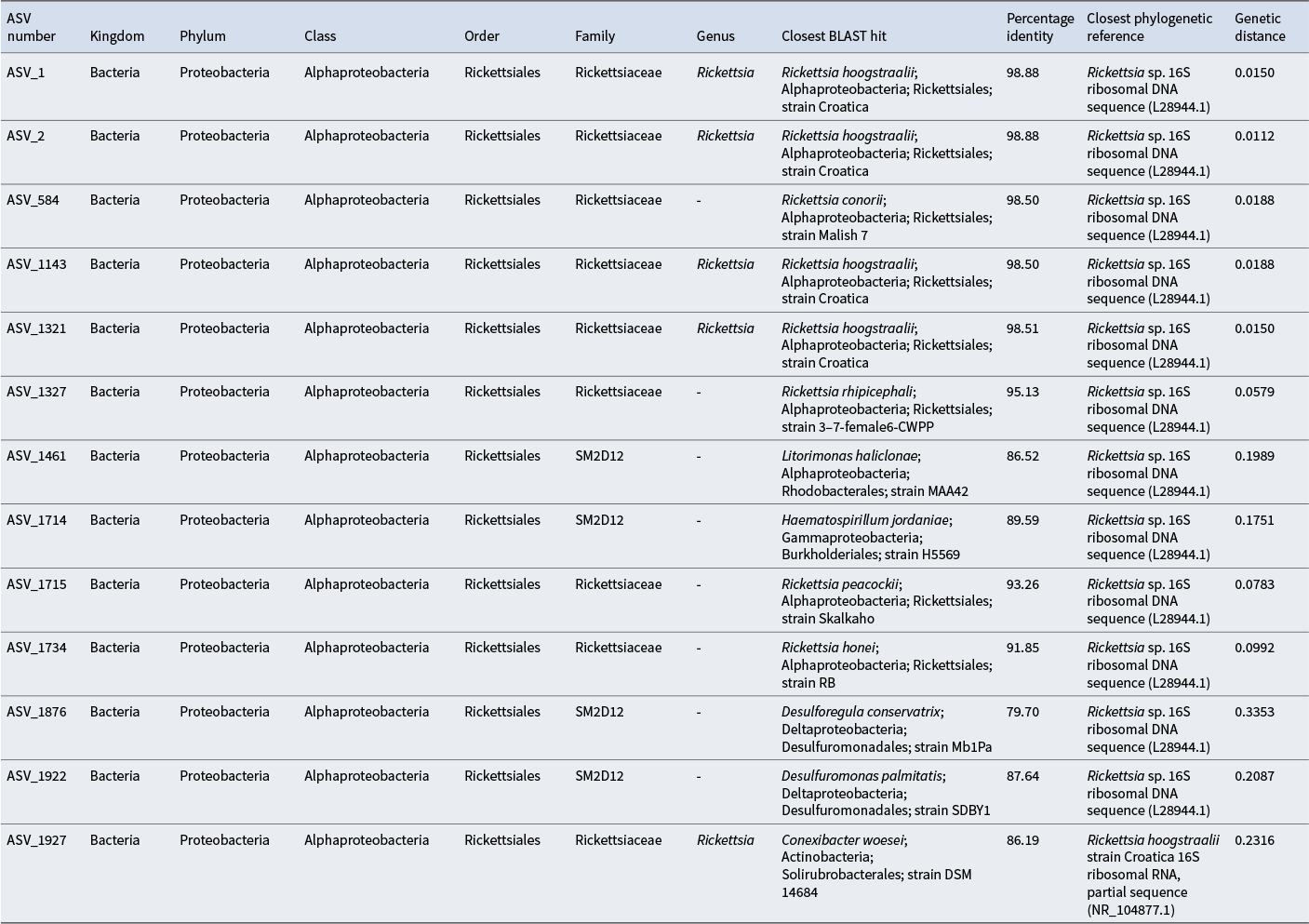
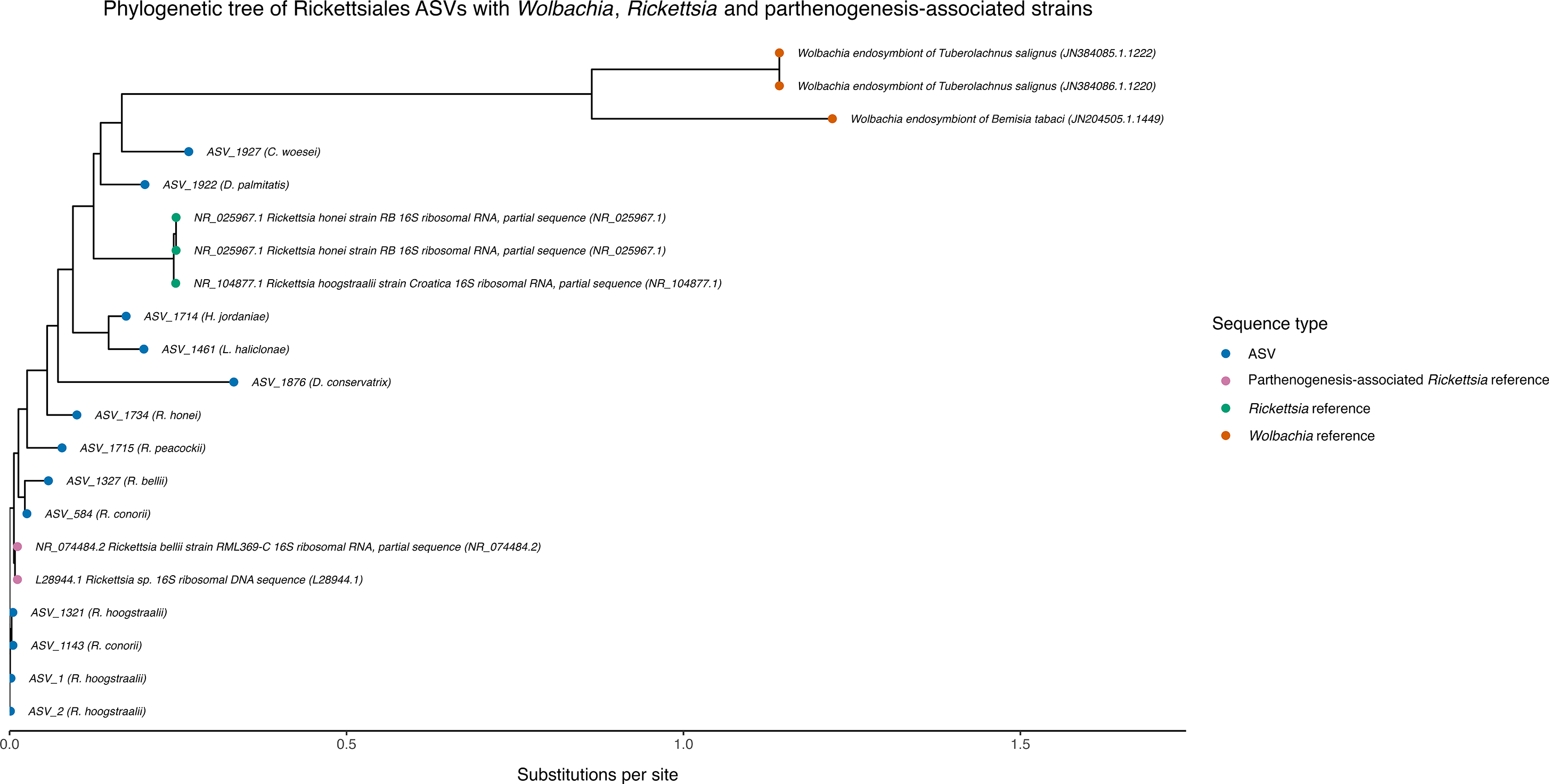
Figure 2. Neighbour-joining phylogenetic tree showing genetic distance values (measured as estimated substitutions per nucleotide site, calculated using maximum likelihood models) between the 13 amplicon sequence variants (ASVs) assigned to the order Rickettsiales in our study and the reference sequences. Tip labels next to ASVs correspond to the closest reference sequence. Shorter branch lengths or lower substitution values represent fewer genetic differences, indicating greater similarity between sequences.
Discussion
The absence of Wolbachia in the microbiome sequences obtained from E. murphyi builds upon the findings of Serga et al. (Reference Serga, Kovalenko, Maistrenko, Deconninck, Shevchenko and Iakovenko2024), which suggest that Wolbachia may be entirely absent from Antarctic terrestrial invertebrates. We can find only one putative record of Wolbachia being reported in Antarctic soil: an environmental DNA study of soils at Mars Oasis on Alexander Island (72°S) in the southern Maritime Antarctic (Pearce et al. Reference Pearce, Newsham, Thorne, Calvo-Bado, Krsek and Laskaris2012). It is plausible that the earlier sequencing methods used at this time misassigned Wolbachia at this site, especially considering invertebrate taxa that could potentially host Wolbachia, such as mites, springtails and nematodes, are generally present at low densities at Mars Oasis (Convey & Smith Reference Convey and Smith1997, Maslen & Convey Reference Maslen and Convey2006). As noted by Serga et al. (Reference Serga, Kovalenko, Maistrenko, Deconninck, Shevchenko and Iakovenko2024), there are no known records of any of these groups anywhere in Antarctica, so its presence at the extreme southern limit of the Maritime Antarctic would seem surprising.
