Introduction
Disruptive behavior, i.e., defiance towards authority, irritability, disobedience, and verbal or physical aggression (DSM-V; American Psychiatric Association, 2013), increases the risk of developing more severe behavior problems later in life, including antisocial and criminal behavior (Campbell et al., Reference Campbell, Shaw and Gilliom2000; Costello et al., Reference Costello, Foley and Angold2006). Parents play a key role in shaping their children’s disruptive behavior (Pinquart, Reference Pinquart2017). While parenting is often regarded as a purely environmental factor, studies using genetically sensitive designs have shown that individual differences in parenting are at least partly influenced by genetic factors (Klahr & Burt, Reference Klahr and Burt2014; Runze et al., Reference Runze, Bakermans-Kranenburg, Cecil, van IJzendoorn and Pappa2023; Wertz et al., Reference Wertz, Belsky, Moffitt, Belsky, Harrington, Avinun, Poulton, Ramrakha and Caspi2019, Reference Wertz, Moffitt, Agnew-Blais, Arseneault, Belsky, Corcoran, Houts, Matthews, Prinz, Richmond-Rakerd, Sugden, Williams and Caspi2020). Genetically informed research on adolescents and adults has demonstrated that both parental genes and behavior, as well as the genetic makeup of their offspring, contribute to variations in externalizing behaviors (Kretschmer et al., Reference Kretschmer, Vrijen, Nolte, Wertz and Hartman2022; Kuo et al., Reference Kuo, Poore, Barr, Chirico, Aliev, Bucholz, Chan, Kamarajan, Kramer, McCutcheon, Plawecki and Dick2022; Luo et al., Reference Luo, Pappa, Cecil, Jansen, van IJzendoorn and Kok2021 Teeuw et al., Reference Teeuw, Mota, Klein, Blankenstein, Tielbeek, Jansen, Franke and Hulshof Pol2023). These genetic influences may operate directly—through the child’s genetic predisposition affecting their behavior—or indirectly, through gene-environment correlations (rGE). Building on this foundation, the current study investigates how both direct genetic effects and rGE contribute to the development of disruptive behavior in at-risk parent-child pairs. Externalizing behavior is typically less severe and associated with milder consequences than disruptive behavior, which often intensifies over time and leads to financial and personal burdens in adulthood for the individuals and society at large (Mesman et al., Reference Mesman, Bongers and Koot2001; Rissanen et al., Reference Rissanen, Kuvaja-Köllner, Elonheimo, Sillanmäki, Sourander and Kankaanpää2021; Rivenbark et al., Reference Rivenbark, Odgers, Caspi, Harrington, Hogan, Houts, Poulton and Moffitt2018; Scott et al., Reference Scott, Knapp, Henderson and Maughan2001). By focusing on disruptive behavior, rather than broader externalizing behaviors, this research aims to provide a more nuanced understanding of the interplay between genes and the environment in shaping early developmental trajectories in young children of at-risk families. Studying disruptive behaviors is critical not only because they remain underexplored in the context of gene-environment interplay, but also because early identification of genetic and environmental contributors may inform targeted interventions, reducing the long-term societal and individual costs associated with these behaviors.
Parental genes and their children’s genes might be directly associated with the environments that parents create or that children experience, a phenomenon known as gene-environment correlation (rGE, Plomin et al., Reference Plomin, DeFries and Loehlin1977; Scarr & McCartney, Reference Scarr and McCartney1983). One type of gene-environment correlation is passive rGE (e.g., Scarr & McCartney, Reference Scarr and McCartney1983), where parents provide both genes and an environment that aligns with those genes. For example, parents with a genetic tendency for certain behaviors (e.g., disruptive or harsh parenting styles) may unintentionally create environments that reflect these traits. Genetic nurture is a specific form of passive rGE where the parents’ genes influence the environment, impacting the child’s development even if the child doesn’t inherit those traits (Kong et al., Reference Kong, Thorleifsson, Frigge, Vilhjalmsson, Young, Thorgeirsson, Benonisdottir, Oddsson, Halldorsson, Masson, Gudbjartsson, Helgason, Bjornsdottir, Thorsteinsdottir and Stefansson2018). For example, parents with a higher genetic propensity for disruptive behavior may be more likely to engage in harsh and less supportive and warm parenting. This harsh parenting behavior may foster the development of disruptive behavior in their children, regardless of the child’s own genetics (Jaffee et al., Reference Jaffee, Belsky, Harrington, Caspi and Moffitt2006).
Another type of gene-environment correlation is evocative rGE. This occurs when an individual’s genetically influenced traits or behavior elicits specific responses from others in the environment (e.g., Burt, Reference Burt2008). For example, a child with a genetic predisposition toward disruptive behavior may frequently exhibit impulsivity or aggression. These behaviors can, in turn, evoke a particular response from parents or caregivers, such as harsher discipline or stricter control. The key here is that the child’s genetically influenced behavior triggers a reaction from the environment, which then reinforces or alters the child’s experiences and development.
In the past, research has been focused on behavioral and molecular genetic models to investigate both parental and child genetics in the association between parenting and, primarily, externalizing child behavior—a broader construct encompassing some parts of disruptive child behavior along with less severe traits, such as attention deficits.
