Management Implications
Hybridization between native and introduced subspecies of Phragmites (reed) is a significant concern for natural resource managers in North America. The polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) assays based on the NRT2 and PaGT4 markers described in this report add to the tools available to differentiate North American native from introduced Phragmites and provide a simple and rapid method to screen for hybrids between them. We also demonstrate that the commonly used chloroplast DNA (cpDNA) PCR-RFLP tests do not differentiate between all newly introduced lineages, so investigators should use nucleotide sequencing in such cases.
Introduction
In North America, the introduced subspecies of the common reed [Phragmites australis (Cav.) Trin. ex Steud.] greatly concerns resource managers, because it can dominate ecosystems and form dense monocultures. In contrast, the native subspecies [Phragmites australis (Cav.) Trin. ex Steud. ssp. americanus Saltonst., P.M. Peterson & Soreng] is not invasive and should not be targeted for control. Like many invasive organisms, Phragmites raises concern over the possibility of hybridization between native and introduced populations, which can lead to a more aggressive invasive population and/or extinction of the native form (Saltonstall et al. Reference Saltonstall, Castillo and Blossey2014). Thus, natural resource managers need to be able to identify both the introduced subspecies and potential hybrids between native and introduced populations.
Researchers have observed that hybrids in the field display phenotypes in common with both parental populations (Saltonstall et al. Reference Saltonstall, Castillo and Blossey2014; Williams et al. Reference Williams, Lambert, Long and Saltonstall2019), but morphology alone cannot differentiate a potential hybrid from an atypical member of either parental subspecies. Although researchers can typically distinguish P. australis subspecies by a combined set of morphological traits, within each subspecies there is a range of phenotypes and the ranges can overlap. Additionally, traits used to distinguish between groups exhibit regional differences (Lambert et al. Reference Lambert, Saltonstall, Long and Dudley2016; McTavish et al. Reference McTavish, Smith, Mechanda, Smith and Bourchier2023). Therefore, atypical specimens of either subspecies may be misidentified as possible hybrids.
Managers focused on detection and eradication of invasive Phragmites frequently employ molecular tools to assist in identifying individual populations to subspecies. Chloroplast DNA (cpDNA) haplotypes that differentiate the three most common subspecies present in North America can be clearly assayed by a simple but powerful polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) test (Saltonstall Reference Saltonstall2003a). However, to identify hybrids, one must use genetic markers based on nuclear DNA polymorphisms. The original markers described by Saltonstall and used for these purposes were microsatellite markers. Because two of the subspecies in question share alleles for the microsatellites, a panel of multiple markers must be used to identify likely hybrids that have a set of marker alleles shared by but distinct from the two parental species (Saltonstall Reference Saltonstall2003b). Now that there is a reference genome for Phragmites (Oh et al. Reference Oh, Kowalski, Quach, Wijesinghege, Tanford, Dassanayake and Clay2022), researchers can also develop panels of single-nucleotide polymorphisms (SNPs) and genotype specimens by genomic sequencing.
We previously reported a nuclear gene marker, NRT2-Δ4, which users can genotype with a PCR-RFLP assay for a simple screen for potential hybrids between the North American native and introduced haplotype M (Wendell et al. Reference Wendell, Huang, Gryspeerd and Freeland2021). At the time, the range of samples was limited, and we did not address other introduced haplotypes or the Gulf Coast type [Phragmites australis (Cav.) Trin. ex Steud. ssp. berlandieri (E. Fourn.) Saltonst. & Hauber]. Here we report findings for this marker to show that it distinguishes native P. australis ssp. americanus from multiple introduced haplotypes and the Gulf Coast type. We also describe a PCR-RFLP assay for a diagnostic SNP linked to the PaGT4 locus previously identified by Saltonstall (Reference Saltonstall2003b). The two markers together can both identify the P. australis ssp. americanus subspecies and screen for hybrids. In the course of this work, we have also identified the first case of P.a. australis haplotype AS in North America.
