Introduction
The freshwater systems of southern Africa hold highly diverse fish communities (Skelton Reference Skelton2024); however, the role of these fishes as intermediate and definitive hosts to trematodes remains poorly studied (Scholz et al. Reference Scholz, Vanhove, Smit, Jayasundera and Gelnar2018). Knowledge is especially depauperate regarding heteroxenous (i.e., with multiple-host life cycles) parasitic species. Freshwater fishes play an essential ecological role in freshwater systems, supporting trophic and ecological diversity and, as such, ensure links within food webs and interactions across trophic levels (Dunne et al. Reference Dunne, Lafferty, Dobson, Hechinger, Kuris, Martinez, McLaughlin, Mouritsen, Poulin, Reise, Stouffer, Thieltges, Williams and Zander2013; Poulin Reference Poulin2014). These interactions provide an integral platform for life cycle completion for organisms such as digenetic trematodes, of which several representatives utilise molluscs as first intermediate hosts, fishes as second intermediate (or definitive) hosts, and piscivorous birds as definitive hosts. Records of adult trematodes are sparse in southern African freshwater fishes, with only a handful of species known from the region (Beverley-Burton Reference Beverley-Burton1962; Bray and Hendrix Reference Bray and Hendrix2007; Curran et al. Reference Curran, Dutton, Warren, Du Preez and Bullard2021; Douëllou Reference Douëllou1992b; Dumbo et al. Reference Dumbo, Dos Santos and Avenant-Oldewage2019a; Dumbo et al. Reference Dumbo, Dos Santos and Avenant-Oldewage2019b; Jansen van Rensburg et al. Reference Jansen van Rensburg, van As and King2013; Kudlai et al. Reference Kudlai, Scholz, Smit, Scholz, Vanhove, Smit, Jayasundera and Gelnar2018; Warren et al. Reference Warren, Jacobs, Dutton, Netherlands, Du Preez and Bullard2024). In South Africa, adult trematodes are represented by a mere five species across the families Gorgoderidae (two species), Allocreadiidae, Cephalogonimidae, and Orientocreadiidae (one each) (Boomker Reference Boomker1984; Dos Santos et al. Reference Dos Santos, Gilbert, Avenant-Oldewage and Dumbo2021; King et al. Reference King, Smit, Baker and Luus-Powell2018; Prudhoe and Hussey Reference Prudhoe and Hussey1977; Truter et al. Reference Truter, Hadfield and Smit2023a; Truter et al. Reference Truter, Hadfield and Smit2023b). The first species to be reported was Phyllodistomum vanderwaali Prudhoe & Hussey, 1977, who described this gorgoderid species from the urinary bladder of North African catfish, Clarias gariepinus (Burchell) (Clariidae) from the Olifants River, Transvaal (now Limpopo Province) (Prudhoe and Hussey Reference Prudhoe and Hussey1977). Boomker (Reference Boomker1984) subsequently described Phyllodistomum bavuri Boomker, 1984 from the same fish host collected from the Bangu River, Kruger National Park. King et al. (Reference King, Smit, Baker and Luus-Powell2018) described Emoleptalea nwanedi King, Smit, Baker & Luus-Powell, 2018 (Cephalogonimidae) from silver catfish, Schilbe intermedius Rüppell (Schilbeidae) from the Nwanedi-Luphephe Dam in Limpopo Province, and Dos Santos et al. (Reference Dos Santos, Gilbert, Avenant-Oldewage and Dumbo2021) described Allocreadium apokryfi Dos Santos, Gilbert, Avenant-Oldewage & Dumbo, 2021 from smallmouth yellowfish, Labeobarbus aeneus (Burchell) (Cyprinidae) from a site downstream of the Vaal Dam, Gauteng Province. Truter et al. (Reference Truter, Hadfield and Smit2023a) reported a species of Orientocreadium Tubangui, 1931 (Orientocreadiidae) from C. gariepinus from two dam and river systems in central South Africa. The species was identified as being Orientocreadium batrachoides Tubangui, 1931; the status of this species in southern Africa is uncertain, identifications (e.g., by Dumbo et al. Reference Dumbo, Dos Santos and Avenant-Oldewage2019b) being made based on morphological similarity and in the absence of molecular sequence data from the type-locality (Philippines).
In contrast to this depauperate adult fauna, the richness of intermediate stages (both first-intermediate stages and metacercariae) is highly diverse. Studies on freshwater first intermediate-stage infections have a long and productive history in southern Africa, with key work predating much of that on adult trematodes (e.g., Cawston Reference Cawston1917; Cawston Reference Cawston1920; Faust Reference Faust1919; Faust Reference Faust1920; Porter Reference Porter1938) and continuing to this day (e.g., Mudavanhu et al. Reference Mudavanhu, Schols, Goossens, Nhiwatiwa, Manyangadze, Brendonck and Huyse2024; Outa and Avenant-Oldewage Reference Outa and Avenant-Oldewage2024a; Outa and Avenant-Oldewage Reference Outa and Avenant-Oldewage2024b; Outa et al. Reference Outa, Bhika and Avenant-Oldewage2024). Foundational work such as that by Porter (Reference Porter1938) has demonstrated that southern African freshwater molluscs harbour a rich fauna of trematode first intermediate stages, though most species have yet to be matched to definitive or other intermediate stages. This knowledge is still deficient in many aspects, with no records of first intermediate stages for highly prevalent families such as clinostomids, cryptogonimids, and diplostomids in the region. Knowledge regarding second intermediate stages in the region is also patchy. To date, records of intermediate stages representing at least 15 putative species across two trematode families (Clinostomidae and Diplostomidae) have been documented from fish hosts in southern Africa (Barson and Avenant-Oldewage Reference Barson and Avenant-Oldewage2006; Barson et al. Reference Barson, Bray, Ollevier and Huyse2008; Grobbelaar et al. Reference Grobbelaar, Van As, Butler and Van As2014; Hoogendoorn et al. Reference Hoogendoorn, Smit and Kudlai2019; Hoogendoorn et al. Reference Hoogendoorn, Smit and Kudlai2020; Madanire-Moyo and Barson Reference Madanire-Moyo and Barson2010; Madanire-Moyo et al. Reference Madanire-Moyo, Luus-Powell and Olivier2010; Madanire-Moyo et al. Reference Madanire-Moyo, Luus-Powell and Olivier2012; Moema et al. Reference Moema, King, Rakgole and Baker2013; Moema et al. Reference Moema, King and Rakgole2019; Olivier et al. Reference Olivier, Luus-Powell and Saayman2009; Smit et al. Reference Smit, Vanhove, Moyo and Luus-Powell2023, see Table 2 for clinostomid records). Most reports of metacercariae from the region lack either morphological or molecular data. Although the use of molecular sequencing to identify and link intermediate stages is increasing in the region (e.g., Hoogendoorn et al. Reference Hoogendoorn, Smit and Kudlai2019; Hoogendoorn et al. Reference Hoogendoorn, Smit and Kudlai2020; Moema et al. Reference Moema, King and Rakgole2019), its utilisation remains limited. Perceived difficulty in identification of underdeveloped stages, with limitations in reliable morphological characters for taxonomic placement, further hinder the linking of life cycle stages to definitive hosts. This has led to a depauperate comparative molecular data repository for species of the region and Africa as a whole. In an effort to address the morphological and molecular data void of the parasite fauna of southern African freshwater fishes, we sought to comprehensively investigate and characterise this fauna. In the process of our investigations, we encountered several types of metacercariae belonging to four trematode families: the Clinostomidae, Cryptogonimidae, Diplostomidae, and Strigeidae. Findings concerning the latter two families will be reported in a separate publication. We herewith report our observations of the former two families from four freshwater fish families in southern Africa.
Materials and methods
Host collection and parasite fixation
Sampling of Clarias gariepinus in Zambia was done in 2019. The present manuscript provides molecular data for Clinostomum brieni (Dollfuss, 1950) previously reported in Truter et al. (Reference Truter, Hadfield and Smit2023a, Reference Truter, Hadfield and Smit2023b). The collection of all other host species, namely Chiloglanis sp., Labeo cylindricus Peters and Marcusenius pongolensis (Fowler) occurred in 2023 as part of a larger aquatic biodiversity project (REFRESH) and parasitological data is reported here for the first time. A list of hosts and sampling localities are presented in Table 1. Methods used for host collection included rod and reel, baited longlines, seine netting, cast netting, and electrofishing. All organs of freshly collected host individuals were screened for trematode infection, including the gills, branchial chambers, eyes, brain, cranial cavity, muscle tissue, and viscera. After removal from the host, free metacercarial stages were rinsed in a 0.9% saline solution, and encysted or encapsulated metacercariae were excysted using fine insect needles and rinsed, and all individuals were heat fixed and stored in 96% molecular grade ethanol.
Table 1. Localities where respective hosts were collected from in South Africa and Zambia
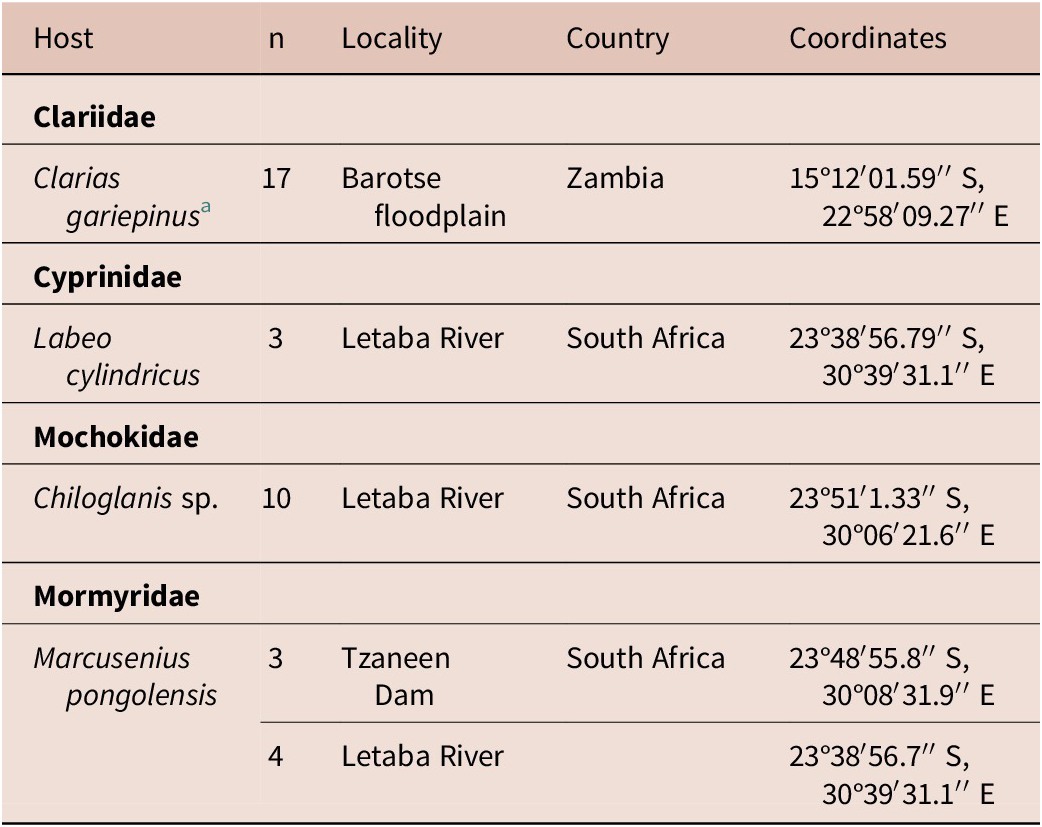
a Same C. gariepinus individuals reported on in Truter et al. (Reference Truter, Hadfield and Smit2023a, Reference Truter, Hadfield and Smit2023b).
Morphological and molecular analyses
The general morphology of whole individuals was initially studied to identify different morphotypes using a Nikon Eclipse Ni (Nikon, Tokyo, Japan) compound microscope equipped with differential interference contrast. Photomicrographs and measurements were obtained using the computerised digital camera system and NIS-Elements BR 4.60© software for image analysis. A selection of individuals representing each morphotype was used to prepare hologenophores (Pleijel et al. Reference Pleijel, Jondelius, Norlinder, Nygren, Oxelman, Schander, Sundberg and Thollesson2008) and permanent mounts stained with either Mayer’s hematoxylin or acetocarmine; permanent mounts were prepared using standard protocols for each respective stain (Georgiev et al. Reference Georgiev, Biserkov and Genov1986; Yong et al. Reference Yong, Cribb and Cutmore2021). Photomicrographs of all representatives not prepared as hologenophores were used as photohologenophores (Achatz et al. Reference Achatz, Martens, Kudlai, Junker, Boe and Tkach2022) and whole specimens were subsequently molecularly analysed. Hologenophores and other vouchers were deposited in the parasitological collection of the National Museum, Bloemfontein, South Africa (NMB). Photohologenophore records were stored in the electronic parasitological collection of the Water Research Group, North-West University, South Africa. Measurements are given in micrometres (μm) unless stated otherwise, with means following ranges in parentheses.