In childhood and adolescence, children’s polygenic scores (PGS) of externalizing behaviors (and, in one study, aggression) predicted externalizing behavior (Kretschmer et al., Reference Kretschmer, Vrijen, Nolte, Wertz and Hartman2022; Kuo et al., Reference Kuo, Poore, Barr, Chirico, Aliev, Bucholz, Chan, Kamarajan, Kramer, McCutcheon, Plawecki and Dick2022; Luo et al., Reference Luo, Pappa, Cecil, Jansen, van IJzendoorn and Kok2021; Teeuw et al., Reference Teeuw, Mota, Klein, Blankenstein, Tielbeek, Jansen, Franke and Hulshof Pol2023) which is an indication of direct genetic effects on externalizing behavior.
Findings for genetic nurture effects have been mixed. In a prospective-longitudinal study on more than 2000 Dutch adolescents, followed between the ages of 11 and 29, a parental PGS of externalizing behavior did not significantly predict family dysfunction (Kretschmer et al., Reference Kretschmer, Vrijen, Nolte, Wertz and Hartman2022). Also, in terms of genetic nurture, parents’ PGS were not associated with adolescents’ externalizing behaviors, while controlling for child PGS (Kretschmer et al., Reference Kretschmer, Vrijen, Nolte, Wertz and Hartman2022). Kuo et al. (Reference Kuo, Poore, Barr, Chirico, Aliev, Bucholz, Chan, Kamarajan, Kramer, McCutcheon, Plawecki and Dick2022) studied an at-risk sample of around 1000 12–17 year old US adolescents and found a significant association between parental PGS of externalizing behavior and lower parent-child closeness, but not parental involvement. Also, their study showed that parents’ PGS did predict adolescent externalizing behavior. In a study using data from a cohort study with more than 30,000 mother-father-child trios, genetic transmission effects (i.e. genetic material inherited from parents) but not genetic nurture effects were found for conduct problems in children between 8 and 14 years old (Frach et al., Reference Frach, Barkhuizen, Allegrini, Ask, Hannigan, Corfield, Andreassen, Dudbridge, Ystrom, Havdahl and Pingault2024).
A number of studies utilizing parent–offspring adoption designs (i.e., comparing associations between adopted children and their biological versus adoptive parents to disentangle genetic and environmental influences) found support for evocative rGE wherein adolescent aggression, adolescent externalizing behaviors or child impulsivity predicted negative parenting behaviors (Marceau et al., Reference Marceau, Horwitz, Narusyte, Ganiban, Spotts, Reiss and Neiderhiser2013; Narusyte et al., Reference Narusyte, Andershed, Neiderhiser and Lichtenstein2007, Reference Narusyte, Neiderhiser, Andershed, D’Onofrio, Reiss, Spotts, Ganiban and Lichtenstein2011; Sellers et al., Reference Sellers, Harold, Thapar, Neiderhiser, Ganiban, Reiss, Shaw, Natsuaki and Leve2020). Studies using molecular genetic methods also have found support for evocative rGE: Kretschmer and colleagues (2022) found that adolescents’ PGS of externalizing behaviors were associated with more family dysfunction via adolescents’ externalizing behavior—evidence for an evocative gene-environment correlation (Kretschmer et al., Reference Kretschmer, Vrijen, Nolte, Wertz and Hartman2022).
Notably, almost all studies focused on adolescents and externalizing behavior; there is less knowledge about the role of gene-environment correlations in disruptive behavior of young children.
The present study
In the present cross-sectional study, we investigated the association between (1) genetic variants associated with disruptive behavior in one parent and the parenting they provide and (2) genetic variants associated with disruptive behavior in children and the parenting they experience (evocative rGE). We also investigated genetic nurture (3), in which parental genotype predicts children’s disruptive behavior over and above genetic transmission, via parenting behavior that in turn predicts children’s disruptive behavior.
Our polygenic score of disruptive behavior (PGS-DB) was based on a recent GWAS of 13 externalizing phenotypes (Baselmans et al., Reference Baselmans, Hammerschlag, Noordijk, Ip, van der Zee, de Geus, Abdellaoui, Treur and van’t Ent2021). We selected the meta-analyzed phenotype GWAS data for disruptive behavior including summary statistics of six phenotypes; aggression, angry outbursts, extreme irritability, irritability, irritable for two days, and attention deficit/hyperactivity disorder (ADHD), to achieve a close match between polygenic score and phenotype (which included the intensity of oppositional behavior, aggressive behavior, attention problems or hyperactivity and conduct problems). Genetic data of disruptive behavior from this GWAS consists of an effective sample size of 523,150 participants of several ages, ranging from early to middle childhood to middle and old age. To better compare our results to previous research, as supplementary analysis, we also computed a polygenic score of externalizing behaviors based on a GWAS that covered the following seven phenotypes: attention-deficit/hyperactivity disorder, problematic alcohol use, lifetime cannabis use, reverse-coded age at first sexual intercourse, number of sexual partners, general risk tolerance and lifetime smoking initiation (Karlsson Linnér et al., Reference Karlsson Linnér, Mallard, Barr, Sanchez-Roige, Madole, Driver, Poore, de Vlaming, Grotzinger, Tielbeek, Johnson, Liu, Rosenthal, Ideker, Zhou, Kember, Pasman, Verweij, Liu and Dick2021). This polygenic score has been used in previous research (e.g., Kretschmer et al., Reference Kretschmer, Vrijen, Nolte, Wertz and Hartman2022) but encompasses broader phenotypes which are not yet seen in young children.