Materials and Methods
Plant Genetic Materials
The Phragmites samples used in this work are listed in Table 1. Both North American native and introduced populations have multiple lineages that are distinguished by cpDNA haplotypes (Saltonstall Reference Saltonstall2016), and the samples used in this study were selected to represent a wide range of haplotypes and geographic locations. Those noted as “this work” were collected by the Washington State Department of Agriculture (WSDA), Plant Pathology and Molecular Diagnostics Laboratory. These include the first identification of haplotype AS in North America and three herbarium specimens collected from the population of haplotype AS on Sand Island, WA, and submitted to the University of Washington Herbarium (accession nos. 297-461762, 298-461763, 299-461764). Other samples were donated by researchers (Lambert Lab). The remaining samples were submitted to the Wendell lab for genetic identification by natural resources managers or private individuals with permission given to use them in further study. Every sample was collected in a separate envelope to avoid cross-contamination and upon receipt was stored in a sealed container with desiccant. Except for the haplotype P sample, at least two samples of the same haplotype from the same site were tested.
Table 1. Haplotypes and sampling locations of Phragmites samples used in this study.
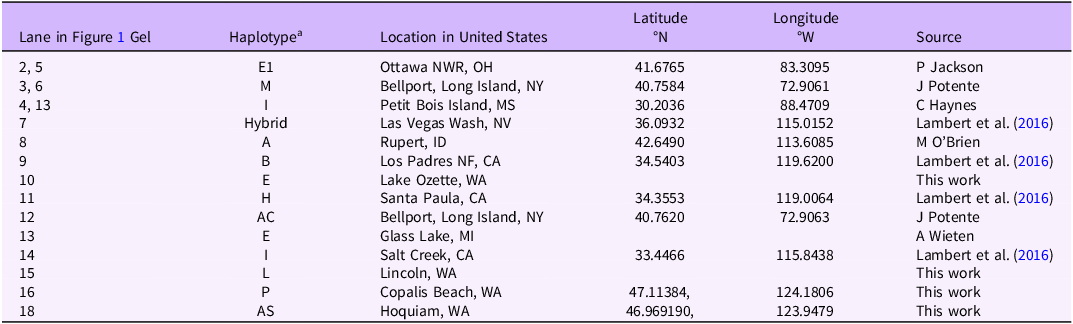
a Haplotypes A, B, AC, E, E1, and H are subspecies P. australis ssp. americanus. Haplotypes AS, L, M, and P are introduced types. Haplotype I is subspecies P. australis ssp. berlandieri (Gulf Coast type).
DNA was extracted from leaf tissue using commercially available kits, either a DNeasy Plant Mini Kit (Qiagen, Valencia, CA) in the WSDA Lab, or a Nucleospin Plant II Mini Kit (Macherey-Nagel, Allentown, PA) in the Wendell lab. Both kits were found to be satisfactory in extraction of genomic DNA from Phragmites tissue.
Chloroplast Haplotype Determination
To identify cpDNA haplotypes, we performed nucleotide sequencing of the trnT-trnL and rbcL-psaI regions using previously described primers (Saltonstall Reference Saltonstall2002). In some cases, to increase the quality of sequencing reads, we designed alternative primers that were specific to Phragmites and had a higher GC content: primers trnT-Pa CCGATGACCCTCGCATTACA and trnL-Pa TAGCGTCTACCGATTTCGCC for the trnT-trnL region and rbcL-Pa GGGTGGCTTTAGAAGCCTGT and psaI-Pa GAGAAAATTTGGCCCCGTCG for the rbcL-psaI region. These new primers were used for PCR and sequencing under the same conditions as the traditional primers. After PCR, amplicons were prepared for sequencing using ExoSAP-II Express PCR Product Clean-up Reagent (Thermo Fisher, Waltham, MA). Sequencing reactions were performed using the BigDye Terminator v. 3.1 Cycle Sequencing Kit, purified using the BigDye XTerminator Purification Kit, and read in a SeqStudio Genetic Analyzer (Thermo Fisher).
To identify cpDNA haplotypes, we aligned sequences to reference sequences following the guide provided by Saltonstall (Reference Saltonstall2016).
Genotyping the NRT2-Δ4 Marker
The genotype for the NRT2-Δ4 marker was assayed by PCR-RFLP using the same PCR and electrophoresis conditions as previously described by Lindsay et al. (Reference Lindsay, Freeland, Gong, Guan, Harms, Kowalski, Lance, Oh, Sartain and Wendell2023a).