Genomic DNA from whole specimens (photohologenophores) and hologenophores were extracted using the PCRBIO Rapid Extract PCR Kit (PCRBiosystems, Analytical Solutions, Randburg, South Africa). Buffer volume adjustments were as follows for the extraction reaction: 10 μl 5× PCRBIO Rapid Extract Buffer A, 5 μl 10× PCRBIO Rapid Extract Buffer B, and 70 μl of PCR grade water to a final volume of 85 μl. Final reaction dilution after incubation, as per manufacturer instruction, was done with 200 μl PCR grade water instead of 900 μl to obtain DNA at a higher concentration. The Polymerase Chain Reactions (PCR) for all three gene regions, 28S rDNA, ITS1-5.8S-ITS2 (ITS1–2), and cytochrome oxidase I (COI) mtDNA were adapted in volume and in the thermal cycling profiles compared to the reference literature and are provided below. The PCR reactions for the partial 28S rDNA and ITS1–2 regions were made in a final volume of 25 μl, consisting of 12.5 μl DreamTaq PCR Master Mix (2×) (Thermo Fisher Scientific, Waltham, Massachusetts, USA), 1.25 μl of each primer (10 μM), 7–8 μl PCR grade water, and 2 μl and 3 μl of DNA supernatant, respectively. While the final reaction volume for the COI region was performed at a final volume of 20 μl with 4 μl of DNA supernatant. Amplification of the 28S rDNA region was performed using the primer set Digl2 (5’- AAG CAT ATC ACT AAG CGG -3’) (Tkach et al. Reference Tkach, Snyder and Swiderski2001) and 1500R (5’- GCT ATC CTG AGG GAA ACT TCG -3’) (Snyder and Tkach Reference Snyder and Tkach2001). The thermal cycling profile was as follows: initial denaturation 95°C for 5 min, 40 cycles of amplification at 95°C for 30 sec, 55°C for 30 sec, 72°C for 2 min and final extension at 72°C for 7 min. Primers D1F (5’- AGG AAT TCC TGG TAA GTG CAA G -3’) and D2R (5’- CGT TAC TGA GG GAA TCC TGG T -3’) (Galazzo et al. Reference Galazzo, Dayanandan, Marcogliese and McLaughlin2002) were used for the ITS1–2 rDNA region. Thermal cycling conditions were as follows: initial denaturation 95°C for 3 min, 40 cycles of amplification at 94°C for 1 min, 56°C for 1 min, 72°C for 2 min, and final extension at 72°C for 5 min. The following primer sets were used for the COI mtDNA region: Dice1F (5’- ATT AAC CCT CAC TAA ATT WCN TTR GAT CAT AAG -3’) and Dice14R (5’- TAA TAC GAC TCA CTA TAC CHA CMR TAA ACA TAT GAT G -3’) (Van Steenkiste et al. Reference Van Steenkiste, Locke, Castelin, Marcogliese and Abbott2015), or MPlatCOX1dF (5’- TGT AAA ACG ACG GCC AGT TTW CIT TRG ATC ATA AG -3’) and MPlatCOX1dR (5’- CAG GAA ACA GCT ATG ACT GAA AYA AYA IIG GAT CIC CAC C -3’) (Moszczynska et al. Reference Moszczynska, Locke, McLaughlin, Marcogliese and Crease2009). Amplification of COI mtDNA region was done using one of the following thermal cycling conditions: initial denaturation of 95°C for 2 min, 40 cycles of amplification at 94°C for 30 sec, 50°C for 30 sec, 72°C for 1 min, and final extension at 72°C for 10 min, or alternatively, an initial denaturation at 94°C for 4 min, 40 cycles at 94°C for 40 sec, 51°C for 40 sec, 72°C for 1 min, and final extension at 72°C for 10 min was used. PCR products were visualised on 1% agarose gel using SafeView™ Classic (Applied Biological Materials Inc, Richmond, Canada). The 28S PCR products were sequenced using the amplification primer set (Digl2 and 1500R) and internal primers 300F (5’- CAA GTA CCG TGA GGG AAA GTT G -3’) (Littlewood et al. Reference Littlewood, Curini-Galletti and Herniou2000) and ECD2 (5’- CTT GGT CCG TGT TTC AAG ACG GG -3’) (Tkach et al. Reference Tkach, Littlewood, Olson, Kinsella and Swiderski2003), while the ITS1–2 and COI gene regions were sequenced with the same primers used in the PCR reaction. PCR amplicons were sent to Inqaba Biotechnical Industries (Pty) Ltd, Pretoria, South Africa for purification and sequencing.
Phylogenetic analyses
Novel sequences of the partial 28S and ITS1–2 rDNA and COI mtDNA gene regions were generated for all clinostomid specimens. For the cryptogonimid specimens, only partial 28S rDNA sequences were generated and analysed; COI mtDNA sequence data were generated but not analysed due to a lack of public data with which to compare for the family from the region. Newly generated sequences were visually inspected, and consensus sequences assembled using Geneious® 2025.0.3 (Kearse et al. Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock, Buxton, Cooper, Markowitz, Duran, Thierer, Ashton, Meintjes and Drummond2012). All sequences were subjected to a BLAST search to identify congeners for inclusion in the subsequent phylogenetic analyses (Supplementary Tables 1–4). Sequences of clinostomid and cryptogonimid metacercariae were aligned with those of their respective families available on GenBank. Alignments were performed under default parameters using MAFFT version 7.490 (Katoh and Standley Reference Katoh and Standley2013). Each alignment was visually inspected, and final trimming was done using GBlocks v0.91b under the least stringent criteria (Castresana Reference Castresana2000; Dereeper et al. Reference Dereeper, Guignon, Blanc, Audic, Buffet, Chevenet, Dufayard, Guindon, Lefort, Lescot, Claverie and Gascuel2008). For partial 28S and ITS1–2 rDNA alignments of the Clinostomidae, analyses were focused on the genus Clinostomum Leidy, 1856, with species of Ithyoclinostomum Witenberg, 1925 and Odhneriotrema Travassos, 1928 designated as outgroup taxa and Euclinostomum Travassos, 1928 as ingroup taxa. Selected species of Diplostomidae (Bolbophorus sp.) were designated as outgroup taxa for the COI mtDNA analyses of the Clinostomidae. Selected species of Heterophyidae and Opisthorchiidae were used as outgroup taxa in the partial 28S rDNA analyses of the Cryptogonimidae. Optimal phylogenetic model selection for all alignments was determined in jModeltest version 2.1.10 (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012; Guindon and Gascuel Reference Guindon and Gascuel2003). Based on the Akaike Information Criterion (AIC), the generalised time-reversible model GTR+I+Γ was selected. Each alignment was subjected to a Bayesian Inference (BI) and Maximum Likelihood (ML) analyses using MrBayes v3.2.7a (Ronquist et al. Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Hohna, Larget, Liu, Suchard and Huelsenbeck2012) and RAxML 8.2.12 (Stamatakis Reference Stamatakis2014) implemented in the CIPRES portal (Miller et al. Reference Miller, Pfeiler and Schwartz2010). Each BI analysis was run (ngen = 10 ,000,000), two chains with four MCMC chains with a sample frequency of 1,000 and sample burnin of 30%. For each dataset, 100 bootstrap pseudoreplicates were run. Uncorrected p-distances for each alignment were generated using MEGA X (Kumara et al. Reference Kumara, Stecher, Li, Knyaz and Tamura2018) and numbers of base-pair differences were determined in Geneious® 2025.0.3.
Results
Clinostomid and cryptogonimid metacercariae were recovered from four host species collected during the present study (Table 1). Metacercariae were found in various organs of the fish hosts. All representatives of the Clinostomidae corresponded with the genus Clinostomum (Figures 1a–c) and were free in the branchial chambers of C. gariepinus or encysted in the body cavities of suckermouth catfishes, Chiloglanis sp. (Mochokidae), and the mormyrid Marcusenius pongolensis (Peters). Cryptogonimid metacercariae (Figure 6) were encysted in the muscle tissue and fin rays of Redeye labeo, Labeo cylindricus Peters (Cyprinidae). Specimens recovered in the present study correspond to one described and two undescribed species of the genus Clinostomum which have been previously recorded from South Africa and elsewhere on the African continent, while that of the Cryptogonimidae do not correspond to any known taxa with available molecular data. Final alignment lengths were 1,240 bp and 840 bp for the partial 28S rDNA region of the Clinostomidae and Cryptogonimidae, respectively, and 996 bp and 529 bp for the ITS1–2 rDNA and COI mtDNA regions respectively for the Clinostomidae. Sequence data for the partial 28S and ITS1–2 rDNA and COI mtDNA gene regions (Figures 2–4) were obtained for the species of Clinostomidae found during the present study. Tree topologies for BI and ML analyses of the partial 28S rDNA region were identical, but not so for the ITS1–2 rDNA and COI mtDNA regions (see Figures 3a–b and Figures 4a–b). Tree topologies for the partial 28S rDNA gene region (Figure 5) of the Cryptogonimidae were 100% congruent.
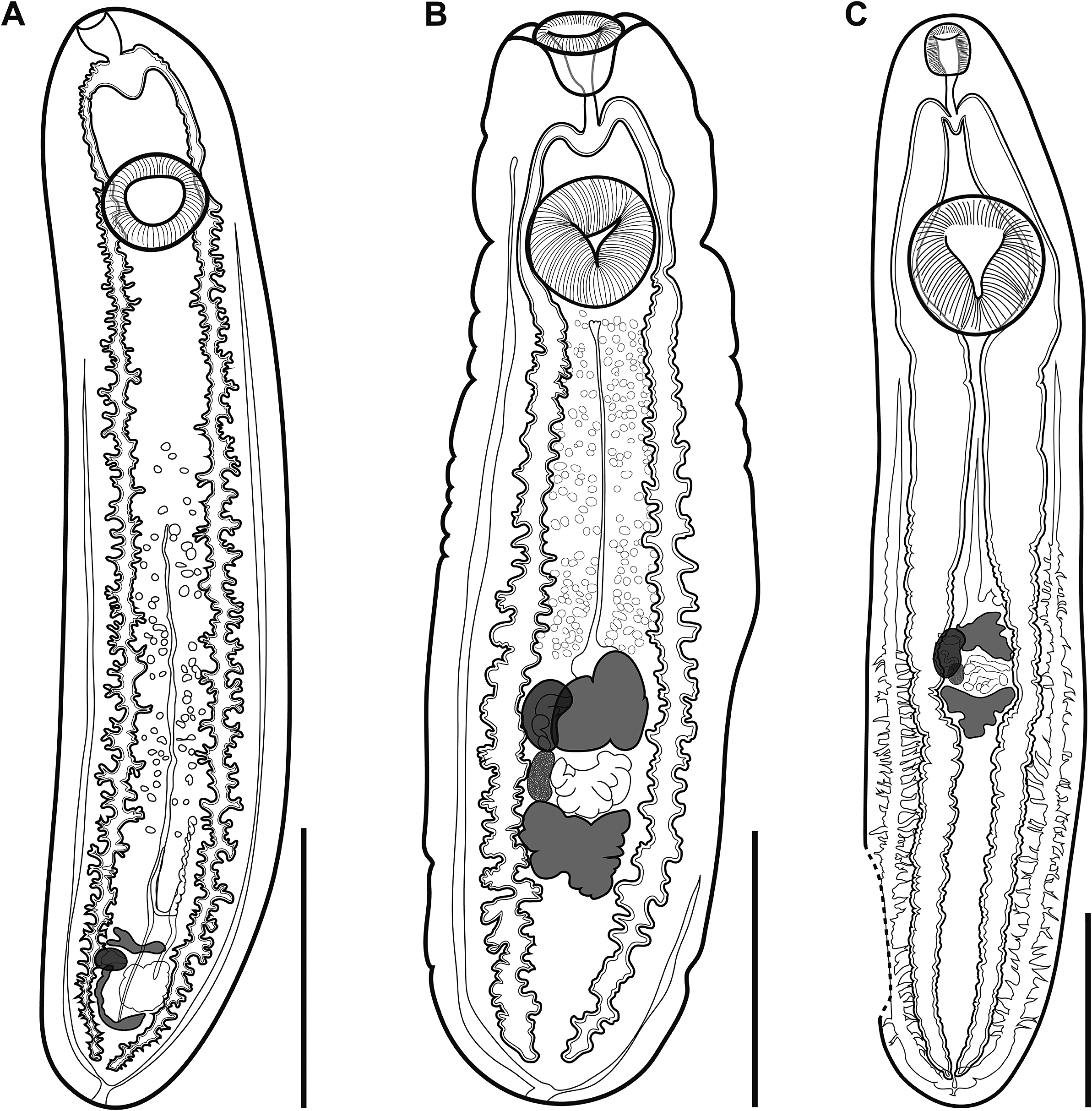
Figure 1. (A), Clinostomum brieni from the branchial chambers of Clarias gariepinus from the Barotse floodplain, Zambia (NMB P 1071); (B), Clinostomum sp. ‘morphotype 2’ sensu Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) encysted in the body cavity of Marcusenius macrolepidotus from the Letaba River, South Africa (NMB P 1077); (C), Clinostomum sp. ‘morphotype 3’ sensu Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) encysted in the body cavity of Chiloglanis sp. from the Letaba River, South Africa (NMB P 1078). Scale bars: 2 mm (A, B); 1 mm (C).