We expected to find (1) a correlation between parenting behavior (i.e., harsh and warm-supportive) and disruptive child behavior, (2) a correlation between children’s PGS-DB and child disruptive behavior, (3) gene-environment correlations indicated by significant association between parents’ PGS-DB and the parenting they provide and from children’s PGS-DB to the parenting they receive (evocative rGE), (4) genetic nurture, indicated by significant direct association between parents’ PGS-DB and disruptive child behavior—while controlling for genetic transmission from parent to child (4a), or the link between parents’ PGS-DB and their children’s disruptive behavior environmentally mediated by parenting (4b).
Methods
Participants
This study used cross-sectional baseline data of the larger ORCHIDS project (Chhangur et al., Reference Chhangur, Weeland, Overbeek, Matthys and de Castro2012), a randomized controlled trial (RCT) of the Incredible Years parenting program. Others have described the intervention and its effects in full (Chhangur et al., Reference Chhangur, Weeland, Overbeek, Matthys and de Castro2012; Overbeek et al., Reference Overbeek, Van Aar, De Castro, Matthys, Weeland, Chhangur and Leijten2021; Weeland et al., Reference Weeland, Chhangur, van der Giessen, Matthys, de Castro and Overbeek2017). The original RCT was preregistered with the Netherlands Trial Register (#3594, ORCHIDS, www.trialregister.nl) and was approved by a relevant medical-ethical review board (METC, #11-320/K). All participants provided written informed consent. Families (i.e., one parent and their child) were recruited in two cohorts for logistical reasons (September-October 2012 and 2013) based on a screening for above average disruptive behavior (>75th percentile on the Eyberg Child Behavior Inventory, ECBI, Eyberg & Pincus, Reference Eyberg and Pincus1999; Weeland et al., Reference Weeland, van Aar and Overbeek2018). Exclusion criteria were intellectual disability of the parent and/or child (IQ ≤ 70) and not mastering the Dutch language. Half of the parents were randomly assigned to receive the intervention, however intervention effects did not bias the current analysis which was based on pretest data. Out of the 387 families in the original sample, 296 (77%) parents agreed to provide saliva samples at 2.5 years follow-up for DNA extraction, and a total of 288 samples of parents and children passed quality controls (see Figure S1). Parents (92% mothers) were between 27.07 and 49.27 years old (M = 38.10, SD = 4.84). Based on reported birth countries of all four grandparents, 75.3% of children had all grandparents born in European countries. The remaining children had grandparents born in Asian (1.4%), African (2.1%), and South American (2.1%) countries, or a mix of these regions (19.1%). Although we did not collect information on race, ancestry or ethnicity, genetic research demonstrated that the Dutch population is predominantly of European descent (Byrne et al., Reference Byrne, van Rheenen, Veldink and McLaughlin2020). Most fathers (91%) and mothers (93%) were born in the Netherlands. Children (48% girls) were between 3.61 and 8.61 years old (M = 6.26, SD = 1.31) and the majority was born in the Netherlands (97%). Among the parents, 53.31% completed university or college degree, 42.16% completed secondary education and/or vocational education), 3.82% completed primary education, and 0.70% gave ambiguous or other answers (e.g., “I don’t know”). Families included in the current study did not significantly differ from families that were not included in terms of sex of the child, sex of the parent, age of the child, age of the parent, harsh parenting, warm-supportive parenting, and disruptive child behavior (see Table S1).
Measures
Eyberg child behavior inventory
The intensity of disruptive child behavior was reported by parents with a Dutch translation of the Intensity subscale of the Eyberg Child Behavior Inventory (ECBI; Eyberg & Pincus, Reference Eyberg and Pincus1999; Weeland et al., Reference Weeland, van Aar and Overbeek2018). Parents reported the frequency of disruptive behavior by using a 7-point Likert scale (1 = never, 7 = always) on a total of 36 items (e.g., “Does not obey house rules,” “Has temper tantrums,” and “Whines”). The ECBI has sufficient psychometric properties, is widely used internationally to assess the effectiveness of caregiver-training programs, and accurately discriminates between children with and without conduct-related disorders (Abrahamse et al., Reference Abrahamse, Junger, Leijten, Lindeboom, Boer and Lindauer2015; Leijten et al., Reference Leijten, Gardner, Landau, Harris, Mann, Hutchings, Beecham, Bonin and Scott2017; Rich & Eyberg, Reference Rich and Eyberg2001). Internal consistency was α = .85. As recommended, we computed a sum score (Eyberg & Pincus, Reference Eyberg and Pincus1999).
Parenting practices
Parenting practices were measured using the Parent Practice Inventory (PPI; Webster-Stratton et al., Reference Webster-Stratton, Reid and Hammond2001). This self-report consists of several sections, each including multiple items (7-point Likert scale: 1 = not likely at all/never, 7 = likely/always) related to parents’ frequency of responses to parenting situations . To assess warm-supportive parenting we used the praise and incentive dimensions scale (11 items, e.g., “Giving a hug or compliment”) combined with the positive verbal scale (9 items, e.g., “In an average week, how often do you praise or reward your child for doing a good job at home or school?”). To assess harsh parenting, we used the harsh and inconsistent discipline scale (15 items, e.g., “Threatening but not punishing”) combined with the punishment scale (6 items, e.g., “Slapping or hitting when misbehavior occurs”). The internal consistency of both dimensions, i.e., harsh parenting and warm-supportive parenting, was α = .75 and α = .72, respectively.