Genotyping an SNP Linked to the PaGT4 Microsatellite
We developed a PCR-RFLP test to determine genotype for an C/A SNP at the PaGT4 locus previously identified by Saltonstall (Reference Saltonstall2003b). The “C” allele in the P. australis ssp. americanus subspecies fortuitously creates the sequence CGCG, which is cut by the restriction endonuclease BstUI (and its isoschizomers AccII, BspFNI, Bsh1236I, BstFNI, and MvnI), while the “A” allele is uncut. PCR was performed using the primers PaGT4-F TGCTCCCTGCCAGTTTCTTG and PaGT4-R TATCCACCCTTCGAAGGCAC (Saltonstall Reference Saltonstall2003b) following the same cycle conditions previously described for the NRT2-Δ4 marker. Amplicons were digested with BstUI (New England Biolabs, Ipswich, MA) and visualized with gel electrophoresis following the same procedures used for the NRT2-Δ4 marker (Lindsay et al. Reference Lindsay, Guan, Harms, Cronin, Meyerson and Lance2023b).
Genotyping the trnLb and rbcL Region Markers
We tested specimens using the PCR-RFLP assays of the trnLb and rbcL regions as described by Saltonstall (Reference Saltonstall2003a). These assays were developed to identify haplotypes present in North America which can differentiate between P. australis ssp. americanus, P. australis ssp. berlandieri, and introduced haplotype M (Table 2).
Table 2. Expected results of polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) markers a .

a Summary of restriction enzyme digestion results previously published.
Results and Discussion
The nuclear DNA marker NRT2-Δ4 was previously shown to differentiate P. australis ssp. americanus from P.a. australis haplotype M and serve as a nuclear marker that could identify hybrids (Wendell et al. Reference Wendell, Huang, Gryspeerd and Freeland2021). However, the initial range of tested samples was limited, and the Gulf Coast type P. australis ssp. berlandieri was not included in the study. Here we show that the Gulf Coast type and introduced haplotypes L, P, and AS are all homozygous for the “uncut” allele as we previously reported for haplotype M, while five different P. australis ssp. americanus haplotypes collected coast to coast are all homozygous for the “cut” allele (Figure 1A). Thus, this marker is reliable to differentiate all described haplotypes of P. australis ssp. americanus from the other lineages tested, and because it is a nuclear locus, it is also suitable to screen for hybrids with P. australis ssp. americanus.
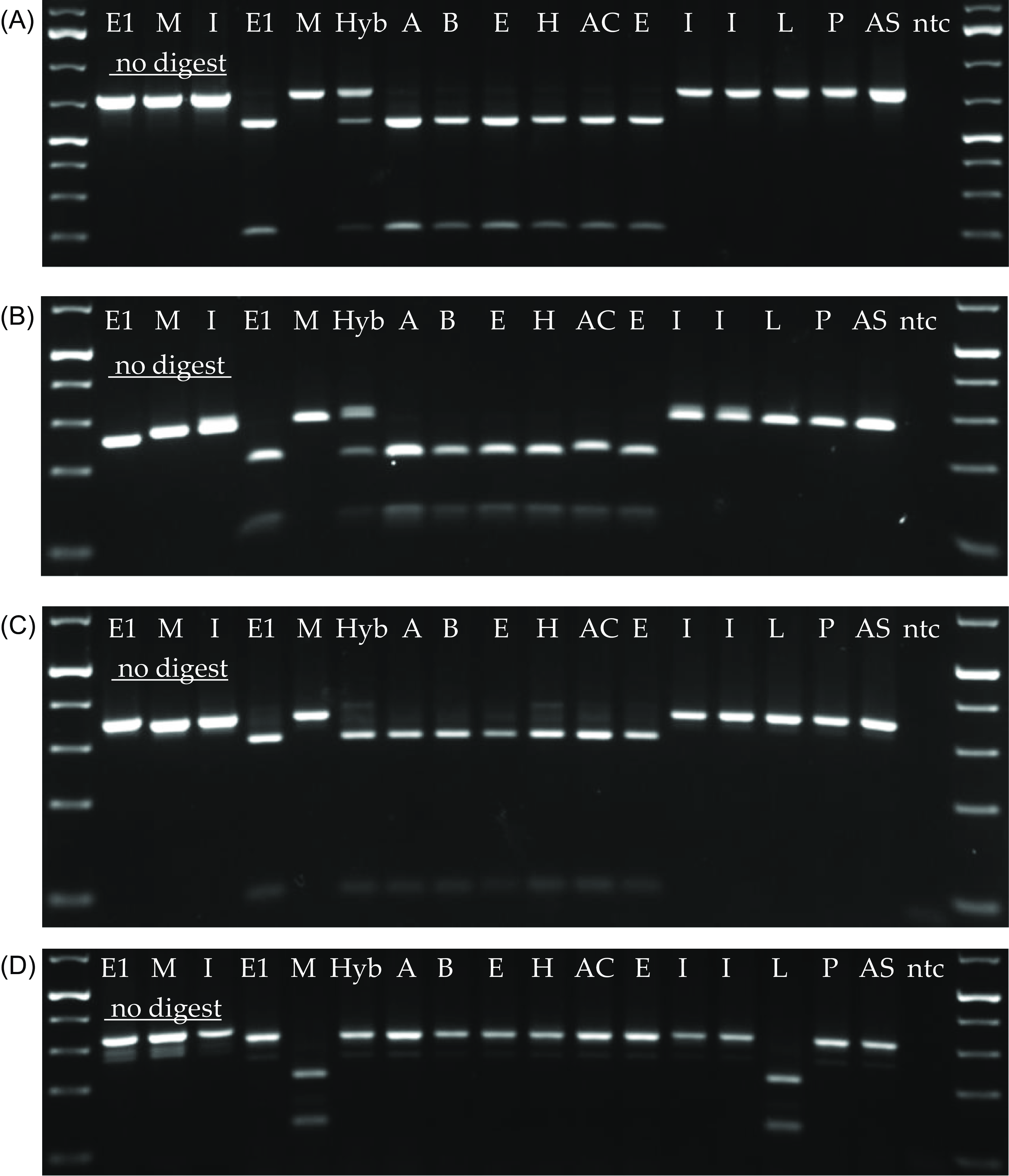
Figure 1. Polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) assays of (A) NRT2Δ4, (B) paGT4 SNP, (C) trnLb, and (D) rbcL genetic markers. Those marked “no digest” are samples not treated with restriction enzyme, showing that a single band is produced by PCR. Lanes are labeled with chloroplast DNA (cpDNA) haplotype, and specimens are described in Table 1. For each haplotype and location (except P and the hybrid), at least two samples were tested and were found to give identical results. ntc, no template control for contamination.