Figure 2. Phylogenetic tree based on the 1,240 bp alignment of the 28S rDNA gene region of the Clinostomidae, with our species analysed against available sequence data from Old World and New World taxa. Tree topology presented based on the Bayesian Inference (BI) analysis. Nodal support values indicate posterior probabilities from the BI and bootstrap values from the ML analyses. Dashes indicate values below 75. Species from the present study presented in bold. IG = ingroup; BRA = Brazil; CHN = China; EGY = Egypt; FRA = France; IND = India; ITA = Italy; JPN = Japan; KEN = Kenya; MEX = Mexico; USA = United States of America; VNM = Vietnam; ZAF = South Africa.
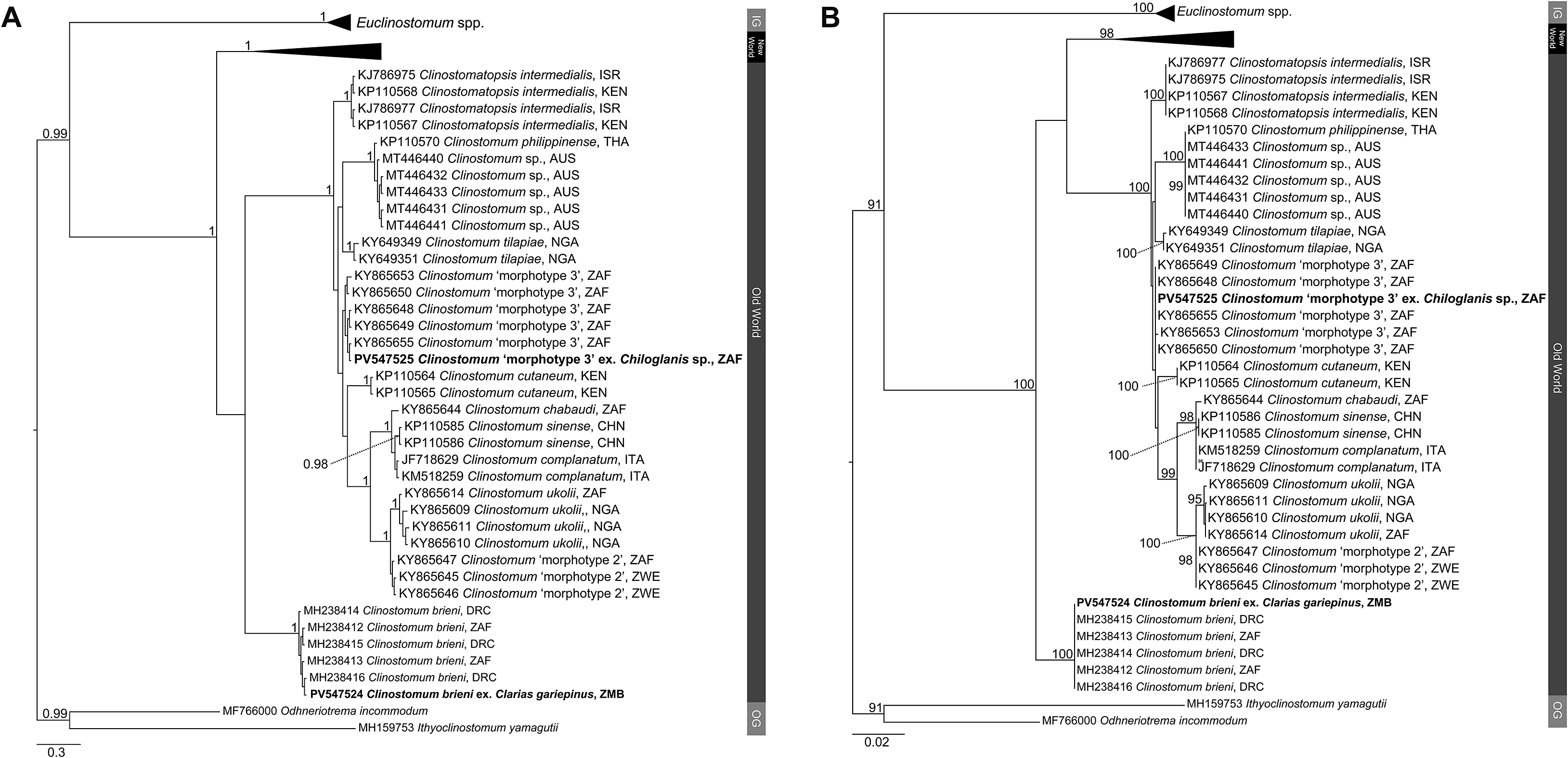
Figure 3. Phylogenetic trees based on the 996 bp alignment of the ITS1–2 rDNA gene region of the Clinostomidae, with our species analysed against available sequence data from Old World and New World taxa. Tree topologies presented based on (A), Bayesian Inference (BI) and (B), Maximum likelihood (ML) analysis. Nodal support values indicate posterior probabilities from the BI and bootstrap values from the ML analyses. Species from the present study presented in bold. OG = Outgroup; BOL = Bolivia; BRA = Brazil; CAN = Canada; DRC = Democratic Republic of the Congo; CHN = China; ISR = Israel; ITA = Italy; MEX = Mexico; NGA = Nigeria; KEN = Kenya; THA = Thailand; PER = Peru; USA = United States of America; ZAF = South Africa; ZMB = Zambia; ZWE = Zimbabwe.
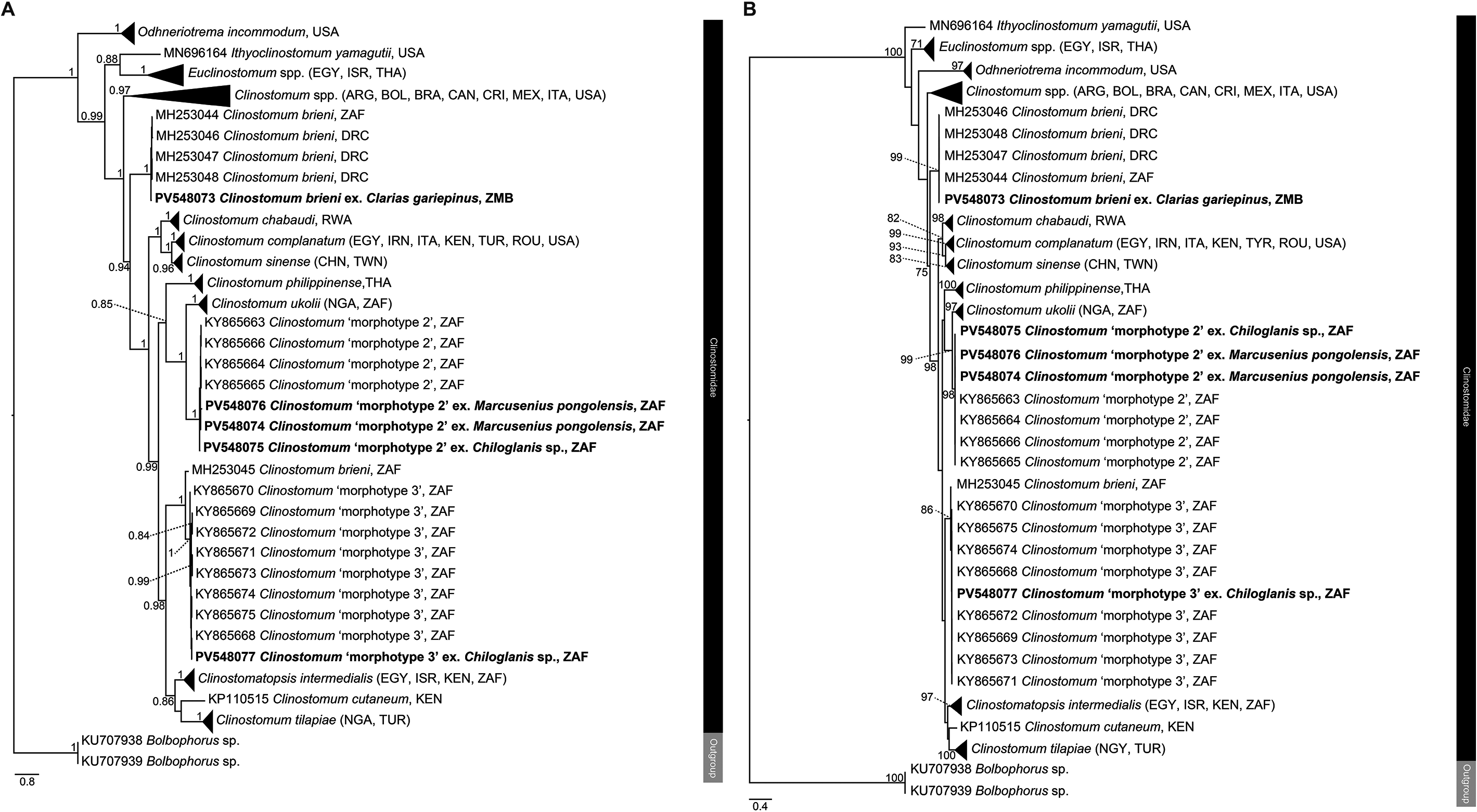
Figure 4. Phylogenetic trees based on the 529 bp alignment of the COI mitochondrial region of the Clinostomidae, with our species analysed against available sequence data from Old World and New World taxa. Tree topology presented based on (A), Bayesian Inference (BI) and (B), Maximum likelihood (ML) analysis. Nodal support values indicate posterior probabilities from the BI and bootstrap values from the ML analyses. Dashes indicate values below 75. Species from the present study presented in bold. ARG = Argentina; BOL = Bolivia; BRA = Brazil; CAN = Canada; DRC = Democratic Republic of the Congo; CRI = Costa Rica; CHN = China; EGY = Egypt; IRN = Iran; ISR = Israel; ITA = Italy; MEX = Mexico; NGA = Nigeria; KEN = Kenya; THA = Thailand; TUR = Turkey; TWN = Taiwan; ROU = Romania; USA = United States of America; ZAF = South Africa; ZMB = Zambia.