Genotyping and computation
Saliva was collected via passive drool with the Oragene-DNA OG-600 container (DNA Genotek, Canada) following manufacturer’s instructions and stored at room temperature. Genomic DNA was extracted from saliva and bisulfite converted with the ZYmo EZ DNA methylation kit (Zymo research, Irvine, CA, USA). Genotyping was performed on saliva samples using the Infinium iSelect GSA array (Illumina, San Diego CA, USA). Genotypes were called using Illumina’s GenomeStudio software and quality control was performed using PLINK v1.90b6.17 (Purcell et al., Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender, Maller, Sklar, de Bakker, Daly and Sham2007). Ten principal components (PCs) were estimated separately for the parent and the child in PLINK. PCs were estimated separately for parents and children, as these groups were genotyped and imputed independently. This approach ensured proper control for population stratification within each sample. Participants were excluded if genetic quality controls failed i.e., (1) the sample call rate for this person was low (< 95%) (n child = 2 and n parent = 0), (2) the parent-child genetic relatedness did not match the expected familial relation (proportion of IBD in PLINK >0.2; n child = 0 and n parent = 0), (3) there was a phenotype-genotype gender mismatch (n child = 2 and n parent = 5), or (4) a person was identified as an ethnic outlier based on a principal component analysis to account for population stratification (n child = 11 and n parent = 10). There were 10 children and 9 parents for whom the heterozygosity value was ± 3 SD from the mean. We decided not to remove them because the values were strongly shifted to the mean (i.e., high median) in such a way that it was easy to fail this criterium. Besides, none of the heterozygosity rates were above .04, indicating no sign of low sample quality.
Before imputation, single nucleotide polymorphisms (SNPs) were removed when (1) they had a low call rate (i.e., genotype missingness was <95%) or, in other words, SNPs that are missing in a large proportion of individuals (n child = 10,623 SNPs and n parent = 6,220 SNPs), (2) minor allele frequency (MAF) was low (< 5%) (n child = 407,136 and n parent = 409,292 SNPs), or (3) if they were not in Hardy-Weinberg equilibrium (p<1e-6, n child = 421 SNPs and n parent = 84). To impute the genotypes, we made use of the Michigan Imputation Server (Das et al., Reference Das, Forer, Schönherr, Sidore, Locke, Kwong, Vrieze, Chew, Levy, McGue, Schlessinger, Stambolian, Loh, Iacono, Swaroop, Scott, Cucca, Kronenberg, Boehnke and Fuchsberger2016) and imputed the genotypes using the HapMap2, build GRCh37/hg19 as reference panel. Monomorphic SNPs (with MAF <0.1%) and SNPs with low imputation quality (R 2 <0.3) were excluded, resulting in 2,377,699 SNPs for the children and 1,720,570 for the parents.
Polygenic score computation
We computed two polygenic scores. First, a polygenic score for disruptive behavior (PGS-DB) was based on the most recent meta-analytic genome-wide association study (GWAS) summary statistics based on six disruptive behavior phenotypes in 523,150 individuals (Baselmans et al., Reference Baselmans, Hammerschlag, Noordijk, Ip, van der Zee, de Geus, Abdellaoui, Treur and van’t Ent2021). Phenotypes from the original studies were aggression, angry outbursts, extreme irritability, irritability, irritable for two days, and ADHD. Participants ranged from early childhood to old age. Second, for the supplementary analysis, we used a polygenic score of externalizing behaviors (PGS-EXT) based on multivariate genomic analyses of the following seven phenotypes: attention-deficit/hyperactivity disorder, problematic alcohol use, lifetime cannabis use, reverse-coded age at first sexual intercourse, number of sexual partners, general risk tolerance and lifetime smoking initiation (Karlsson Linnér et al., Reference Karlsson Linnér, Mallard, Barr, Sanchez-Roige, Madole, Driver, Poore, de Vlaming, Grotzinger, Tielbeek, Johnson, Liu, Rosenthal, Ideker, Zhou, Kember, Pasman, Verweij, Liu and Dick2021). Given that these discovery samples were predominantly of European ancestry, we acknowledge the potential for mismatch in genetic ancestry between the discovery samples and our target population, which may include, albeit few, individuals of diverse ancestries. This mismatch may affect the predictive power and generalizability of the polygenic scores for individuals of non-European ancestry. We used LDpred2 to compute the polygenic scores. This method utilizes GWAS summary statistics and LD information from an external LD reference sample (Hapmap3 with independent LD blocks) to calculate the posterior mean effect size of each SNP (Privé et al., Reference Privé, Arbel and Vilhjálmsson2020). We performed standard quality control procedures on the summary statistics following the tutorial of Choi et al. (Reference Choi, Mak and O’Reilly2020).
Statistical analyses
We tested our hypotheses with SEM in R (R Core Team, 2022) using the lavaan package (Rosseel, Reference Rosseel2012). We included a covariance between harsh parenting and warm-supportive parenting because those constructs were expected to be highly correlated. Because the model was saturated—the number of data points equaled the number of estimated parameters—evaluation of model fit was uninformative. Before analyzing, the data were screened to verify the statistical assumptions of structural equation modeling (SEM). Skewness and kurtosis of the variable residuals were examined and showed univariate and multivariate normality (Tabachnick & Fidell, Reference Tabachnick and Fidell2006). Furthermore, we controlled for 10 genetic principal components (PC). Because of the relatively small sample size and relatively large model, we regressed out the effects of the PCs and used the residuals in subsequent analyses. We tested whether potential covariates (parental education, age and sex and child age and sex) were significantly associated with the outcome variable and included significant covariates (i.e., child age and sex, see Table S2). Variables were not standardized beforehand, instead, we reported standardized estimates. We used full information maximum likelihood estimation to handle missing data because data were missing completely at random (MCAR, χ2 (18) = 12.16, p = .839). Path coefficients were assumed to be significantly different from zero if the confidence intervals (CIs) around them did not include 0. Indirect effects were tested using bootstrapping (i.e., 5,000 bootstrap resamples) to obtain robust estimates of the indirect effects and their confidence intervals, as recommended (Preacher & Hayes, Reference Preacher and Hayes2008). We computed indirect effects by multiplying the path coefficients of the direct effects, and assessed significance using the percentile method. Model fit was good (χ2 (9) = 8.06, p = .528, CFI = 1.00, TLI = 1.02, RMSEA <0.001).