PCR-RFLP analysis of an SNP at the PaGT4 locus (Figure 1B) expands on the previous report by Saltonstall (Reference Saltonstall2003b) and shows that all P. australis ssp. americanus samples tested are homozygous for an allele that is “cut” by the enzyme BstUI. Therefore, we can infer that they are all homozygous for the “C” allele of that SNP. All non–P. australis ssp. americanus samples tested, consisting of haplotypes M, I, L, P, and AS, are homozygous for the “uncut” allele and are thus fixed for the “A” allele. A hybrid of P. australis ssp. americanus and haplotype M shows both the “cut” and “uncut” expected of a heterozygote (Figure 1B).
The current Phragmites genome assembly (version lpPhrAust1.1, accession no. PRJEB64034) places NRT2 on chromosome 5 and PaGT4 on chromosome 6. Because they are independently assorting, using them in combination provides two independent tests to screen for hybridization between native P. australis ssp. americanus and introduced Eurasian haplotypes. However, they cannot differentiate between P.a. australis and P. australis ssp. berlandieri.
The occurrence of hybridization between native and introduced Phragmites in North America appears to be rare but needs to be surveilled. To date, there are only four published reports of confirmed hybrids in the field (Paul et al. Reference Paul, Vachon, Garroway and Freeland2010; Saltonstall et al. Reference Saltonstall, Castillo and Blossey2014, Reference Saltonstall, Lambert and Rice2016; Wu et al. Reference Wu, Murray and Heffernan2015), and there have been several comprehensive studies that tested for hybrids but found none (Kettenring and Mock Reference Kettenring and Mock2012; Saltonstall Reference Saltonstall2011; Tippery et al. Reference Tippery, Pesch, Murphy and Bautzmann2020; Wani et al. Reference Wani, Shah, Tekeu, Reshi, Atangana and Khasa2020). Given the apparent scarcity of hybrids, it would be beneficial to screen for their presence with a low-cost and simple method. For example, the Wendell lab provides genetic testing of Phragmites using the Saltonstall (Reference Saltonstall2003a) PCR-RFLP test as a service to natural resource managers who are having difficulty determining subspecies based on morphology alone. Adding the PaGT4 SNP and NRT2-Δ4 PCR-RFLP tests described in this paper allows us to screen all samples for evidence of hybridization with little extra effort. Any potential hybrids would then be confirmed by analysis with microsatellite markers or an SNP array.