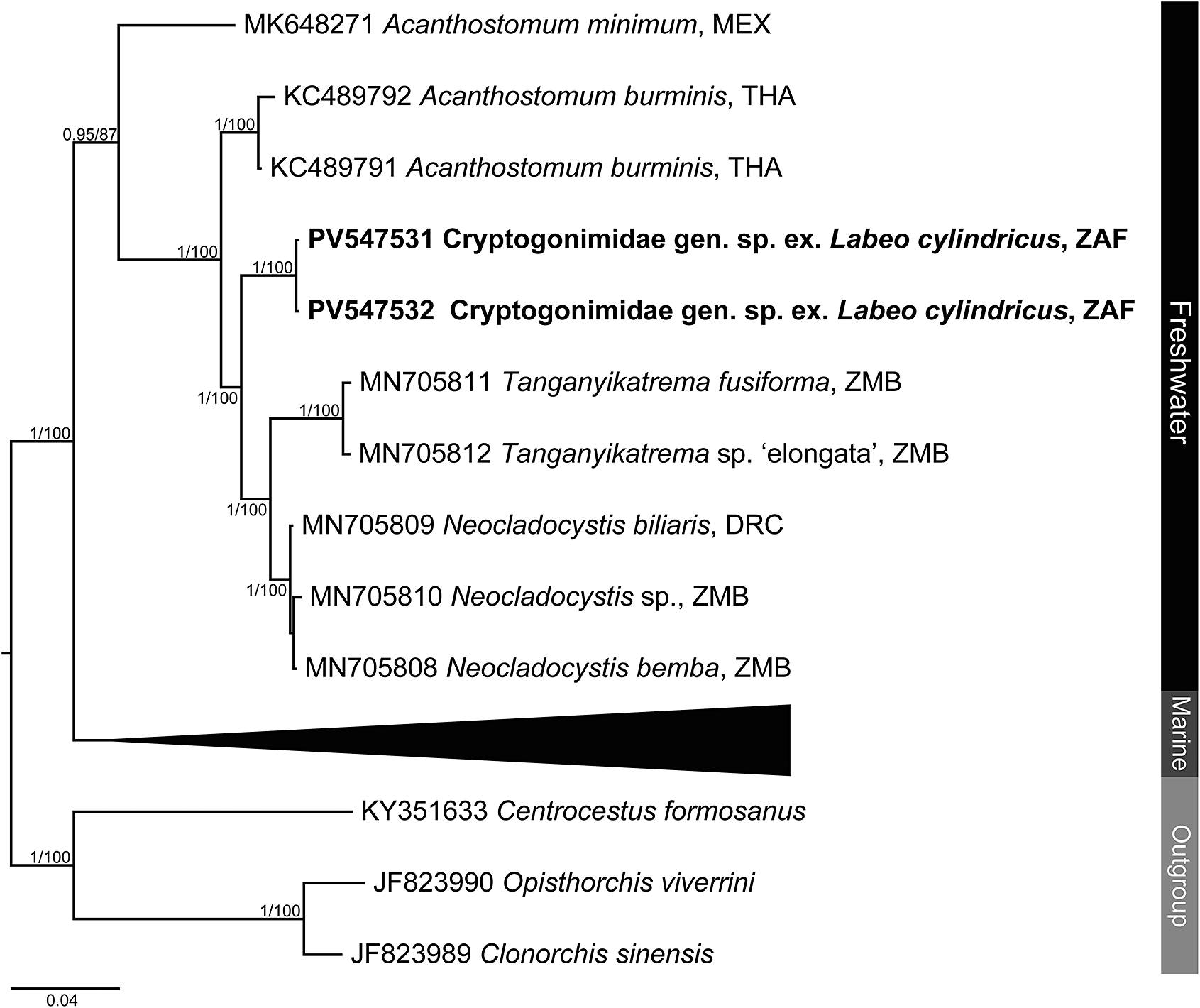
Figure 5. Phylogenetic tree based on the 840 bp alignment of the 28S rDNA region of the Cryptogonimidae. Tree topology presented based on the Bayesian inference analysis. Nodal support values indicate posterior probabilities from the BI and bootstrap values from the ML analyses. Dashes indicate values below 75. DRC = Democratic Republic of the Congo; MEX = Mexico; THA = Thailand; ZAF = South Africa; ZMB = Zambia.
Taxonomy
Superfamily Schistosomatoidea Stiles & Hassall, 1898
Family Clinostomidae Lühe, 1901
Genus: Clinostomum Leidy, 1856
Clinostomum brieni (Dollfus, 1950) (Figure 1a)
Host: Clarias gariepinus (Burchell) (Siluriformes: Clariidae).
Locality: Barotse floodplain, Southwestern Province, Zambia (15° 12′ 1" S, 22° 58′ 9" E).
Prevalence: Three out of 17 fishes (18%) infected by 1–2 metacercariae.
Site of infection: Free in branchial chambers.
Voucher material: One hologenophore (NMB P 1070), five voucher specimens, all slides mounted (NMB P 1071–1075).
Representative DNA sequence data: 28S rDNA – one sequence of 1,229 bp length (GenBank PV547526); ITS1–2 – one sequence of 1,222 bp length (GenBank PV547524); COI – one sequence of 539 bp length (GenBank PV548073).
Morphology
With features of species. Body 4,589–9,582 (6,865) long, maximal breadth 908–1,681 (1,297), 5.0–5.7 (5.3) times longer than broad. Oral sucker 175–408 × 136–336 (296 × 233), 1.2–1.4 (1.3) times longer than broad. Ventral sucker 518–758 × 512–822 (641 × 642), 0.9–1.1 (1.0) times longer than broad, anterior edge 472–806 (650) from posterior edge of oral sucker. Forebody (pre-ventral sucker portion of body) 759–1,187 (982) or 12.4–17.1% (14.6%) of total body length. Hindbody 3,217–7,637 (5,237) or 70.1–79.7% (75.6%) of total body length. Caeca distinctly diverticulated, extend to near posterior extremity. Nascent testes, ovary, and cirrus-sac form genital complex in posterior hindbody, 5,455–8,026 (6,622) or 83.8–84.9% (84.3%) of total body length from anterior extremity, 381–543 (480) or 5.7–6.5% (6.1%) of total body length from posterior extremity. Testis anlages distinctly lobed, tandem, anterior testis 180–348 (281) from posterior testis. Anterior testis 61–151 × 277–618 (119 × 426), posterior testis 49–151 × 206–416 (94 × 338). Ovary anlage reniform, dextral in genital complex, between and slightly overlapping testes, 106–271 × 53–167 (192 × 87), 1.6–4.1 (2.5) times longer than broad. Cirrus-sac anlage reniform, dextral in genital complex, overlaps ovary and testicular margins, 139–268 × 105–190 (181 × 134), 0.9–2.0 (1.4) times longer than broad.
Remarks
Clinostomum brieni was described by Dollfus (Reference Dollfus1950) (as Clinostomoides brieni) from an adult infecting a Goliath Heron, Ardea goliath Cretzschmar (Aves: Ardeidae) from Kadia, Belgian Congo [now the southern Democratic Republic of the Congo (DRC)]. This species is among the most frequently recorded of the African clinostomids, having been reported multiple times from localities across the continent, including Botswana, the DRC, Ghana, Rwanda, and Zimbabwe (Table 2). Most recently, Caffara et al. (Reference Caffara, Locke, Halajian, Luus-Powell, Benini, Tedesco, Kasembele and Fioravanti2019) reported this species from localities in the greater Lubumbashi region, DRC, and from Phalaborwa, South Africa. With the exception of the type-description, all subsequent records are of metacercariae recovered from catfishes of the genus Clarias Scapoli. Despite these numerous records, the latter study is the only one to be supported by complementary molecular sequence data, the only other such data coming from specimens putatively ascribed to this species from Manipur, northeast India (Athokpam et al. Reference Athokpam, Jyrwa and Tandon2016). The record from Zambia is the first of this species from this country and is broadly morphologically consistent with all previous reports.
Table 2. List of records of Clinostomum species from Africa; Clinostomatopsis intermedialis is included as its junior synonym, Clinostomum phalocrocoracis, is still widely recognised in literature. Unless otherwise noted, all hosts are fishes bearing metacercariae. Entries marked ‘*’ represent type-records from original descriptions

Notes:
a Dubois (Reference Dubois1931) gives the host as Phalacrocorax levaillanti, a name that does not exist. We cite the host as Phalacrocorax sp.
b Calhoun et al. (Reference Calhoun, Leslie, Riepe, Achatz, McDevitt-Galles, Tkach and Johnson2019), in their supplementary table, ascribe these records to Echi et al. (Reference Echi, Okafor and Eyo2009b), but that paper does not contain any mention of them.
c Skrjabin (1947) gives the location of discovery as Angola; this is not mentioned in Dubois (Reference Dubois1930), who only gives the location as ‘Africa’.
d Dollfus (Reference Dollfus1950) noted these specimens, collected by P. Brien, but did not describe them; Manter and Pritchard (Reference Manter and Pritchard1969) subsequently identified these as Clinostomum tilapiae on the basis of two of Brien’s specimens.
Analyses of the partial 28S, ITS1–2 rDNA and COI mtDNA datasets for the Clinostomidae support the morphological identification of the specimen as being C. brieni (Figures 2–4). In both the ITS1–2 rDNA and COI mtDNA analyses, the sequence from the specimen in this study formed a clade with others of C. brieni from South Africa and DRC, with no differences in ITS1–2 rDNA and only 1–2 bp differences in the COI mtDNA region (Supplementary Tables 5–7). Caffara et al. (Reference Caffara, Locke, Halajian, Luus-Powell, Benini, Tedesco, Kasembele and Fioravanti2019), who provided most of the available COI mtDNA sequences with which to compare, also generated one sequence of C. brieni which fell distant to all other conspecifics, instead forming a clade with Clinostomum ‘morphotype 3’ of Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) and explained this as most likely due to hybridisation. The same topology was observed in the present analyses.
The partial 28S rDNA sequence for this taxon was not comparable with others from Africa, as no other study had generated sequence data for this gene region from the continent. The sequence generated in the present study formed a clade with the only sequence on GenBank identified as belonging to C. brieni, a sequence obtained from a specimen infecting the clariid catfish Heteropneustes fossilis (Bloch) from India (Athokpam and Tandon Reference Athokpam and Tandon2016). The genetic difference between the specimen from this study and the one from India (23 bp difference and a p-distance of 1.9%), in combination with the lack of divergence in the other analysed gene regions between our sequence and others from Africa, strongly suggest that the taxon from India is not C. brieni and is instead a closely related but new species. Analysis of 18S rDNA sequence data of the latter taxon by Caffara et al. (Reference Caffara, Locke, Halajian, Luus-Powell, Benini, Tedesco, Kasembele and Fioravanti2019), wherein it also forms a clade with C. brieni from Africa, further supports this inference.
Clinostomum sp. ‘morphotype 2’ (sensu Caffara et al. Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) (Figure 1b)
Hosts: Chiloglanis sp. (Siluriformes: Mochokidae); Marcusenius pongolensis (Peters, 1852) (Osteoglossiformes: Mormyridae)
Localities: Tzaneen Dam, Limpopo, South Africa (23° 48′ 55′′ S, 30° 08′ 31′′ E) – M. pongolensis; Letaba River, Limpopo, South Africa (23° 51′ 1′′ S, 30° 06′ 21′′ E) – Chiloglanis sp.
Prevalence: 67% (2/3), IF = 1 – M. pongolensis; 10% (1/10), IF = 1 – Chiloglanis sp.
Site of infection: Encysted in body cavity.
Voucher material: Two hologenophores, mounted, NMB P 1076–1077.
Representative DNA sequence data: 28S rDNA – one sequence of 1,237 bp length ex. Chiloglanis sp. (GenBank PV547527); two identical sequences ex. Marcusenius pongolensis, 1,199 and 1,266 bp length submitted to GenBank (GenBank PV547528, PV547529); COI – one sequences of 668 bp length ex. Chiloglanis sp.; 671 and 703 bp ex. M. pongolensis (GenBank PV548074, PV548075)
Morphology
With features of taxon. Body 7,981 long, maximal breadth 2,130, 3.7 times longer than broad. Oral sucker 545 × 683, 0.8 times longer than broad. Ventral sucker 994 × 971, 1.0 times longer than broad, anterior edge 649 from posterior edge of oral sucker. Forebody 1,211 or 15.2% of total body length. Hindbody 5,885 or 73.7% of total body length. Caeca distinctly diverticulated, extend to near posterior extremity. Nascent testes, ovary, and cirrus-sac form genital complex in mid-hindbody, 4,837 or 60.6% of total body length from anterior extremity, 1,497 or 18.8% of total body length from posterior extremity. Testis anlages distinctly lobed, anterior much less so than posterior; tandem, anterior testis 367 from posterior testis. Anterior testis 866 × 728, posterior testis 638 × 824. Ovary anlage reniform, dextral in genital complex, between testes, 385 × 183, 2.1 times longer than broad. Cirrus-sac anlage reniform, dextral in genital complex, overlaps ovary and anterior testicular margins, 547 × 242, 2.3 times longer than broad.
Remarks
Attempts to generate ITS1–2 rDNA sequence data for these specimens was unsuccessful. The COI mtDNA sequences, however, showed that three specimens, from the mormyrid Marcusenius pongolensis and the mochokid catfish Chiloglanis sp., were nearly or 100% identical to those referred to by Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) as Clinostomum ‘morphotype 2’ (Figures 3a–b). One specimen from M. pongolensis was genetically identical to those from the same host from northeastern South Africa, while the other two specimens differed from those of Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) by 1–2 bp (0.2%), showing intraspecific variation of 2–4 bp (0.3–0.6%) (Supplementary Tables 5 and 7). Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) did not generate 28S rDNA sequence data for this taxon. The partial 28S rDNA analyses inferred in the present study indicate sequences obtained in the present study formed a clade basal to that containing species of Clinostomum from elsewhere in Africa (Egypt and Kenya), Vietnam, and Australia. Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) obtained specimens of this taxon from two species of Marcusenius Gill (Mormyridae) from Zimbabwe and far northeastern South Africa. The record from Chiloglanis sp. constitutes a new host record for this taxon. The specimens from the present study do not strongly resemble those of Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) in terms of genital complex morphology, lacking the strong digitate lobulation of the testes and with the cirrus pouch overlapping the anterior testis (Figure 1b). In those regards, the newly obtained specimens more closely resemble those of Clinostomum ‘morphotype 1’, which has since been described as Clinostomum ukolii Caffara, Locke, Echi, Halajian, Luus-Powell, Benini, Tedesco & Fioravanti, 2020 (Caffara et al. Reference Caffara, Locke, Echi, Halajian, Luus-Powell, Benini, Tedesco and Fioravanti2020).