The absence of Wolbachia in E. murphyi does not support the hypothesis that it plays a role in parthenogenesis in this species, although this finding does not rule out the possibility that a different, perhaps undescribed bacteria could influence the insect’s reproductive system. In particular, we identified representatives of the genus Rickettsia (a close relative of Wolbachia) in our database, some members of which are known to induce thelytokous parthenogenesis in parasitoid wasps (Hagimori et al. Reference Hagimori, Abe, Date and Miura2006, Giorgini et al. Reference Giorgini, Bernardo, Monti, Nappo and Gebiola2010) and cytoplasmic incompatibility in true bugs (Owashi et al. Reference Owashi, Arai, Adachi-Hagimori and Kageyama2024). Furthermore, our phylogenetic analysis and associated low genetic distances revealed that most of our ASVs assigned to Rickettsiales are most closely related to Rickettsia sp. (accession L28944.1), which has been shown to induce parthenogenesis in the larval endoparasitoid N. formosa (Hagimori et al. Reference Hagimori, Abe, Date and Miura2006), although this is a parasitoid within the Hymenoptera rather than the Diptera. Even though the ASVs are closely related to this Rickettsia sp., the similarity values suggest that they probably represent distinct Rickettsia taxa; in particular, ASV_1876 was highly divergent and may be a novel Rickettsia lineage (Yarza et al. Reference Yarza, Yilmaz, Pruesse, Glöckner, Ludwig and Schleifer2014). As this is the first study that has assessed some of the bacterial communities associated with E. murphyi, this result is not unexpected. Nonetheless, the phylogenetic analyses confirm that Wolbachia is not involved in the parthenogenetic life history of E. murphyi, but they do not discount the notion that other species within the Rickettsiales may facilitate this reproductive mechanism. To test this hypothesis, future studies could utilize antibiotic treatments to target Rickettsia taxa in order to determine whether this group is involved in altering reproduction (Giorgini et al. Reference Giorgini, Bernardo, Monti, Nappo and Gebiola2010), isolate and culture the specific bacteria to carry out more detailed phylogenetic analysis (Owashi et al. Reference Owashi, Arai, Adachi-Hagimori and Kageyama2024) and conduct microscopy to track the bacteria from adult to egg in order to infer possible function (Hagimori et al. Reference Hagimori, Abe, Date and Miura2006). There are other possible causes for parthenogenesis, including spontaneous differential gene expression (Sperling et al. Reference Sperling, Fabian, Garrison and Glover2023), developmental changes and alterations to cell cycle mechanisms (Sperling & Glover Reference Sperling and Glover2023), as well as strong selective forces leading to the evolution of obligate asexual reproduction (Chown & Convey Reference Chown and Convey2016, Leihy & Chown Reference Leihy and Chown2020).
Various potential explanations for the absence of Wolbachia in Antarctic invertebrates and insects, including whether this observation is simply the result of a lack of targeted research, were raised by Serga et al. (Reference Serga, Kovalenko, Maistrenko, Deconninck, Shevchenko and Iakovenko2024). For example, as Wolbachia is known to be sensitive to temperature (Charlesworth et al. Reference Charlesworth, Weinert, Araujo and Welch2019), it is possible that the polar regions are simply too cold for Wolbachia to persist within its hosts, which is consistent with observations that some widely distributed species host Wolbachia at lower but not higher latitudes (Chrostek et al. Reference Chrostek, Martins, Marialva and Teixeira2021). However, research has yet to directly investigate whether low temperature impacts Wolbachia survival (Serga et al. Reference Serga, Kovalenko, Maistrenko, Deconninck, Shevchenko and Iakovenko2024). Polar terrestrial environments in Antarctica are also generally nutrient-limited (Lambrechts et al. Reference Lambrechts, Willems and Tahon2019), which may mean that it is too costly to the host to harbour Wolbachia in these regions, as suggested by Serga et al. (Reference Serga, Kovalenko, Maistrenko, Deconninck, Shevchenko and Iakovenko2024). Further research on the bacterial diversity (particularly representatives of Rickettsia) associated with E. murphyi, including expansion of the taxonomic databases representing Antarctic microbes, is required to understand how parthenogenesis developed and persists in this midge.
Supplementary material
To view supplementary material for this article, please visit http://doi.org/10.1017/S0954102025000197.
Acknowledgements
The authors are very grateful to Monica Aquilino for sample collection.
Financial support
O.D.M. Brayley and S. Liu are funded by NERC CENTA2 grant NE/S007350/1. O.D.M. Brayley’s PhD is also supported by the British Antarctic Survey. S.A.L. Hayward is supported by a Leverhulme Research Fellowship RF-2024-396/2. P. Convey is supported by NERC core funding to the British Antarctic Survey ‘Biodiversity, Evolution and Adaptation’ team. S.A.L. Hayward, P. Convey and K. McCready were also supported by NSFGEO-NERC grant NE/T009446/1. N. Teets is supported by National Science Foundation Grant OPP-1850988 and USDA National Institute of Food and Agriculture Hatch Project 1010996.
Competing interests
The authors declare none.
Author contributions
PC conceived the study concept. ODMB and SL carried out the DNA extractions. ODMB analysed the data and wrote the initial draft. KM and SL assisted with the bioinformatic analysis. PC, SALH, YC, SU and NT contributed to the manuscript. All authors gave final approval for manuscript submission.