Results
Table 1 displays all univariate means and standard deviations and bivariate correlations for each variable. Bivariate correlations indicated significant, albeit small, positive associations between (1) disruptive child behavior and children’s own PGS-DB (r = .13) and (2) disruptive child behavior and harsh parenting (r = .21), and, as expected, (3) a small negative association between warm-supportive parenting and harsh parenting (r = −.15), and (4) a strong positive association between children’s polygenic score for disruptive behavior and parents’ polygenic score for disruptive behavior (r = .50).
Table 1. Descriptive statistics of and correlations between the polygenic scores, characteristics, parenting behaviors, and disruptive child behavior
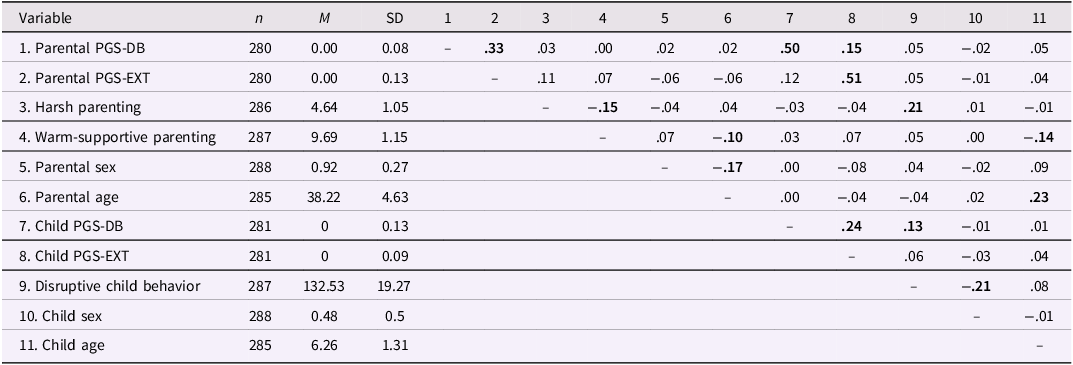
Note. PGS-DB = polygenic score of disruptive behavior; PGS-EXT = polygenic score of externalizing behavior; bold estimates are significant (p < .05).
Harsh parenting
Path coefficients are shown in Table 2 and Figure 1. The model explained 12.5% of the variance in disruptive child behavior. Harsh parenting was, as we expected, positively associated with disruptive child behavior, indicating that harsh parents were more likely to have children with higher levels of disruptive behavior (β = 0.23, 95% CI [0.13, 0.32], p < .001). Also as expected, children’s PGS-DB was positively associated with their disruptive behavior (β = 0.15, 95% CI [0.01, 0.29], p = .031), and parents’ PGS-DB was directly correlated with children’s PGS-DB (β = 0.50, 95% CI [0.42, 0.59], p < .001).
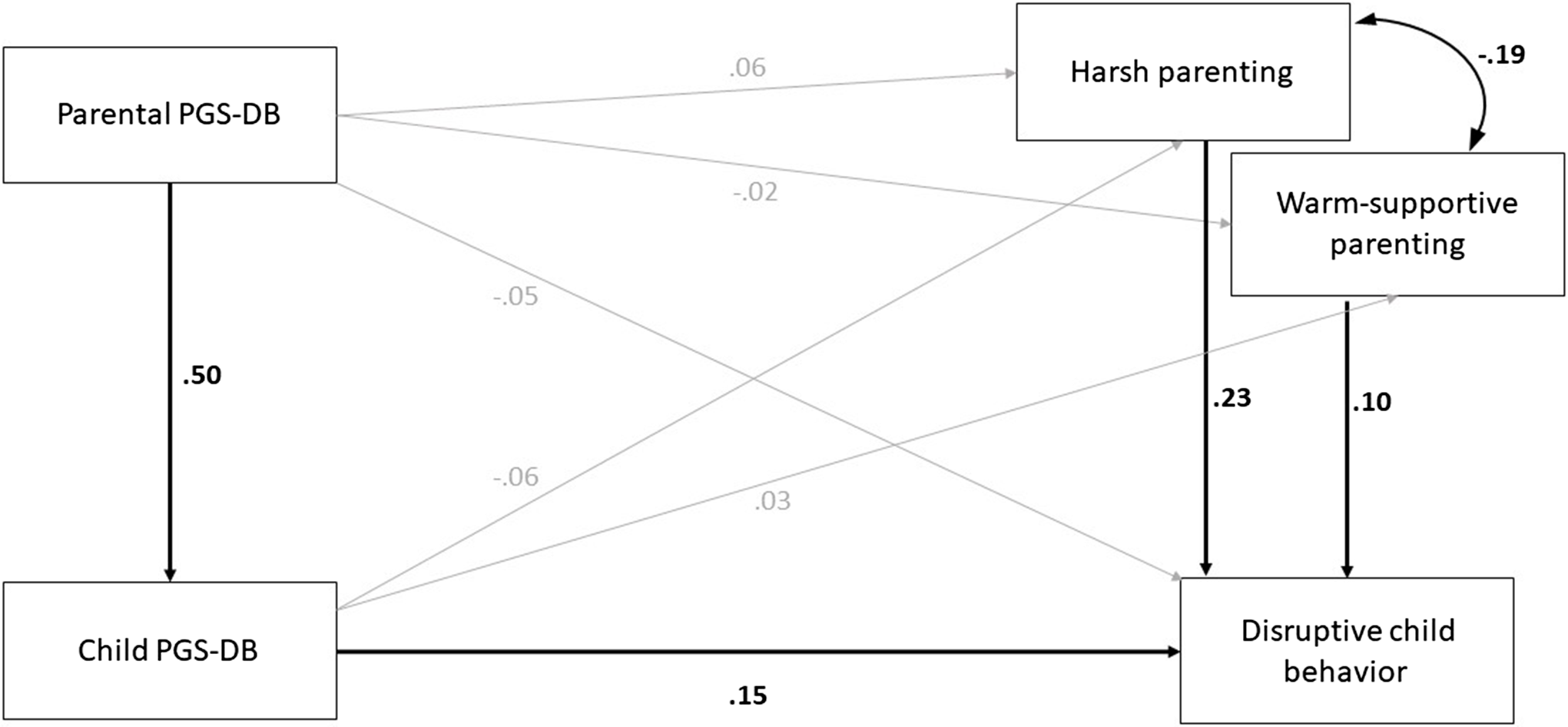
Figure 1. Estimated model of gene-environment correlation, genetic confounding, and genetic nurture for disruptive child behaviors. Note. standardized regression coefficients are presented. Significant estimates are in bold and have bold arrows. PGS-DB = polygenic score of disruptive behaviors.