Clinostomum sp. ‘morphotype 3’ (sensu Caffara et al. Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) (Figure 1c)
Host: Chiloglanis sp. (Siluriformes: Mochokidae)
Locality: Letaba River, Limpopo, South Africa (23° 51’ 1” S, 30° 06’ 21” E)
Prevalence: One out of 10 fishes (10%) infected with two metacercariae.
Site of infection: Encysted in body cavity.
Voucher material: One hologenophore, mounted (NMB P 1078).
Representative DNA sequence data: 28S rDNA – one sequence of 1,204 bp length (GenBank PV547530); ITS1–2 rDNA – one sequence of 1,161 bp length (GenBank PV547525); COI mtDNA– one sequence of 587 bp length (GenBank PV548077).
Morphology
With features of taxon. Body 3,673–6,162 (4,918) long, maximal breadth 1,113–1,155 (1,134), 3.3–5.3 (4.3) times longer than broad. Oral sucker 249–254 × 240–245 (252 × 243), 1.0–1.1 times longer than broad. Ventral sucker 688–770 × 665–764 (729 × 715), 1.0 times longer than broad, anterior edge 489–689 (589) from posterior edge of oral sucker. Forebody 788–1,020 (904) or 16.6–21.5% (19.0%) of total body length. Hindbody 2,207–3,262 (2,735) or 52.9–60.1% (56.5%) of total body length. Caeca not distinctly diverticulated, extend to near posterior extremity. Nascent testes, ovary, and cirrus-sac form genital complex in mid-hindbody, 2,216–3,478 (2,847) or 56.4–60.3% (58.4%) of total body length from anterior extremity, 630–2,176 (1,403) or 17.2–35.3% (26.2%) of total body length from posterior extremity. Testis anlages distinctly lobed, tandem, anterior testis 167–293 (230) from posterior testis. Anterior testis 191–268 × 313–336 (230 × 325), posterior testis 191–276 × 335–414 (234 × 375). Ovary anlage reniform, dextral in genital complex, between testes, 118–126 × 74–153 (122 × 114), 0.8–1.6 (1.2) times longer than broad. Cirrus-sac anlage reniform, dextral in genital complex, overlaps ovary and anterior testicular margins, 278–282 × 130–160 (280 × 145), 1.8–2.1 (2.0) times longer than broad.
Remarks
Both the ITS1–2 rDNA and COI mtDNA analyses indicated that specimens recovered from Chiloglanis sp. from Letaba River, South Africa conformed with Clinostomum ‘morphotype 3’ of Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017). This taxon was recovered by Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) from catfishes of the same genus (specifically Chiloglanis pretoriae van der Horst) as well as the amphiliid catfish Anoplopterus sp. ‘southern stargazer’ from rivers in northeast South Africa. In both analyses, specimens from the present study were within the range of intraspecific variation obtained by Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) (0–1 bp difference in ITS1–2 rDNA, 0–4 bp difference in COI mtDNA). As with Clinostomum ‘morphotype 2’ (see above), novel partial 28S rDNA sequence data was produced for this taxon. Analyses of each respective gene region produced conflicting topologies with respect to the molecular phylogenetic position of Clinostomum ‘morphotype 3’. In analyses of the partial 28S rDNA dataset, Clinostomum ‘morphotype 3’ formed a clade with sequences of Clinostomum cutaneum Paperna, 1964 from Kenya and Egypt, sister to Clinostomatopsis intermedialis (Lamont, 1920) (Gustinelli et al. Reference Gustinelli, Caffara, Florio, Otachi, Wathuta and Fioravanti2010; Hamouda and Younis Reference Hamouda and Younis2021) (Figure 2). [As an aside, Clinostomatopsis intermedialis has been extensively reported in literature as Clinostomum phalacrocoracis Dubois, 1931; the synonymy of this species with Neutraclinostomum intermedialis (Lamont, 1920) by Feizullaev and Mirzoeva (Reference Feizullaev and Mirzoeva1983) and subsequent reclassification as a species of Clinostomatopsis (after the synonymising of the two genera by Kanev et al. Reference Kanev, Radev, Fried, Gibson, Jones and Bray2002) does not appear to be widely accepted]. Our COI mtDNA analyses produced a similar topology, with sequences of Clinostomum ‘morphotype 3’ from the present study and Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) forming a clade with an anomalous sequence of Clinostomum brieni (see section for C. brieni above), sister to [C. intermedialis (C. cutaneum + C. tilapiae)], with strong support at all nodes (Figure 4). This topology conflicts strongly with that produced by the ITS1–2 rDNA analyses, which placed Clinostomum ‘morphotype 3’ distant to Cl. intermedialis and C. tilapiae, instead forming a clade sister to one that included several other African and Asian Clinostomum species, but with poor nodal resolution (Figure 3). Morphologically, the specimen from the present study conforms reasonably well with those described by Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) but with some minor variation. The anterior testis depicted in that paper seems markedly less lobed than that of the newly obtained specimen but is likely within the continuum of variation for this taxon. Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017) described the caeca of morphotype 3 as digitated; the specimen from the present study shows very slight digitation (Figure 1c).
Superfamily Plagiorchioidea Looss, 1899
Family Cryptogonimidae Ward, 1917
Cryptogonimidae gen. sp. (Figure 6a–d)
Host: Labeo cylindricus Peters, 1852 (Cypriniformes: Cyprinidae)
Locality: Letaba River, Limpopo, South Africa (23°38’56.79” S, 30°39’31.1” E)
Site of infection: Encysted in fin rays.
Prevalence: Three out of three individuals (100%), IF = 68–167.
Voucher material: Photohologenophore (Figure 6a–d)
Representative DNA sequence data: 28S rDNA – two identical sequences, 1,158 and 1,235 bp in length, (GenBank PV547531 and PV547532); COI – two sequences 695 and 770 bp in length (GenBank PV548078 and PV548079).
Morphology
Measurements based on three excysted specimens. Body 347–397 (372) long, maximal breadth 83–90 (87), 4.2–4.4 (4.3) times longer than broad. Oral sucker robust, subtriangular to infundibuliform, 77–115 × 67–88 (100 × 77), 1.1–1.3 times longer than broad, bearing 22–23 (23) circumoral spines. Spines robust, straight, undivided, 18–27 (23) long. Ventral sucker subspherical, 45–51 × 48–54 (49 × 51), 0.9–1.0 (1.0) times longer than broad. Ventrogenital sac not apparent. Forebody 242–253 (248) or 64–70% (67%) of total body length. Hindbody 100–130 (115) or 25–37% (31%) of total body length. Digestive tract not traceable, though hints of pharynx and caecal bifurcation observed pre-ventral sucker. Genital anlages not visible in any specimens.
Remarks
Analyses of the partial 28S rDNA region for species of Cryptogonimidae places the newly obtained sequence data within a clade formed by other African freshwater species of cryptogonimids (Figure 5 and Supplementary Table 8) of the genera Neocladocystis Manter & Pritchard, 1969 and Tanganyikatrema Kmentová, Georgieva & Bray in Kmentová, Bray, Koblmüller, Artois, De Keyzer, Gelnar, Vanhove & Georgieva, 2020, but also including a taxon identified by Jayawardena et al. (Reference Jayawardena, Tkach, Navaratne, Amerasinghe and Rajakaruna2013) as Acanthostomum burminis (Bhalerao, 1926), a species which infects anuran amphibians as metacercariae and natricine snakes as adults. The stout oral spines of the metacercariae from the present study (Figure 6a–b, d), are more in common with species of Acanthostomum Looss, 1899 and Proctocaecum Baugh, 1957 than any of the other freshwater African cryptogonimid species, therefore it is highly likely that it corresponds to a species of one of these two genera. The genus Acanthostomum was recovered as polyphyletic in the 28S rDNA analysis. This is perhaps illustrative of the fact that defining this and other acanthostomine genera has long been problematic. Both Acanthostomum and Proctocaecum have had complex histories, with species of many genera (including the latter) previously being classified in the former (Brooks Reference Brooks1980; Lamothe-Argumedo and Ponciano-Rodriguez Reference Lamothe-Argumedo and Ponciano-Rodriguez1986). A lack of molecular sequence data for the type-species of the Acanthostomum and for any species of Proctocaecum compounds the difficulties in delineating the genera. Both genera include species which infect fishes and semi-aquatic reptiles; those which infect the latter are known to infect amphibians as metacercariae (see Jayawardena et al. Reference Jayawardena, Tkach, Navaratne, Amerasinghe and Rajakaruna2013). The newly obtained metacercarial specimens possess 22–23 oral spines (Figure 6a, d), a feature of several species of Proctocaecum: P. absconditum, P. productum (Odhner, 1902), and P. vicinum. The fact that the metacercaria was found in a fish means it is perhaps more likely to be a fish (specifically a catfish) rather than reptile-infecting species, but the lack of knowledge regarding the freshwater cryptogonimid fauna of the region precludes any firm conclusions.
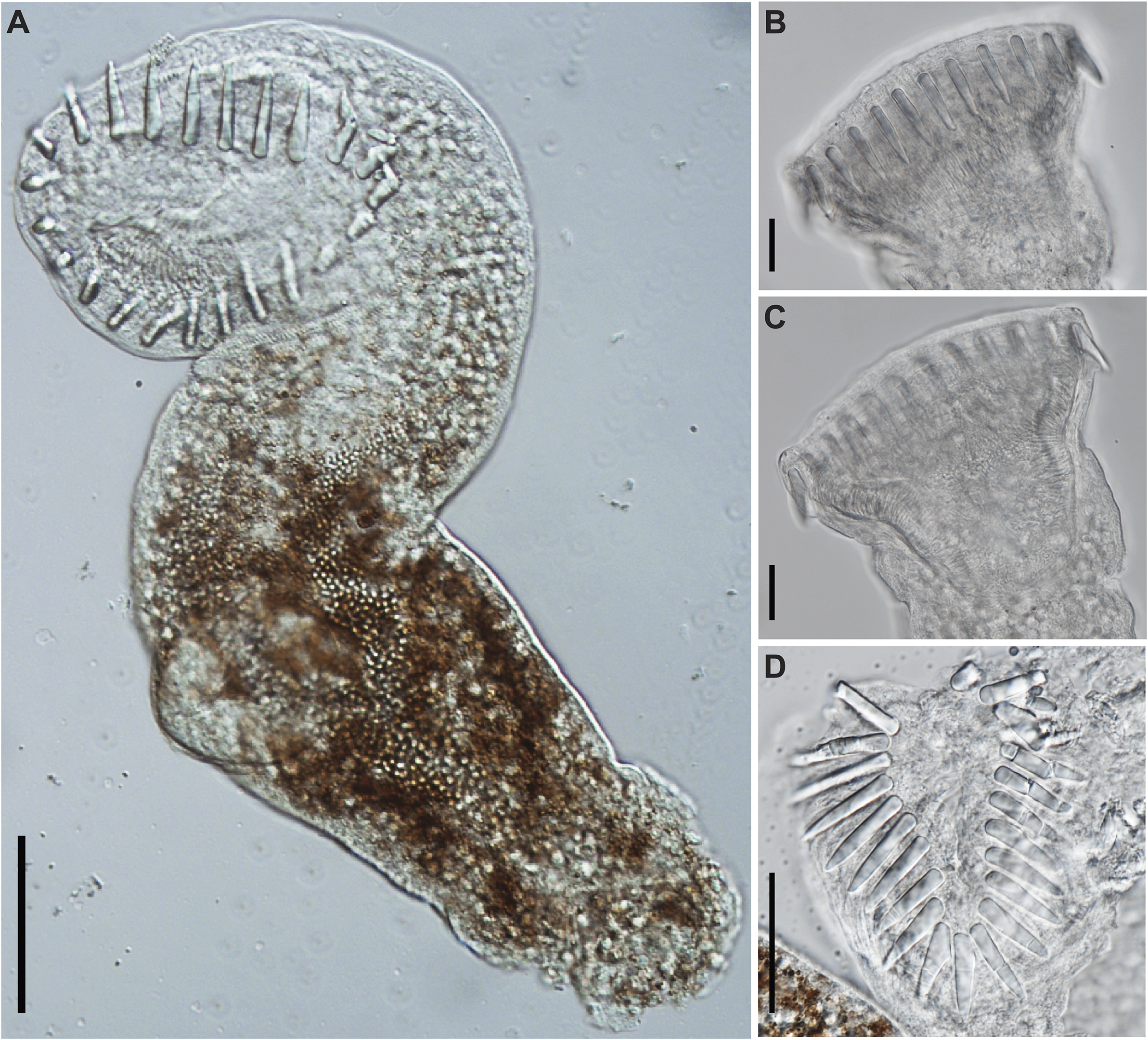
Figure 6. Photomicrographs of Cryptogonimidae gen. sp. found encysted on the fin rays of Labeo cylindricus from the Letaba River, South Africa (photohologenophore; GenBank PV547531). (A), Whole body, ventral view; circumoral spines, lateral view; (B), subtriangular oral sucker (C), and apical view (D). Scale bars: 50 μm (A); 20 μm (B, C, D).
