Crossref Citations
This article has been cited by the following publications. This list is generated based on data provided by
Crossref.
Vilor-Tejedor, Natàlia
Alemany, Silvia
Cáceres, Alejandro
Bustamante, Mariona
Pujol, Jesús
Sunyer, Jordi
and
González, Juan R.
2018.
Strategies for integrated analysis in imaging genetics studies.
Neuroscience & Biobehavioral Reviews,
Vol. 93,
Issue. ,
p.
57.
Bougeard, Stéphanie
Niang, Ndèye
Verron, Thomas
and
Bry, Xavier
2018.
Current multiblock methods: Competition or complementarity? A comparative study in a unified framework.
Chemometrics and Intelligent Laboratory Systems,
Vol. 182,
Issue. ,
p.
131.
Garali, Imene
Adanyeguh, Isaac M
Ichou, Farid
Perlbarg, Vincent
Seyer, Alexandre
Colsch, Benoit
Moszer, Ivan
Guillemot, Vincent
Durr, Alexandra
Mochel, Fanny
and
Tenenhaus, Arthur
2018.
A strategy for multimodal data integration: application to biomarkers identification in spinocerebellar ataxia.
Briefings in Bioinformatics,
Vol. 19,
Issue. 6,
p.
1356.
Jun, Inyoung
Choi, Wooree
and
Park, Mira
2018.
Multi-block Analysis of Genomic Data Using Generalized Canonical Correlation Analysis.
Genomics & Informatics,
Vol. 16,
Issue. 4,
p.
e33.
Min, Eun Jeong
Safo, Sandra E
Long, Qi
and
Stegle, Oliver
2019.
Penalized co-inertia analysis with applications to -omics data.
Bioinformatics,
Vol. 35,
Issue. 6,
p.
1018.
Guigui, N.
Philippe, C.
Gloaguen, A.
Karkar, S.
Guillemot, V.
Lofstedt, T.
and
Frouin, V.
2019.
Network Regularization in Imaging Genetics Improves Prediction Performances and Model Interpretability on Alzheimer’s Disease.
p.
1403.
Karkar, S.
Gloaguen, A.
Le Guen, Y.
Pierre-Jean, M.
Dandine-Roulland, C.
Le Floch, E.
Philippe, C.
Tenenhaus, A.
and
Frouin, V.
2019.
Multivariate Haplotype Analysis Of 96 Sulci Opening For 15,612 UK-Biobank Sujects.
p.
392.
Smolinska, Agnieszka
Engel, Jasper
Szymanska, Ewa
Buydens, Lutgarde
and
Blanchet, Lionel
2019.
Data Fusion Methodology and Applications.
Vol. 31,
Issue. ,
p.
51.
Leung, Marcus H. Y.
Tong, Xinzhao
Bastien, Philippe
Guinot, Florent
Tenenhaus, Arthur
Appenzeller, Brice M. R.
Betts, Richard J.
Mezzache, Sakina
Li, Jing
Bourokba, Nasrine
Breton, Lionel
Clavaud, Cécile
and
Lee, Patrick K. H.
2020.
Changes of the human skin microbiota upon chronic exposure to polycyclic aromatic hydrocarbon pollutants.
Microbiome,
Vol. 8,
Issue. 1,
Min, Eun Jeong
and
Long, Qi
2020.
Sparse multiple co-Inertia analysis with application to integrative analysis of multi -Omics data.
BMC Bioinformatics,
Vol. 21,
Issue. 1,
Varghese, Rency S.
Zhou, Yuan
Barefoot, Megan
Chen, Yifan
Di Poto, Cristina
Balla, Abdalla Kara
Oliver, Everett
Sherif, Zaki A.
Kumar, Deepak
Kroemer, Alexander H.
Tadesse, Mahlet G.
and
Ressom, Habtom W.
2020.
Identification of miRNA-mRNA associations in hepatocellular carcinoma using hierarchical integrative model.
BMC Medical Genomics,
Vol. 13,
Issue. 1,
Hwang, Heungsun
and
Cho, Gyeongcheol
2020.
Global Least Squares Path Modeling: A Full-Information Alternative to Partial Least Squares Path Modeling.
Psychometrika,
Vol. 85,
Issue. 4,
p.
947.
Wozniak, Jacob M.
Mills, Robert H.
Olson, Joshua
Caldera, J.R.
Sepich-Poore, Gregory D.
Carrillo-Terrazas, Marvic
Tsai, Chih-Ming
Vargas, Fernando
Knight, Rob
Dorrestein, Pieter C.
Liu, George Y.
Nizet, Victor
Sakoulas, George
Rose, Warren
and
Gonzalez, David J.
2020.
Mortality Risk Profiling of Staphylococcus aureus Bacteremia by Multi-omic Serum Analysis Reveals Early Predictive and Pathogenic Signatures.
Cell,
Vol. 182,
Issue. 5,
p.
1311.
Fan, Ziling
Zhou, Yuan
and
Ressom, Habtom W.
2020.
MOTA: Network-Based Multi-Omic Data Integration for Biomarker Discovery.
Metabolites,
Vol. 10,
Issue. 4,
p.
144.