In the course of this work, we identified an invasive population of P.a. australis in North Bay, Grays Harbor County, WA, that cpDNA sequencing identified as haplotype AS. We initially genotyped 12 samples using Saltonstall’s PCR-RFLP assay (Saltonstall Reference Saltonstall2003a), and all yielded undigested products for both HhaI and RsaI enzymes in the trnLb and rbcL regions. (One representative is shown in Figure 1C and 1D.) Following Saltonstall’s (Reference Saltonstall2003) native versus non-native interpretation guide, these results suggested that the samples belonged to the Gulf Coast lineage. Because there are no reports of the Gulf Coast lineage in Washington State, we sequenced a subset of the samples (n = 8) in both the trnT-trnL and rbcL-psaI chloroplast regions. The trnT-trnL sequences obtained for all eight samples were identical and matched a previously described haplotype T1. The rbcL-psaI sequences were also identical and matched a previously described haplotype R26. Following Saltonstall’s guidelines for haplotype assignment (Saltonstall Reference Saltonstall2016), they were identified as the combined haplotype AS, which has not been previously reported in North America. Phragmites haplotype AS was first described as an unpublished haplotype with GenBank accessions KP994334 (rbcl-psaI) and KP994329 (trnT-trnL) (Saltonstall Reference Saltonstall2016) collected from the Yangtze River delta, Shanghai, in eastern China.
The markers we present fill a niche in the collection of genetic tools available for Phragmites identification. A recent review by Lindsay et al. (Reference Lindsay, Freeland, Gong, Guan, Harms, Kowalski, Lance, Oh, Sartain and Wendell2023a) provides a comprehensive description of all published methods that have been applied to Phragmites identification with a side-by-side comparison of the benefits and disadvantages and their management implications. cpDNA analysis is frequently used to differentiate North American subspecies and introduced lineages, and previously described methods include direct nucleotide sequencing, RNase H-dependent PCR, PCR-RFLP, and quantitative PCR (see also Lindsay et al. Reference Lindsay, Guan, Harms, Cronin, Meyerson and Lance2023b). However, as new introductions of invasive Phragmites haplotypes are detected throughout North America, resource managers should be aware that non-native P.a. australis lineages such as haplotype AS (and also haplotype P) may produce erroneous results when being interpreted with cpDNA PCR-RFLP tests that they were not previously validated with (Figure 1C and 1D). Therefore, cpDNA sequencing is called for if introduced haplotypes other than M are a concern. Only nuclear DNA can identify hybrids, and the available methods are microsatellites and PCR-RFLP. The results we present here further support the power of nuclear DNA–based PCR-RFLP tools by expanding the number of independent markers and haplotypes tested. RFLP-PCR has the advantage of being faster and less expensive than microsatellite analysis, although we still recommend it as a screening tool, and any potential hybrids identified should be confirmed with microsatellites. As nucleotide sequencing becomes less expensive, and now that there is a P. australis reference genome assembly (Oh et al. Reference Oh, Kowalski, Quach, Wijesinghege, Tanford, Dassanayake and Clay2022), nucleotide sequencing of nuclear DNA may become a method of choice, as it has been applied to identification in other plant species (Tippery et al. Reference Tippery, Olson and Wendtlandt2021). However, we expect that the PCR-RFLP method will remain relevant as a quick low-cost test.
Acknowledgments
For collection and processing of Phragmites samples from Washington State, we thank Greg Haubrich, McKenzie Watson, Chad Phillips, and Sapphitah Dickerson. For collection of Phragmites tissue samples from elsewhere, we thank Abigale Bristol, Cody Haynes, Phoebe Jackson, Mark O’Brien, Adam Lambert, John Potente, Alex Wietan, and Trevor Zook.
Funding statement
This work was funded in part by the Oakland University Research Committee.
Competing interests
The authors declare no conflicts of interest.





