Introduction
The family Juglandaceae is one of the most iconic groups of nut trees (Zhang et al. Reference Zhang, Ree, Salamin, Xing and Silvestro2022) and holds significant scientific and conservation value (Song et al. Reference Song, Fragnière, Meng, Li, Bétrisey, Corrales, Manchester, Deng, Jasińska, Văn Sâm and Kozlowski2020). Notable genera within this family include Carya, Juglans, Pterocarya, and Platycarya (Stanford et al. Reference Stanford, Harden and Parks2000). Among them, the genus Juglans is the most representative and comprises the majority of fruit-bearing and timber-yielding species (Trouern-Trend et al. Reference Trouern-Trend, Falk, Zaman, Caballero, Neale, Langley, Dandekar, Stevens and Wegrzyn2020). The genus Juglans consists of 21 species and is classified into four taxonomic sections (i.e., Cardiocaryon, Juglans, Rhysocaryon, and Trachycaryon) based on the morphological characteristics of its flowers and leaves (Manning Reference Manning1960). One of these species, namely Juglans neotropica traditionally known as black walnut, is native to Colombia, western Venezuela, Ecuador, and Peru (Manning Reference Manning1960).
Juglans neotropica is a woody angiosperm that can reach heights of 25–30 metres (Nieto and Rodríguez Reference Nieto and Rodríguez2010). This species is characterized by having furrowed bark on its trunk and monoecious nature (Ramírez and Kallarackal Reference Ramírez and Kallarackal2021). The altitudinal distribution range of the black walnut J. neotropica is between 500 and 3300 m.a.s.l. (Reynel and Marcelo Reference Reynel and Marcelo2009). This species has been reported in the following regions of Peru: Amazonas, Cajamarca, Cusco, Huancavelica, Junín, La Libertad, Lambayeque, and Pasco (Manning Reference Manning1960, Hurtado Manrique et al. Reference Hurtado Manrique, Jurado Teixeira, Ramos Llica and Calixto Cotos2015). The black walnut J. neotropica is recognized as one of the most ecologically, environmentally, and economically valuable tree species (Vanegas and Rojas Reference Vanegas and Rojas2018) due to its prominent role in the recovery of degraded soils and its contribution to improving air quality and water conditions in the ecosystems (Vanegas and Rojas Reference Vanegas and Rojas2018).
In Peru, the black walnut J. neotropica is locally known as nogal and is used as a medicinal and commercial tree due to its fruits with high nutritional value and pigments used in textile products (Woll et al. Reference Woll, Reynel, Palacios, Hermoza and Chavez2023). In the Amazonas region, this nogal tree is also highly valued for their timber quality. For instance, J. neotropica is part of the official list of forest species used for timber purposes (SERFOR 2019), with extraction figures of 2457.21 m3/year for sawn wood and 5416.64 m3/year for round wood (SERFOR 2019). Currently, the black walnut J. neotropica is listed as endangered species (IUCN, 2023) due to rapid reductions in its distribution as a result of anthropogenic activities. These activities generate a loss of genetic variability (Anabat et al. Reference Anabat, Riahi, Sheidai and Koohdar2020), preventing it from responding to natural selection and limiting its ecological recovery (Janat Gul et al. Reference Gul, Saeed, Ahmed, Khan, Basit, Rehman and Zahid2021).
Various studies have been conducted on different species of Juglans regarding its genetic improvement, potential commercial use, restoration capabilities for degraded ecosystems, genetic diversity for conservation purposes, and spatial monitoring (Ross-Davis et al. Reference Ross-Davis, Ostry and Woeste2008, Vischi et al. Reference Vischi, Chiabà, Raranciuc, Poggetti, Messina, Ermacora, Cipriani, Paffetti, Vettori and Testolin2017, Gaisberger et al. Reference Gaisberger, Legay, Andre, Loo, Azimov, Aaliev, Bobokalonov, Mukhsimov, Kettle and Vinceti2020, Veintimilla et al. Reference Veintimilla, Murillo and Gallo2020). For instance, the distribution patterns, diversity, and structure of different populations of J. regia have been evaluated via nuclear microsatellites (Magige et al. Reference Magige, Fan, Wambulwa, Milne, Wu, Luo, Khan, Wu, Qi, Zhu, Maity, Khan, Gao and Liu2022). However, this study has found limitations in the dispersion and discontinuous geographical distribution of the species, which has made it difficult to interpret genetic patterns (Gaisberger et al. Reference Gaisberger, Legay, Andre, Loo, Azimov, Aaliev, Bobokalonov, Mukhsimov, Kettle and Vinceti2020). Strikingly, most of the genetic studies carried out in Juglans have focused on specimens distributed in the Northern Hemisphere (Ma et al. Reference Ma, Huang, Song, Chang and Pei2020, Zhou et al. Reference Zhou, Hu, Ebrahimi, Liu, Woeste, Zhao and Zhang2021). In contrast, in the Southern Hemisphere, only two specimens of Juglans, which are distributed mainly in Argentina (J. australis) and Ecuador (J. neotropica), were sequenced (Aradhya et al. Reference Aradhya, Potter, Gao and Simon2007).