Discussion
Surveys in southern Africa recovered three distinct clinostomid morphotypes, all represented by metacercariae found in fishes. Molecular sequence data generated for all three morphotypes validated the presence of three disparate taxa, all with affinities to the genus Clinostomum. The first of these, from the clariid catfish Clarias gariepinus from Zambia, is ascribed to Clinostomum brieni and the first partial 28S rDNA sequence data is provided for this species from this region. The other two putative species match to two as-yet undescribed Clinostomum species, known as Clinostomum ‘morphotype 2’ and ‘morphotype 3’, respectively, first reported by Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017). A first general morphological account and the first partial 28S rDNA sequence data are provided for these. These taxa were collected from the same hosts and general region (Limpopo and Mpumalanga provinces in northeastern South Africa) as Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017); however, the discovery of Clinostomum ‘morphotype 2’, hitherto only known from mormyrid fishes, from a mochokid catfish (a species of Chiloglanis) represents a new host record for this taxon. Despite the very low infection rates of clinostomid specimens found in this study, the few specimens obtained provided ample material to illustrate their morphological distinctness and generate comparative molecular data. A novelty to the freshwater fauna in South Africa is the first account of cryptogonimid metacercariae from a cyprinid and this record is accompanied by sequence data of the 28S rDNA and COI mtDNA gene regions.
Clinostomidae
The family Clinostomidae (often inaccurately iterated Clinostomatidae) is a small one, comprising 89 species in six genera; of these, most species are of the genus Clinostomum Leidy, 1856 (60 species plus three of uncertain status) and Euclinostomum Travassos, 1928 (20 species) (WoRMS; accessed 03/2025). Species of Clinostomum and Euclinostomum, along with Nephrocephalus Odhner, 1902, have been recorded from Africa (Scholz et al. Reference Scholz, Vanhove, Smit, Jayasundera and Gelnar2018). An additional genus, Clinostomoides Dollfus, 1950, was proposed by Dollfus (Reference Dollfus1950) to accommodate Clinostomoides brieni Dollfus, 1950, described from an adult infecting a Goliath Heron, Ardea goliath Cretzschmar (Aves: Ardeidae) from southern region of the DRC. Species of this genus, including C. brieni, have since been reclassified as species of Clinostomum on the basis of molecular work by Caffara et al. (Reference Caffara, Locke, Halajian, Luus-Powell, Benini, Tedesco, Kasembele and Fioravanti2019).
Clinostomid taxonomy has undergone several major shifts which have done little to resolve much of the confusion surrounding the family. Most notably, several genera were synonymised with the largest clinostomid genus, Clinostomum, and several authors (Feizullaev and Mirzoeva Reference Feizullaev and Mirzoeva1983; Ukoli Reference Ukoli1966a) performed mass-synonymisations of multiple taxa with just one [Clinostomum complanatum (Rudolphi, 1814)] in attempts to resolve the issue of tenuous descriptions. Subsequent studies, however, have shown that many such efforts were over-reaching. On one hand, several species formerly synonymised with C. complanatum have since been revalidated (Caffara et al. Reference Caffara, Davidovich, Falk, Smirnov, Ofek, Cummings, Gustinelli and Fioravanti2014; Dzikowski et al. Reference Dzikowski, Levy, Poore, Flowers and Paperna2004; Matthews and Cribb Reference Matthews and Cribb1998). For example, Dzikowski et al. (Reference Dzikowski, Levy, Poore, Flowers and Paperna2004) demonstrated using 18S rDNA data that Clinostomum marginatum (Rudolphi, 1819), which was synonymised with C. complanatum by Baer (Reference Baer1933), was actually a separate species. On the other hand, other studies have also validated some of these synonymies. Species of Clinostomoides and Ithyoclinostomum Witenberg, 1926, for example, have been demonstrated to be, in fact, species of Clinostomum (Caffara et al. Reference Caffara, Locke, Halajian, Luus-Powell, Benini, Tedesco, Kasembele and Fioravanti2019; Simões et al. Reference Simões, Alves, López-Hernández, Couto, Moreira and Pinto2022).
Several issues surround the identification of species of Clinostomum in Africa, chief among them being the fact that many authors have made tenuous identifications, often based on nothing more than superficial gross morphology. Manter and Pritchard (Reference Manter and Pritchard1969), for example, identified metacercariae collected from Oreochromis niloticus (Linnaeus) in Rwanda as Clinostomum macrosomum Jaiswal, 1957, a species hitherto only known from India, entirely on the basis of its ‘very large size’; C. macrosomum was described from metacercariae (Jaiswal Reference Jaiswal1957) and has never been encountered since.
Eight studies have incorporated molecular sequencing as part of an integrated approach in verifying species identification and systematics of clinostomids in Africa (Caffara et al. Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017; Caffara et al. Reference Caffara, Locke, Halajian, Luus-Powell, Benini, Tedesco, Kasembele and Fioravanti2019; Caffara et al. Reference Caffara, Locke, Echi, Halajian, Luus-Powell, Benini, Tedesco and Fioravanti2020; Hamouda and Younis Reference Hamouda and Younis2021; Locke et al. Reference Locke, Caffara, Marcogliese and Fioravanti2015; Mahdy et al. Reference Mahdy, Abdelsalam, Abdel-Maogood, Shaalan and Salem2021; Mahdy et al. Reference Mahdy, Abdelsalam and Salem2023; Salem et al. Reference Salem, Abdel-Maogood, Abdelsalam and Mahdy2021) and only 16 out of 103 published studies incorporated or generated samples from definitive hosts. In the absence of molecular sequencing, the ambiguous and highly variable morphology of many species has further complicated efforts to resolve clinostomid taxonomy, and many species have been confused with one another. For instance, in Africa, specimens of Clinostomatopsis intermedialis (formerly Neutraclinostomum intermedialis and Clinostomum phalacrocoracis) have been confused with Clinostomum tilapiae (Grobler et al. Reference Grobler, Mokgalong and Saayman1999). This disparity between depth of knowledge of Clinostomum intermediate stages and that of definitive stages is reflected across the wider world, with the majority of records being made from intermediate stages and, in some cases, species described from them in total absence of knowledge of the adult (see Caffara et al. Reference Caffara, Locke, Echi, Halajian, Luus-Powell, Benini, Tedesco and Fioravanti2020 for example). Although such an approach is traditionally problematic, it has been defended by the likes of Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Luus-Powell, Benini, Tedesco and Fioravanti2020), who point out that morphological features characteristic of adult stages, such as egg size/distribution and vitellaria, are not taxonomically discriminatory for clinostomids, that ontogenetic development of other organs minimally impacts their interpretation, that molecular phylogenetics support morphological interpretations of species delineations, and that, contextually, all indications are that the African clinostomid fauna is rather limited and has minimal overlap with the rest of the world. We largely agree with these arguments, though we are still of the opinion that matching known intermediate stages with adults should still be a key goal. We do, however, acknowledge that lack of access to definitive hosts (birds and reptiles) due to ethical and other constraints hampers the attainment of this goal.
The twin issues of historic overly conservative systematic interpretations and lack of molecular sequence data means many species identifications for clinostomids in Africa remain untested. For example, conservatively interpreting many records as being those of C. complanatum may well have concealed as-yet uncharacterised richness (Caffara et al. Reference Caffara, Locke, Echi, Halajian, Luus-Powell, Benini, Tedesco and Fioravanti2020). A handful of studies have molecularly validated the presence of this species in north Africa (Egypt) (Mahdy et al. Reference Mahdy, Abdelsalam, Abdel-Maogood, Shaalan and Salem2021, Salem et al. Reference Salem, Abdel-Maogood, Abdelsalam and Mahdy2021), but until parasitological assessments more broadly incorporate molecular sequencing protocols to aid identification, the status of ‘C. complanatum’ in Africa, particularly sub-Saharan Africa, remains uncertain.
Cryptogonimidae
Of the 80 recognised cryptogonimid genera, 30 are partly or wholly represented in freshwater fishes. Freshwater fish-infecting cryptogonimids have most strongly radiated in the Americas, with species of 17 genera so far recorded from that region. Species of seven genera have been reported from African freshwater systems: Acanthostomum Looss, 1899; Brientrema Dollfus, 1950; Grandifundilamena Bray, Kmentová & Georgieva in Kmentová, Bray, Koblmüller, Artois, De Keyzer, Gelnar, Vanhove & Georgieva, 2020; Gymnatrema Morozov, 1955; Neocladocystis Manter & Pritchard, 1969; Proctocaecum Baugh, 1957; and Tanganyikatrema Kmentová, Georgieva & Bray in Kmentová, Bray, Koblmüller, Artois, De Keyzer, Gelnar, Vanhove & Georgieva, 2020 (Table 3; Figure 7a–d). The genus Neocladocystis is the only one of these also represented outside Africa, with Neocladocystis intestinalis (Vaz, 1932) Manter & Pritchard, 1969 being found in South America. Other species, of the predominantly marine genera Siphodera Linton, 1910 and Siphoderina Manter, 1934, have occasionally been reported from estuarine or marine-adjacent freshwater systems, and are not considered further.
Table 3. List of records of freshwater cryptogonimid species from Africa. Hosts are fishes unless otherwise noted. Entries marked ‘*’ represent type-records from original descriptions
Notes:
a This species was proposed in a thesis and never formally published. It should therefore be regarded as invalid.
b It is unclear where the localities of ‘Maka’ and ‘Maka Lombo’ provided by Dollfus (Reference Dollfus1950) are. They could refer to the town of Kalombo, which is in the vicinity of the lakes of the Upemba Depression, where Brien (the collector of these specimens) was collecting at that time.
c Specimens were found in the bottom of jar containing mixed fish species; the type-host is therefore unknown.