Table 2. Unstandardized and standardized direct and indirect effects using a polygenic score of disruptive behaviors
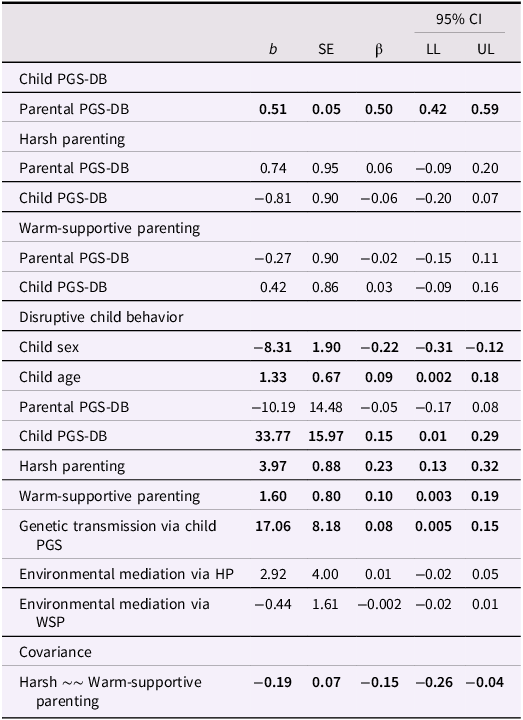
Note. CI = confidence interval; LL = lower limit; UL = upper limit; PGS-DB = polygenic score of disruptive behaviors. HP = Harsh parenting; WSP = Warm-supportive parenting; Significant estimates (p < .05) are in bold.
We did not find evidence for evocative gene-environment correlations, as the path from children’s PGS-DB to harsh parenting (β = −0.06, 95% CI [−0.20, 0.07], p = .367) was non-significant. To investigate the genetic nurture of disruptive child behavior, we assessed whether parents’ PGS-DB predicted disruptive child behavior, controlling for children’s PGS-DB, but the result was not significant (β = −0.05, 95% CI [−0.17, 0.08], p = .482). Note that the sample size was small and confidence intervals were large preventing strong interpretation of these null results.
We also assessed the two indirect pathways from PGS-DB of parents to disruptive child behavior. Children’s PGS-DB partially mediated the association between parents’ PGS-DB and children’s disruptive behavior (β = 0.08, 95% CI [0.005, 0.15], p = .036). However, we did not find evidence for an environmentally mediated effect of parents’ PGS-DB on disruptive child behavior via harsh parenting (β = 0.01, 95% CI [−0.02, 0.05], p = .455).
Warm-supportive parenting
Surprisingly, warmer, more supportive parenting was associated with more disruptive child behavior (β = 0.10, 95% CI [0.003, 0.19], p = .042). Children did not evoke warmer or more supportive parenting based on their own PGS-DB (β = 0.03, 95% CI [−0.09, 0.16], p = .622), and parents did not engage in less warm or supportive parenting based on their PGS-DB (β = −0.02, 95% CI [−0.15, 0.11], p = .763). Finally, we found no evidence for an environmentally mediated effect from parents’ PGS-DB to disruptive child behavior via warm-supportive parenting β = −0.002, 95% CI [−0.02, 0.01], p = .766).