Górecki, Tomasz
Krzyśko, Mirosław
and
Wołyński, Waldemar
2020.
Generalized canonical correlation analysis for functional data.
Biometrical Letters,
Vol. 57,
Issue. 1,
p.
1.
2020.
The Microbiome in Health and Disease.
Vol. 171,
Issue. ,
p.
309.
Vrijheid, Martine
Basagaña, Xavier
Gonzalez, Juan R.
Jaddoe, Vincent W. V.
Jensen, Genon
Keun, Hector C.
McEachan, Rosemary R. C.
Porcel, Joana
Siroux, Valerie
Swertz, Morris A.
Thomsen, Cathrine
Aasvang, Gunn Marit
Andrušaitytė, Sandra
Angeli, Karine
Avraam, Demetris
Ballester, Ferran
Burton, Paul
Bustamante, Mariona
Casas, Maribel
Chatzi, Leda
Chevrier, Cécile
Cingotti, Natacha
Conti, David
Crépet, Amélie
Dadvand, Payam
Duijts, Liesbeth
van Enckevort, Esther
Esplugues, Ana
Fossati, Serena
Garlantezec, Ronan
Gómez Roig, María Dolores
Grazuleviciene, Regina
Gützkow, Kristine B.
Guxens, Mònica
Haakma, Sido
Hessel, Ellen V. S.
Hoyles, Lesley
Hyde, Eleanor
Klanova, Jana
van Klaveren, Jacob D.
Kortenkamp, Andreas
Le Brusquet, Laurent
Leenen, Ivonne
Lertxundi, Aitana
Lertxundi, Nerea
Lionis, Christos
Llop, Sabrina
Lopez-Espinosa, Maria-Jose
Lyon-Caen, Sarah
Maitre, Lea
Mason, Dan
Mathy, Sandrine
Mazarico, Edurne
Nawrot, Tim
Nieuwenhuijsen, Mark
Ortiz, Rodney
Pedersen, Marie
Perelló, Josep
Pérez-Cruz, Míriam
Philippat, Claire
Piler, Pavel
Pizzi, Costanza
Quentin, Joane
Richiardi, Lorenzo
Rodriguez, Adrian
Roumeliotaki, Theano
Sabin Capote, José Manuel
Santiago, Leonardo
Santos, Susana
Siskos, Alexandros P.
Strandberg-Larsen, Katrine
Stratakis, Nikos
Sunyer, Jordi
Tenenhaus, Arthur
Vafeiadi, Marina
Wilson, Rebecca C.
Wright, John
Yang, Tiffany
and
Slama, Remy
2021.
Advancing tools for human early lifecourse exposome research and translation (ATHLETE).
Environmental Epidemiology,
Vol. 5,
Issue. 5,
p.
e166.
Rebei, Amine
Alentorn, Agusti
Chegraoui, Hamza
Frouin, Vincent
and
Philippe, Cathy
2021.
Contribution Of Imaging-Genetics To Overall Survival Prediction Compared To Clinical Status For Pcnsl Patients.
p.
832.
Avants, Brian B.
Tustison, Nicholas J.
and
Stone, James R.
2021.
Similarity-driven multi-view embeddings from high-dimensional biomedical data.
Nature Computational Science,
Vol. 1,
Issue. 2,
p.
143.
Tanabe, Kazuhiro
Hayashi, Chihiro
Katahira, Tomoko
Sasaki, Katsuhiko
and
Igami, Ko
2021.
Multiblock metabolomics: An approach to elucidate whole-body metabolism with multiblock principal component analysis.
Computational and Structural Biotechnology Journal,
Vol. 19,
Issue. ,
p.
1956.
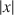 , x2\documentclass[12pt]{minimal}\usepackage{amsmath}\usepackage{wasysym}\usepackage{amsfonts}\usepackage{amssymb}\usepackage{amsbsy}\usepackage{mathrsfs}\usepackage{upgreek}\setlength{\oddsidemargin}{-69pt}\begin{document}$$x^{2}$$\end{document}
, x2\documentclass[12pt]{minimal}\usepackage{amsmath}\usepackage{wasysym}\usepackage{amsfonts}\usepackage{amssymb}\usepackage{amsbsy}\usepackage{mathrsfs}\usepackage{upgreek}\setlength{\oddsidemargin}{-69pt}\begin{document}$$x^{2}$$\end{document}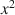 or x4\documentclass[12pt]{minimal}\usepackage{amsmath}\usepackage{wasysym}\usepackage{amsfonts}\usepackage{amssymb}\usepackage{amsbsy}\usepackage{mathrsfs}\usepackage{upgreek}\setlength{\oddsidemargin}{-69pt}\begin{document}$$x^{4}$$\end{document}
or x4\documentclass[12pt]{minimal}\usepackage{amsmath}\usepackage{wasysym}\usepackage{amsfonts}\usepackage{amssymb}\usepackage{amsbsy}\usepackage{mathrsfs}\usepackage{upgreek}\setlength{\oddsidemargin}{-69pt}\begin{document}$$x^{4}$$\end{document}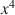 and shrinkage constants 0 or 1, many multiblock component methods are recovered.
and shrinkage constants 0 or 1, many multiblock component methods are recovered.