Additionally, the most widely used molecular markers to infer the phylogeny of numerous Juglans species are the plastid coding markers (rbcL, large subunit of ribulose bisphosphate carboxylase; matK, maturase K), plastid intergenic markers (trnT-trnF, psbA-trnH, trnS-trnfM), and nuclear non-coding marker (ITS, internal transcribed spacer) (Orel et al. Reference Orel, Marchant, Mcleod and Richards2003, Aradhya et al. Reference Aradhya, Potter, Gao and Simon2007). Among these markers, the intergenic trnS-trnfM revealed a better phylogenetic resolution, greater number of sequences, and greater genetic divergences between Juglans species (Aradhya et al. Reference Aradhya, Potter, Gao and Simon2007). For instance, genetic analysis in other Juglandaceae such as Pterocarya used this intergenic marker to identify evolutionary patterns (Lu et al. Reference Lu, Wang, Zheng, Meng, Cao, Song and Kozlowski2024). Despite the great utility of this marker, genetic studies related to the black walnut J. neotropica are currently lacking in Peru.
Genetic diversity studies within the Juglans genus have predominantly focused on Juglans regia (Sun et al. Reference Sun, Hou, Woeste, Zhang, Yue, Yuan and Zhao2019, Ren et al. Reference Ren, Wang, Zhou, Xie, Han, Peng and Liu2023). For instance, low genetic diversity was confirmed in populations from Pakistan and Kyrgyzstan (Gaisberger et al. Reference Gaisberger, Legay, Andre, Loo, Azimov, Aaliev, Bobokalonov, Mukhsimov, Kettle and Vinceti2020, Magige et al. Reference Magige, Fan, Wambulwa, Milne, Wu, Luo, Khan, Wu, Qi, Zhu, Maity, Khan, Gao and Liu2022). Conversely, higher genetic diversity was observed in populations from Uzbekistan (Gaisberger et al. Reference Gaisberger, Legay, Andre, Loo, Azimov, Aaliev, Bobokalonov, Mukhsimov, Kettle and Vinceti2020). These studies suggested that human activities significantly influence the distribution of Juglans regia. Population genetics of Juglans cinerea revealed a high level of genetic diversity, supporting the development of conservation strategies to protect its local populations (Ross-Davis et al. Reference Ross-Davis, Ostry and Woeste2008). Although populations of Juglans hopeiensis exhibited robust genetic structure and were different from other sympatric species within the genus, they face significant threats due to their small size and ongoing hybridization (Hu et al. Reference Hu, Dang, Feng, Woeste and Zhao2017).
Accordingly, understanding the distribution and genetic diversity of Juglans species is crucial (Zhao et al. Reference Zhao, Zhou, Potter, Hu, Feng, Dang, Feng, Zulfiqar, Liu, Zhao and Woeste2018) since it will provide valuable information of its current state and ability to survive against various risk factors (Govindaraj et al. Reference Govindaraj, Vetriventhan and Srinivasan2015, Stojnić et al. Reference Stojnić, Avramidou, Fussi, Westergren, Orlović, Matović, Trudić, Kraigher, Aravanopoulos and Konnert2019). The population genetic analysis of J. neotropica serves multiple critical purposes for both ecological understanding and conservation practice. First, it reveals historical lineages, helping reconstruct post-glacial migration patterns and identify potential refugia in the Amazonas region, which is key for understanding how this species responded to past climatic shifts (Shahi Shavvon et al. Reference Shahi Shavvon, Qi, Mafakheri, Fan, Wu, Vahdati, Al-Shmgani, Wang and Liu2023). Second, quantifying genetic diversity and divergence directly informs conservation priorities, where populations with low diversity may require urgent protection due to their vulnerability to habitat fragmentation (Dexter et al. Reference Dexter, Pennington, Oliveira-Filho, Bueno, Silva de Miranda and Neves2018). Our study thus bridges evolutionary history and actionable conservation strategies for this critically endangered species. For this reason, the present study aimed to evaluate the genetic diversity of the black walnut J. neotropica from eight locations in the Amazonas region. This study addressed the degree of genetic connectivity and identified genetic groups (haplotypes), allowing for the generation of crucial information for further conservation and control strategies.