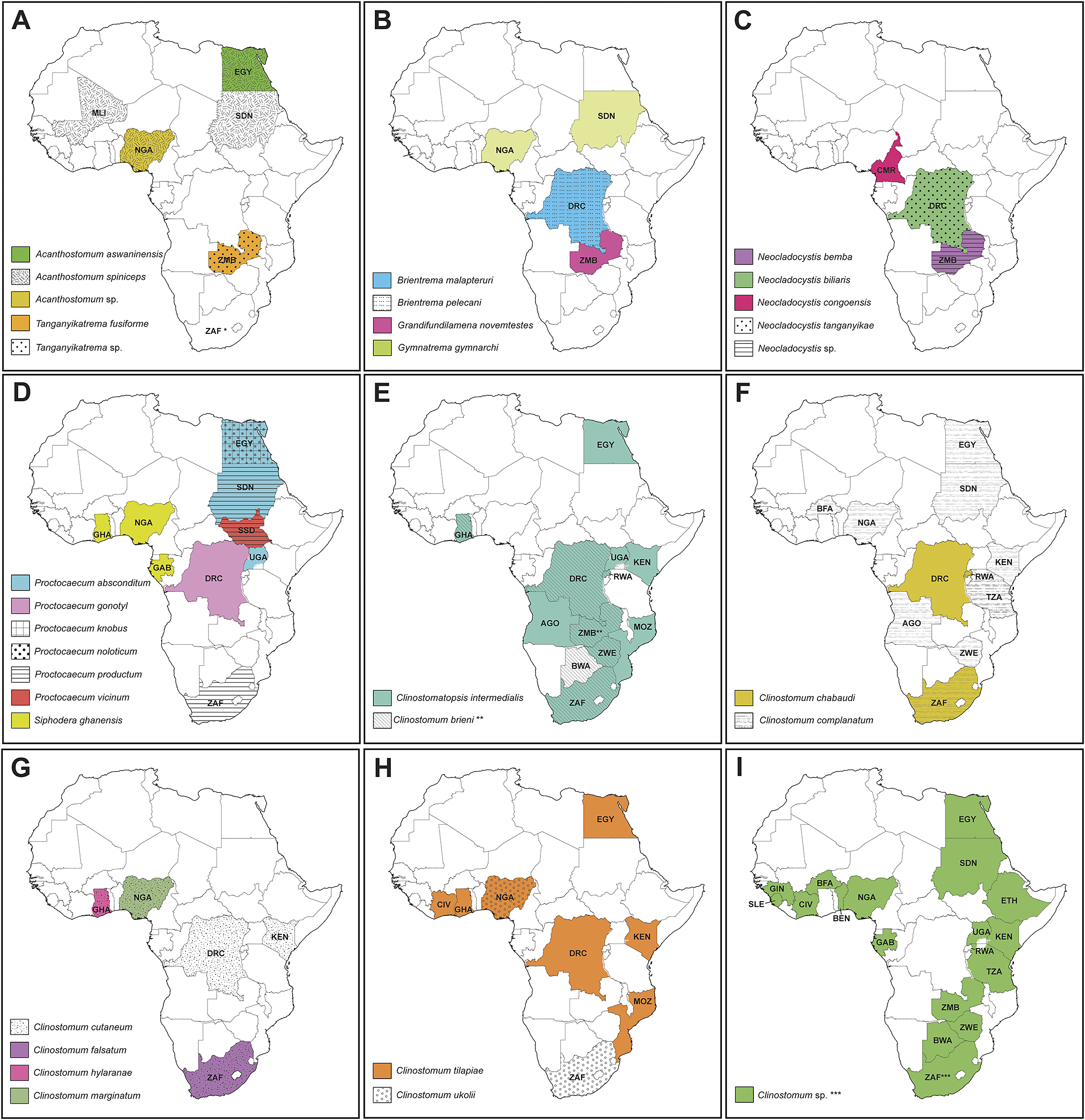
Figure 7. Maps depicting countries with records of species of the Cryptogonimidae (A–D) and Clinostomidae (E–I). Asterisk depicts countries with record data from the current study: * – first record of a species of Cryptogonimidae in South Africa; ** – first report of Clinostomum brieni from Zambia; *** – records of Clinostomum ‘morphotype 2’ and Clinostomum ‘morphotype 3’ from South Africa. AGO = Angola; BEN = Benin; BWA = Botswana; BFA = Burkina Faso; CMR = Cameroon; DRC = Democratic Republic of the Congo; EGY = Egypt; Ethiopia; GAB = Gabon; GHA = Ghana; GIN = Guinea; CIV = Ivory Coast; KEN = Kenya; MLI = Mali; MOZ = Mozambique; NGA = Nigeria; RW = Rwanda; SLE = Sierra Leone; ZAF = South Africa; SSD = South Sudan; SDN = Sudan; TZA = Tanzania; UGA = Uganda; ZMB = Zambia; ZWE = Zimbabwe.
Two species of Acanthostomum have been reported from Africa, including the type-species of the genus, Acanthostomum spiniceps (Looss, 1896), which was originally described (as Distomum spiniceps) from the bagrid catfish Bagrus bajad (Forsskål) (Bagridae) from the Egyptian Nile at Cairo (Looss Reference Looss1896) and has subsequently been reported from this and other bagrid species from this locality numerous times. This species was subsequently reported from catfishes from elsewhere in Africa: Lake Débo, Mali (Dollfus Reference Dollfus1932) and the Sudanese Nile (Khalil Reference Khalil1963), and from Dicentrarchus labrax (L.) (Moronidae) from Idku Lake, Egypt (El-Shahawi and Al-Bassel Reference El-Shahawi and Al-Bassel1992). Multiple authors have also reported it from Nile perch, Lates niloticus (L.) (Latidae) from the Egyptian Nile (Abdel-Gaber et al. Reference Abdel-Gaber, Abdel-Ghaffar, Mehlhorn, Al Quraishy, Morsy and Maher2018; Al-Ghamdi Reference Al-Ghamdi2018; Morsy et al. Reference Morsy, El-Fayoumi and Ali2013;), though one (Al-Ghamdi Reference Al-Ghamdi2018) appears to heavily plagiarise another (Morsy et al. Reference Morsy, El-Fayoumi and Ali2013) and should be disregarded. Acanthostomum spiniceps is a problematic species, having been redescribed multiple times (Issa Reference Issa1964; Looss Reference Looss1901; Moravec Reference Moravec1976; Morsy et al. Reference Morsy, El-Fayoumi and Ali2013) and also reported from marine localities in Europe and South America (Fernandes et al. Reference Fernandes, Pinto and Cohen2002; Pogoreltseva Reference Pogoreltseva1952a; Pogoreltseva Reference Pogoreltseva1952b). The species is quite morphologically variable (Moravec 1976), and despite its apparent familiarity, has never been molecularly sequenced. Hassan et al. (Reference Hassan, Khidr and Samak1990) described Acanthostomum saoudi Hassan, Khidr & Samak, 1990 from D. labrax from off the Egyptian coast. A further species, Acanthostomum aswaninensis Wannas, 1977 was described by Wannas (Reference Wannas1977) in a master’s dissertation and, as far as we are aware, was never formally published. Occasionally, studies have recognised this taxon (Lashien Reference Lashien1993; Taeleb and Lashein Reference Taeleb and Lashien2013) but we regard it as invalid.
The history of the taxonomy of Acanthostomum is inextricably linked to that of the genus Proctocaecum. The latter was proposed by Baugh (Reference Baugh1957), who transferred three species of Acanthostomum whose caeca open as ani close to the posterior extremity to this genus. Brooks (Reference Brooks1980) reassigned a further four Acanthostomum species to Proctocaecum and described one further species. Of the six Proctocaecum species recorded from Africa, one, P. absconditum (Looss, 1901), infects catfishes (Bagridae: Bagrus spp.), two [P. knobus (Issa, 1962) and P. niloticum (Issa, 1962)] infect Nile perch, and three [P. gonotyl (Dollfus, 1950), P. productum and P. vicinum (Odhner, Reference Odhner1902)] infect Nile crocodiles (Crocodylus niloticus Laurenti) as adults (Brooks Reference Brooks1980; Khalil Reference Khalil1963). A listing of Proctocaecum coronarium (Cobbold, 1861>) from Alligator sp. from Sudan by Gohar (Reference Gohar1934) is in error, referring to specimens recovered by Cobbold (Reference Cobbold1861) from an American alligator [Alligator mississippiensis (Duadin) (Reptilia: Alligatoridae)] which died in the London Zoological Society’s menagerie (i.e., London Zoo). Only P. productum has been recorded from southern Africa; Prudhoe and Hussey (Reference Prudhoe and Hussey1977) recovered this species from the intestine of C. niloticus from the Olifants River in South Africa. The remainder of the species were mostly recorded from north and central Africa in the Nile catchment.
Dollfus (Reference Dollfus1950), in his studies on trematodes collected from the Belgian Congo (now Democratic Republic of Congo), described the type-species of Brientrema, Brientrema pelecani Dollfus, 1950, from the Pink-backed Pelican, Pelecanus rufescens Gmelin (Aves: Pelecanidae); it has been noted that this was potentially a case of pseudo-parasitism (Miller and Cribb Reference Miller, Cribb, Gibson, Bray and Jones2008). The other species Brientrema malapteruri Dollfus, 1950 was described by the same author from the electric catfish Malapterurus electricus (Gmelin) (Malapteruridae) and the longsnout distichodus Distichodus lusosso Schilthuis (Distichodontidae) (Dollfus Reference Dollfus1950). Dollfus (Reference Dollfus1950) also described a third cryptogonimid species, Acanthochasmus gymnarchi Dollfus, 1950, from the Nile River by Omdurman, Sudan; this species infects the aba, Gymnarchus niloticus Cuvier (Gymnarchidae). The genus Gymnatrema was proposed by Morozov (Reference Morozov1955) to incorporate A. gymnarchi. A further species, Acanthostomum nigeri Zaidi & Khan, 1977, described from the marine pelagic carangid Parastromateus niger (Bloch) from off Karachi, Pakistan was transferred to Gymnatrema by Brooks (Reference Brooks1980); its status is uncertain, though this classification is almost certainly wrong.
The genus Neocladocystis was proposed by Manter and Pritchard (Reference Manter and Pritchard1969) to separate Cladocystis intestinalis Vaz, 1932 and Cladocystis tanganyikae Prudhoe, 1951 from the then-only other species of that genus, Cladocystis trifolium (Braun, 1901), on the basis of possessing tegumental spines, larger oral and ventral suckers, less extensive vitelline follicles, and unbranched arms of the excretory vesicle; this latter species is now recognised as belonging to the family Opisthorchiidae. Prudhoe (Reference Prudhoe1951) described Neocladocystis tanganyikae from ‘a small bay south of Cape Tembwe’ on the Congolese shore of Lake Tanganyika. The two specimens which formed the basis of description were recovered from the residue of a jar containing several species of cichlid fishes mixed with the killifish Lamprichthys tanganicanus (Boulenger) (Procatopodidae), making the host species impossible to ascertain. Manter and Pritchard (Reference Manter and Pritchard1969) described Neocladocystis congoensis Manter & Pritchard, 1969 from the claroteid catfish Parauchenoglanis monkei (Keilhack) [as Parauchenoglanis guttatus (Lönnberg)], from Ebogo, Cameroon. Most recently, Kmentová et al. (Reference Kmentová, Bray, Koblmüller, Artois, De Keyzer, Gelnar, Vanhove and Georgieva2020) described two species of Neocladocystis, Neocladocystis bemba Georgieva, Kmentová & Bray in Kmentová, Bray, Koblmüller, Artois, De Keyzer, Gelnar, Vanhove & Georgieva, 2020 and Neocladocystis biliaris Georgieva, Kmentová & Bray in Kmentová, Bray, Koblmüller, Artois, De Keyzer, Gelnar, Vanhove & Georgieva, 2020 from three species of Lates Cuvier (Latidae) from Lake Tanganyika. A third species of Neocladocystis was noted by the same authors from Lates angustifrons Boulenger from the same lake on the basis of molecular sequence data, but was not described due to there being only one immature specimen. Kmentová et al. (Reference Kmentová, Bray, Koblmüller, Artois, De Keyzer, Gelnar, Vanhove and Georgieva2020) also proposed Grandifundilamena and Tanganyikatrema for two latid-infecting species from Lake Tanganyika, Grandifundilamena novemtestes Bray, Kmentová & Georgieva in Kmentová, Bray, Koblmüller, Artois, De Keyzer, Gelnar, Vanhove & Georgieva, 2020 and Tanganyikatrema fusiforme Kmentová, Georgieva & Bray in Kmentová, Bray, Koblmüller, Artois, De Keyzer, Gelnar, Vanhove & Georgieva, 2020. A second species of Tanganyikatrema was also reported from L. angustifrons from Lake Tanganyika, but again, was not described due to a lack of material.
Biogeography
Information regarding freshwater cryptogonimids in Africa is too sparse to draw any biogeographic inferences; hence the focus of this aspect of discussion is on the clinostomids. We do note with interest the record by Prudhoe and Hussey (Reference Prudhoe and Hussey1977) of P. productum from northeastern South Africa, a species otherwise only known from the Sudanese Nile. It is hoped future studies in these regions will recollect these taxa and generate molecular sequence data.
In accordance with findings on the global clinostomid fauna in previous studies, results from the current study support the separation between clinostomid species of the ‘Old World’ (Afrotropic, Indo-Malayan, and Palearctic realms) and ‘New World’ (Nearctic and Neotropic realms). This divide is explained by the isolation of avian definitive hosts by oceanic barriers which are rarely crossed by the definitive hosts of clinostomids (see Caffara et al. Reference Caffara, Davidovich, Falk, Smirnov, Ofek, Cummings, Gustinelli and Fioravanti2014; Caffara et al. Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017; Locke et al. Reference Locke, Caffara, Marcogliese and Fioravanti2015). Of the Old World clinostomid fauna, affinities between the Afrotropical, Indomalayan, and Palearctic realms are apparent. The definitive hosts of Old World clinostomids are piscivorous birds of the families Anhingidae (darters), Ardeidae (herons), Pelecanidae (pelicans), and Phalacrocoracidae (cormorants). Birds of these families show a range of dispersal abilities, with some species endemic to single continents but able to undertake infra-continental migrations, while others are naturally distributed and move across multiple biogeographic realms (Afrotropic, Indo-Malayan, and Palearctic). This high dispersal ability among piscivorous bird species undoubtedly explains the wide distributions of some clinostomid species. For example, Cl. intermedialis has been demonstrated to range across most of Africa (with molecularly verified specimens from South Africa, Kenya, and Egypt), the Middle East (Israel), and southern Europe (Italy), and C. tilapiae has been reported (with supporting molecular sequence data) from Nigeria and Turkey. Most dramatically, C. complanatum has been reported from throughout the Old World, with records supported by molecular sequence data from Africa (Kenya, Egypt), Europe (France, Italy, Romania), western Asia (Iran, Turkey), and east Asia (China, Japan), as well as North America (USA, Canada). The definitive host of C. brieni is the Goliath Heron, Ardea goliath. This heron species is widespread and common throughout sub-Saharan Africa and is also found patchily in the Middle East and across the north of the Indian sub-continent (Martínez-Vilalta et al. Reference Martínez-Vilalta, Motis, Christie and Kirwan2020). Although movements in Asia and between continents are unknown, nomadic movements are known within the African populations (Martínez-Vilalta et al. Reference Martínez-Vilalta, Motis, Christie and Kirwan2020). Such movement dynamics are typical of other African bird species which are hosts to Clinostomum spp.