Sensitivity analyses
We repeated our main analyses including a polygenic score of the broader construct externalizing behavior (i.e., PGS-EXT, see Table S3). Results were the same for associations of warm-supportive parenting with polygenic scores and disruptive behavior. Results for association of harsh parenting were similar to the main analysis with a few exceptions (see supplemental materials). Briefly, in this model children’s PGS-EXT did not predict disruptive child behavior (β = 0.07, 95% CI [−0.07, 0.20], p = .328). We did find that higher PGS-EXT scores of parents were associated with more harsh parenting (β = 0.18, 95% CI [0.03, 0.33], p = .016). In this model, we found a significant, albeit very small, genetic nurture —i.e., environmentally mediated —effect of parent’s PGS-EXT on disruptive child behavior via harsh parenting (β = 0.04, 95% CI [0.002, 0.08], p = .041, see Figure S2).
Additionally, we repeated our main analyses using the polygenic score of externalizing behavior for the parent and the polygenic score of disruptive behavior for the child (see Table S4). We did this because the PGS-EXT was constructed in an adult-only sample and the PGS-DB was based on a more developmentally appropriate GWAS. Results were similar with a few exceptions: parent’s PGS-EXT was not associated with children’s PGS-DB. Consequently, the genetic transmission via the PGS of the child was also not significant anymore. Although externalizing and disruptive behaviors are correlated constructs, they do not necessarily share identical genetic determinants which may be reflected in the lack of significant association between the PGS-EXT and PGS-DB.
Discussion
In the present study, we aimed to advance our understanding of gene-environment interplay in disruptive child behavior. Using a polygenic score of disruptive behavior (PGS-DB), we found that parents who report harsher (and more warm-supportive) parenting behavior also report having a child with more disruptive behavior. Moreover, a higher child PGS-DB was associated with more disruptive behavior, consistent with previous research (Kretschmer et al., Reference Kretschmer, Vrijen, Nolte, Wertz and Hartman2022; Kuo et al., Reference Kuo, Poore, Barr, Chirico, Aliev, Bucholz, Chan, Kamarajan, Kramer, McCutcheon, Plawecki and Dick2022; Luo et al., Reference Luo, Pappa, Cecil, Jansen, van IJzendoorn and Kok2021; Teeuw et al., Reference Teeuw, Mota, Klein, Blankenstein, Tielbeek, Jansen, Franke and Hulshof Pol2023). However, using the PGS-DB, no evidence emerged for gene-environment correlations: associations between children’s PGS-DB and parenting, parents’ PGS-DB and parenting, and parents’ PGS-DB and disruptive behavior were not significant.
Interestingly, in a sensitivity analysis using a polygenic score of broader externalizing behavior (PGS-EXT), we found no direct genetic effect (i.e., child genotype predicting phenotypic behavior while controlling for parent genotype), but did find a genetic nurture effect: parent’s higher PGS-EXT predicted more harsh parenting which in turn predicted more disruptive child behavior.
A noteworthy finding was that harsh parenting was associated with disruptive child behavior, while more warm-supportive parenting was associated with disruptive child behavior as well. Disruptive behavior may not be improved by warm-supportive parenting in the presence of harsh parenting or by warm-supportive parenting alone, but in combination with structured, consistent (positive) discipline: A short parenting intervention, in which parents were taught sensitive discipline and conduct problems decreased after the intervention, provides support for this speculation (Runze et al., Reference Runze, Pappa, Van IJzendoorn and Bakermans-Kranenburg2022). Additionally, the association between harsh parenting and disruptive behavior may reflect potential bidirectional effects over time. For example, harsh parenting may–in the long term–exacerbate disruptive behavior, while disruptive behavior may simultaneously provoke harsher parenting responses in the future. This aligns with previous findings of evocative rGE (Kretschmer et al., Reference Kretschmer, Vrijen, Nolte, Wertz and Hartman2022; Marceau et al., Reference Marceau, Horwitz, Narusyte, Ganiban, Spotts, Reiss and Neiderhiser2013; Narusyte et al., Reference Narusyte, Andershed, Neiderhiser and Lichtenstein2007, Reference Narusyte, Neiderhiser, Andershed, D’Onofrio, Reiss, Spotts, Ganiban and Lichtenstein2011; Sellers et al., Reference Sellers, Harold, Thapar, Neiderhiser, Ganiban, Reiss, Shaw, Natsuaki and Leve2020). Finally, it may be the case that in this sample with relatively many families whose children showed mildly elevated—not always clinical-level—disruptive behavior, parents may have upregulated not only harsh but also supportive behaviors. Due to our cross-sectional design, we were unable to shed light on longitudinal, bidirectional effects – an avenue for future research.
The genetic nurture effects revealed in the supplementary analysis is partially consistent with the broader literature. For example, Kuo et al. (Reference Kuo, Poore, Barr, Chirico, Aliev, Bucholz, Chan, Kamarajan, Kramer, McCutcheon, Plawecki and Dick2022) found a similar genetic nurture effect using parental externalizing behaviors instead of parenting behavior. In none of the previously discussed studies, however, a genetic nurture effect with parenting was found (Frach et al., Reference Frach, Barkhuizen, Allegrini, Ask, Hannigan, Corfield, Andreassen, Dudbridge, Ystrom, Havdahl and Pingault2024; Kretschmer et al., Reference Kretschmer, Vrijen, Nolte, Wertz and Hartman2022; Kuo et al., Reference Kuo, Poore, Barr, Chirico, Aliev, Bucholz, Chan, Kamarajan, Kramer, McCutcheon, Plawecki and Dick2022). This may be explained, in part, by different choices of measurement across studies. For instance, Kretschmer et al. (Reference Kretschmer, Vrijen, Nolte, Wertz and Hartman2022) used a broad measure of family dysfunction and Kuo et al. (Reference Kuo, Poore, Barr, Chirico, Aliev, Bucholz, Chan, Kamarajan, Kramer, McCutcheon, Plawecki and Dick2022) assessed constructs such as involvement that focused, for example, on whether the parent would help with schoolwork). These constructs might not fully capture the nature of harsh or warm-supportive parenting.