It is appropriate to include an additional consideration, that of intermediate host specificity. Several species of Clinostomum show stenoxenous host-specificity at the second intermediate stage. For example, C. brieni has only ever been reported from clariid catfishes, having been reported from a number of clariids in Africa and, putatively, India. Other clinostomid species, e.g., C. ukolii and Clinostomum ‘morphotype 3’ of Caffara et al. (Reference Caffara, Locke, Echi, Halajian, Benini, Luus-Powell, Tavakol and Fioravanti2017), infect catfishes across several families; still others, e.g., C. cutaneum, infect several species of cichlid fishes; and additional ones, e.g., Cl. intermedialis and C. tilapiae, infect both catfishes and cichlids but no other fish species. We demonstrated that Clinostomum ‘morphotype 2’, hitherto only known from mormyrid fishes, also infected mochokid catfishes. Clinostomum chabaudi Vercammen-Grandjean, 1960 has been demonstrated to have an interesting form of euryxenicity: metacercariae of this species infect both fishes and frogs as second intermediate hosts (Sinsch et al. Reference Sinsch, Balczun, Scheid and Dehling2021a; Sinsch et al. Reference Sinsch, Dehling, Scheid and Balczun2021b). Nevertheless, it is predicted that most Clinostomum species are stenoxenous, infecting at most two fish families or orders.
The various African freshwater fish lineages showcase a range of particular biogeographic histories, including ancient lineages demonstrating Gondwanan shared ancestry (e.g., lungfishes), radiations which subsequently dispersed to Asia (e.g., knifefishes), and lineages which originated from Asia and subsequently invaded Africa (e.g., synbranchiform air-breathers) (Harrington et al. Reference Harrington, Kolmann, Day, Faircloth, Friedman and Near2023; Inoue et al. Reference Inoue, Kumazawa, Miya and Nishida2009; Skelton Reference Skelton, Lévêque, Bruton and Ssentongo1988). The circumstances inherent to each lineage relevant to us are too complex to recount in detail here. To cite just one example, the clariid family has radiated widely across the Afrotropical and Indo-Malayan realms, having originated from central Asia ~50 mya and subsequently radiated into southeast Asia and Africa during the Lower Miocene (~15 mya) (Agnese and Teugels Reference Agnese and Teugels2005; Otero and Gayet Reference Otero and Gayet2001). Based on the observed genetic divergences of repeated molecular sequencing analyses (see this study; Briosio-Aguilar et al. Reference Briosio-Aguilar, Pinto, Rodríguez-Santiago, López-García, García-Varela and Pérez-Ponce de León2018; Caffara et al. Reference Caffara, Locke, Halajian, Luus-Powell, Benini, Tedesco, Kasembele and Fioravanti2019), it is most plausible that the Indian record of C. brieni instead represents a close relative of C. brieni that has allopatrically speciated following the Pangaean and subsequent Gondwanan breakup (Günther Reference Günther1880; Paugy et al. Reference Paugy, Lévêque and Otero2017; Skelton Reference Skelton2024), with their common ancestor arising from central Asia and diverging ~15 mya. This hypothesis will require further validation through more comprehensive molecular sequence analyses incorporating more data from the Indian sub-continent, as well as re-assessment of other putative Indo-Asian species hitherto regarded as species of Clinostomoides – C. baughi (Pandey, 1988), C. chauhani (Pandey, 1971), C. dollfusi (Agrawal, 1959), C. meerutensis (Pandey & Tyagi, 1986), C. ophicephali Tubangui & Masilungan, 1944, C. pandeyii (Singh & Sharma, 1994), and C. rai (Pandey & Agrawal, 2013). All these species are currently considered species inquirendae but share several morphological and ecological features with C. brieni, particularly genital complex configuration and affinities to clariid intermediate hosts (Caffara et al. Reference Caffara, Locke, Halajian, Luus-Powell, Benini, Tedesco, Kasembele and Fioravanti2019; Pandey and Agrawal Reference Pandey and Agrawal2013).
We predict that a combination of highly dispersive definitive hosts and relatively low intermediate host specificity have served to limit speciation among African clinostomid species. Commonalities between African and other fauna may be explained by dispersal of, and linkages (or absence thereof) between, hosts both ancient (e.g., pre- and post-Gondwanan separation of Africa and India ~148 mya) and relatively recent (e.g., invasion of the continent by Asiatic fish lineages such as clariid catfishes ~15 mya). The presence of a clade of Australian Clinostomum species nested among African taxa is interesting. No definitive conclusions can be made on definitive or intermediate host specificity; however, it would seem that the two realms share closely related species sharing intermediate hosts across a broad geographic range and at least two definitive hosts spanning four biogeographic realms (Matthews and Cribb Reference Matthews and Cribb1998; Shamsi et al. Reference Shamsi, Barton, Day, Masiga, Zhu and McLellan2021a; Shamsi et al. Reference Shamsi, Day, Zhu, McLellan, Barton, Dang and Nowak2021b).
Other affinities to intermediate hosts are apparent based on available data for C. cutaneum, C. tilapiae, Cl. intermedialis (Cichlidae), C. ukolii (Mochokidae) (Table 2) and has been observed at the family level for Nearctic and Neotropical species (see Pérez-Ponce de León et al. Reference Pérez-Ponce de León, García-Varela, Pinacho-Pinacho, Sereno-Uribe and Poulin2016). Clarity on the diversity, affinities, and geographic distribution of clinostomids in the Afrotropical realm compared to the adjacent and geographically connected Indo-Malayan and Palearctic realms await more intensive sampling across geographic ranges and host taxa.
State of work in Africa and future directions
The preponderance of African freshwater cryptogonimid records are biased towards the north of the continent. Most records have been made and species described from the Nile catchment encompassing Egypt and Sudan, west Africa, and the Great Rift Lakes (Table 3). The only records of freshwater cryptogonimids from central and southern Africa include the handful arising from Dollfus’s work in what is now the southeastern DRC (see Dollfus Reference Dollfus1950) and a single record of P. productum from the Olifants River in South Africa (Prudhoe and Hussey Reference Prudhoe and Hussey1977).
The geographical spread of African Clinostomum records is more even, with numerous reports from all regions of Africa including southern Africa (southern DRC, Mozambique, South Africa, Zambia, and Zimbabwe) (see Table 2; Figure 7e–i). This is likely due to several reasons: first, that clinostomids infect fishes of major subsistence importance in Africa (cichlids such as tilapia, as well as bagrid, clariid, and mochokid catfishes); second, being relatively large, brightly coloured, and often encysting on external surfaces (fins, gills, and lining of the mouth), are easily observed during parasitological assessments; and third, are regarded as being of fisheries significance, as their presence often leads to decline in host condition, as well as rejection of product at market (Kabunda and Sommerville Reference Kabunda and Sommerville1984). Species of Clinostomum do have some zoonotic potential and have been reported causing infections in humans (Hara et al. Reference Hara, Miyauchi, Tahara and Yamashita2014; Park et al. Reference Park, Kim, Joo and Kim2009), though these records predominantly originate from regions where eating fresh, raw, freshwater fish is common, a practice that is not prevalent in most of Africa.
The vast majority of clinostomid records in Africa are of second intermediate stages. By comparison, understanding of African clinostomid first intermediate stages is poor; only one first intermediate stage infection, that of Euclinostomum heterostomum (Rudolphi, 1809), has ever been reported, from a bulinid snail, Bulinus globosus (Morelet) from western Nigeria (Dönges Reference Dönges1974). Knowledge of freshwater cryptogonimid intermediate stages is even poorer. As far as we can tell, our study is the first report of freshwater cryptogonimid metacercariae from Africa. This is likely due to the fact that these metacercariae are very small, encysting within fin rays and membranes, and are hence easily overlooked in cursory parasitological assessments. Furthermore, although several life cycles of freshwater cryptogonimids are fully known elsewhere (e.g., Cribb Reference Cribb1986; Ostrowski de Núñez and Gil de Pertierra Reference Ostrowski de Núñez and Gil de Pertierra1991; Sulieman et al. Reference Sulieman, Pengsakul, Guo, Huang and Peng2014; Vélez-Sampedro et al. Reference Vélez-Sampedro, Uruburu and Lenis2022), no first intermediate-stage infections have yet been reported from the continent.
In general, the vast majority of freshwater trematode records in Africa are of intermediate stages, many of which are not identified beyond the family or genus level. Most of these records are not substantiable and will never be relatable to adults, as relatively few studies accession specimens in public collections or provide any more than cursory identifications or overviews. The use of molecular sequence data has been invaluable for disentangling the systematic relationships of freshwater trematode species and will continue to be so, particularly in cases where fine-scale morphological differences are ambiguous or elusive. However, the inability to relate metacercarial specimens from many parts of the world with adult specimens, especially those whose definitive hosts are in higher vertebrates, will continue to hinder resolving the identities and systematics of many species. Nevertheless, we are hopeful that the increasing awareness of the value of molecular sequencing for aiding identification and understanding parasite systematics on the African continent will greatly expand our ability to account for freshwater parasite richness.
Supplementary material
The supplementary material for this article can be found at http://doi.org/10.1017/S0022149X2510045X.
Acknowledgements
The authors acknowledge Machaya Chomba, Kakoma Chinyawezhi (WWF, Zambia), local liaison Fidelis Mutukelwa, Namunda Namunda (Mongu Harbour Master, Zambia), and former SAIAB colleagues Craig Rennie, Dr Takudzwa Comfort Madzivanzira, and Dr Josie South for assistance in obtaining fish in Zambia. The Ministry of Fisheries and Livestock (Department of Fisheries, Mongu, Zambia) and World Wide Fund for Nature (WWF, Zambia) are thanked for their support and permission for joint research in the Upper Zambezi Basin, Zambia. Former and current Water Research Group members including Dr Aline Angelina Costa, Dr Iva Přikrylová, Dr Rodrigo Narciso Bravin, Angela Minnie, and Tshenolo Masilo are acknowledged for assistance in collection of parasitic material. Maditaba Meltaf from the Margaret Smith Library (SAIAB) and Prof Maxwell Barson are thanked for aiding in literature acquisition from across Africa. This is publication number 975 from the NWU Water Research Group.
Financial support
NJS, AC, WJL-P conceptualising, funding acquisition, and management of the larger project (REFRESH—FBIP-211006643719) that this output resulted from. MT collected material, analysed data; RQYY compiled distribution records and illustrations. MT and RQYY wrote the original draft of the manuscript. All authors reviewed and edited the final draft of the manuscript. The use of infrastructure and equipment provided by the NWU-WRG and the NRF-SAIAB Research Platform, and funding channelled through the NRF-SAIAB Institutional Support system is acknowledged. This work forms part of research and researchers supported by the National Research Foundation (NRF) – South African Research Chairs Initiative of the Department of Science and Innovation (DSI) (Inland Fisheries and Freshwater Ecology, Grant no. 110507) and the REFRESH project funded by the Foundational Biodiversity Initiative Programme (FBIP) of the National Research Foundation (NRF) of South Africa (Grant Number 138573). Opinions, findings, conclusions, and recommendations expressed are those of the authors, and the NRF accepts no liability whatsoever in this regard. MT acknowledges funding from NRF-Department of Science and Innovation (DSI) Professional Development Programme (Grant UID 144831) and RQYY funding from the North-West University (NWU) Postdoctoral fellowship programme. WJL-P and WJS acknowledge funding from the DSI-NRF SARChI Chair in Ecosystem Health (Grant UID 101054).
Competing interests
None.
Ethical standard
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals. Ethical approval for the use of animal for research purposes and research permits for fish collection was obtained prior to sampling from the North-West University AnimCare Research Ethics Committees [ethics numbers: NWU-00156-18-A5 (MTruter PhD), NWU-00781-22-A5 (REFRESH)]. Permits for the collection of animals for the purpose of research were obtained from the Department of Economic Development, Environment and Tourism (permit no. ZA/LP/116078) and access to Tzaneen Dam was obtained from the Department Water and Sanitation, Limpopo, South Africa. The Ministry of Fisheries and Livestock (Department of Fisheries, Mongu, Zambia) and World Wide Fund for Nature (WWF, Zambia) coordinated research permission in Zambia.