The discrepancy between results using the PGS-DB and the PGS-EXT may be due to the phenotypes and age groups in the GWAS of the summary statistics that the PGS were constructed from. While the PGS-DB quite closely matched the phenotype under study (i.e., disruptive child behavior) and included childhood samples in its associated GWAS, the PGS-EXT included a broader set of phenotypes encompassing problematic alcohol use, number of sexual partners and lifetime smoking initiation which are less relevant in a child sample. This may explain why in the PGS-EXT model, the link between parental genotype and parenting emerged—as the phenotypes underlying the PGS-EXT were more developmentally appropriate for adults than the PGS-DB. Similarly, this may explain why the link between child genotype and disruptive child behavior did emerge in the PGS-DB model, but not in the PGS-EXT model. Furthermore, the phenotypic composition of the PGS-DB included both aggressive and regulatory components (e.g., ADHD), making it a broader measure than one purely capturing disruptive behavior as defined by the DSM-5. This may also have contributed to differences in findings between the two PGS models. Future research could further disentangle these dimensions by examining polygenic influences on more narrowly defined components of disruptive behavior.
Several limitations of our study warrant mentioning. Overall, the ability to detect a genetic effect, let alone complex gene-environment interplay effects, was still relatively limited in the present study due to the small sample size and subsequent low statistical power. Future studies may increase power by either collecting larger samples or by pooling data from several, comparable, studies. Another limitation is that we collected genetic data from only one parent (mostly the mother). This could bias our estimates of both direct genetic effects and genetic nurture effects (Tubbs et al., Reference Tubbs, Zhang and Sham2020). Specifically, using only one parent’s genotype underestimates the heritability by capturing only half of the child’s genetic variance. The unmeasured genetic contribution of the other parent is not accounted for, which may result in an incomplete estimate of the parental PGS on the child PGS. Moreover, the direct genetic pathway from the child’s PGS to disruptive child behavior may be biased as the unmeasured parent’s genotype could influence the child’s PGS-DB indirectly (via genetic nurture) or through genetic transmission not accounted for in the model. The pathway between the parent’s PGS and parenting behavior could also be biased if the unmeasured parent’s genotype is also associated with parenting behaviors. Given that couples often exhibit assortative mating (i.e., they are genetically more similar than expected by chance), the absence of one parent’s genotype may lead to an inaccurate estimate of genetic nurture effects through harsh parenting. Overall, the absence of (mostly) paternal genotypic data limits our ability to comprehensively disentangle genetic nurture from direct genetic effects, potentially underestimating the interplay between parental genotypes and parenting behaviors. Additionally, adjusting for population stratification using principal components across the full sample rather than within ancestry groups may have introduced residual confounding due to population structure.
Also, we had to rely on parent self-report measures for both parenting and child behavior which may have introduced common-method variance or other response biases (Runze & Van IJzendoorn, Reference Runze and Van IJzendoorn2024). Future studies should incorporate multi-informant approaches to minimize potential biases. While previous research has explored gene-environment interplay effects in families with older children or adolescents, our study provides valuable novel insight into the gene-environment interplay by adding to the scarce knowledge to date regarding the role of genetic predisposition specifically disruptive child behavior in young children using a GWAS based on appropriate phenotypes. Moreover, our sample was comprised of children with higher disruptive behavior problems compared to – often used – normative population samples. Future research should invest in testing the direct and indirect effects of genetic predisposition for other phenotypes on disruptive child behavior, and the potential mediating role of parenting, with models that contain genetic predisposition for both parents. Additionally, it should be established whether PGS that are instead derived from GWAS that already partition indirect and direct genetic effects may have better power to predict genetic nurture effects.
Conclusion
We found associations between children’s early environment (i.e., parenting behavior) and their genetic predisposition on disruptive child behavior, as well as genetic nurture effects when using a polygenic score of externalizing behaviors. However, this depended on whether genetic predisposition was based on a more specific score for disruptive behaviors seen in children or a broader externalizing score. Our findings emphasize the need to include both genetic and environmental data to provide a complete understanding of pathways that shape complex traits such as disruptive child behavior and parenting behavior. It is therefore important to replicate this study with multi-informant, multi-method assessments, trio genetic data, and various PGS for various behavior-related phenotypes.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0954579425100254
Data, Materials and/or Code availability
The data, materials and code that support the findings of this study are available from the corresponding author, upon reasonable request.
Acknowledgements
We thank the families participating in this research. The data necessary to reproduce the analyses presented here are not publicly accessible. Analytic code and materials necessary to reproduce the analyses presented in this paper are not publicly accessible. All data, code and materials are available from the first author. The analyses presented here were not preregistered.
Author contributions
Conceptualization: MN, NC, GO; Methodology: MN, NC, JR; Formal analysis: MN, NC, JR; Resources: GO; Data curation: NC, JR; Writing – Original draft: MN, NC, JR, GO; Writing – Review & Editing: JR, MN, NC, GO; Visualization: MN, JR; Supervision: NC, GO; Project administration: NC, GO; Funding acquisition: GO.
Funding statements
This work was supported by a Vici grant (016.Vici.185.063) awarded to G. Overbeek by the Dutch Science Council (NWO).
Competing interests
The authors declare none.