Material and methods
Collection of samples
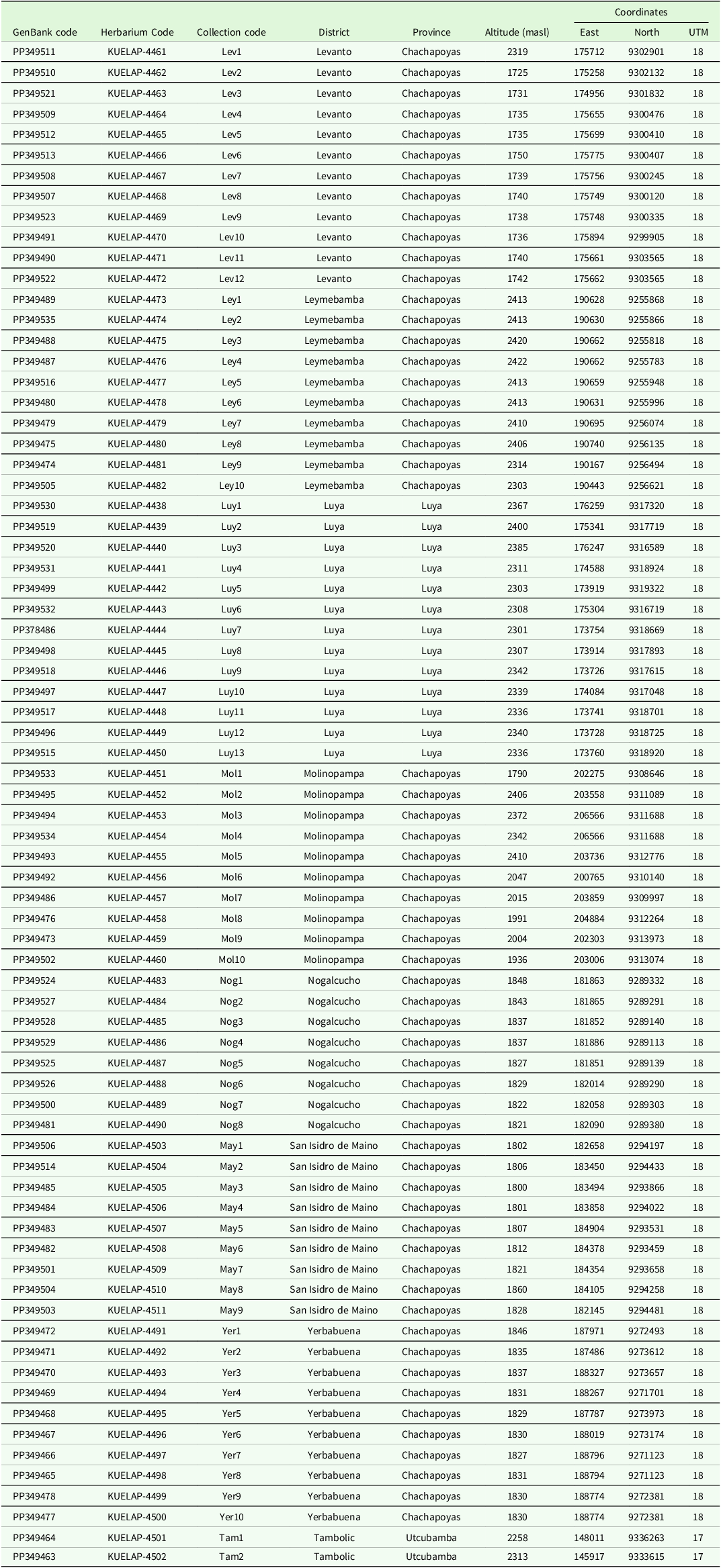
A total of 74 specimens of the black walnut J. neotropica (locally known as nogal) were collected from the following eight locations in the Amazonas region: Levanto (12 specimens), Leymebamba (10 specimens), Luya (13 specimens), Molinopampa (10 specimens), Nogalcucho (8 specimens), San Isidro de Maino (9 specimens), Yerbabuena (10 specimens), and Tambolic (2 specimens) (Figure 1). The identification of these specimens as Juglans neotropica was based on taxonomical keys (Nieto and Rodríguez Reference Nieto and Rodríguez2010, Ramírez and Kallarackal Reference Ramírez and Kallarackal2021) and distribution reports (Manning Reference Manning1960, Hurtado Manrique et al. Reference Hurtado Manrique, Jurado Teixeira, Ramos Llica and Calixto Cotos2015). The collected specimens from each location were at least 15 metres apart (Deng et al. Reference Deng, Liu, Xie, Wei, Xie, Shi and Deng2020). The black walnut J. neotropica trees chosen for collection were young with new leaves and flowers; the leaves were tender with no epiphytes and free of pests and diseases. A wild flora scientific research permit (D000506-2020-MINAGRI-SERFOR-DGGSPFFS) was obtained from the National Forest and Wildlife Service (SERFOR). The collection points were defined by reviewing the distributions of the black walnut J. neotropica in the Amazonas region recorded in the databases of international herbaria, such as the JSTOR Global Plants (https://plants.jstor.org/), the New York Botanical Garden Steere Herbarium (https://sweetgum.nybg.org/science/), the Global Biodiversity Information Facility (https://www.gbif.org/) and Tropicos database of the Missouri Botanical Garden (http://www.tropicos.org). Additionally, each collection point was georeferenced, and each collected sample was deposited in the herbarium KUELAP of the Universidad Nacional Toribio Rodríguez de Mendoza (Table 1).
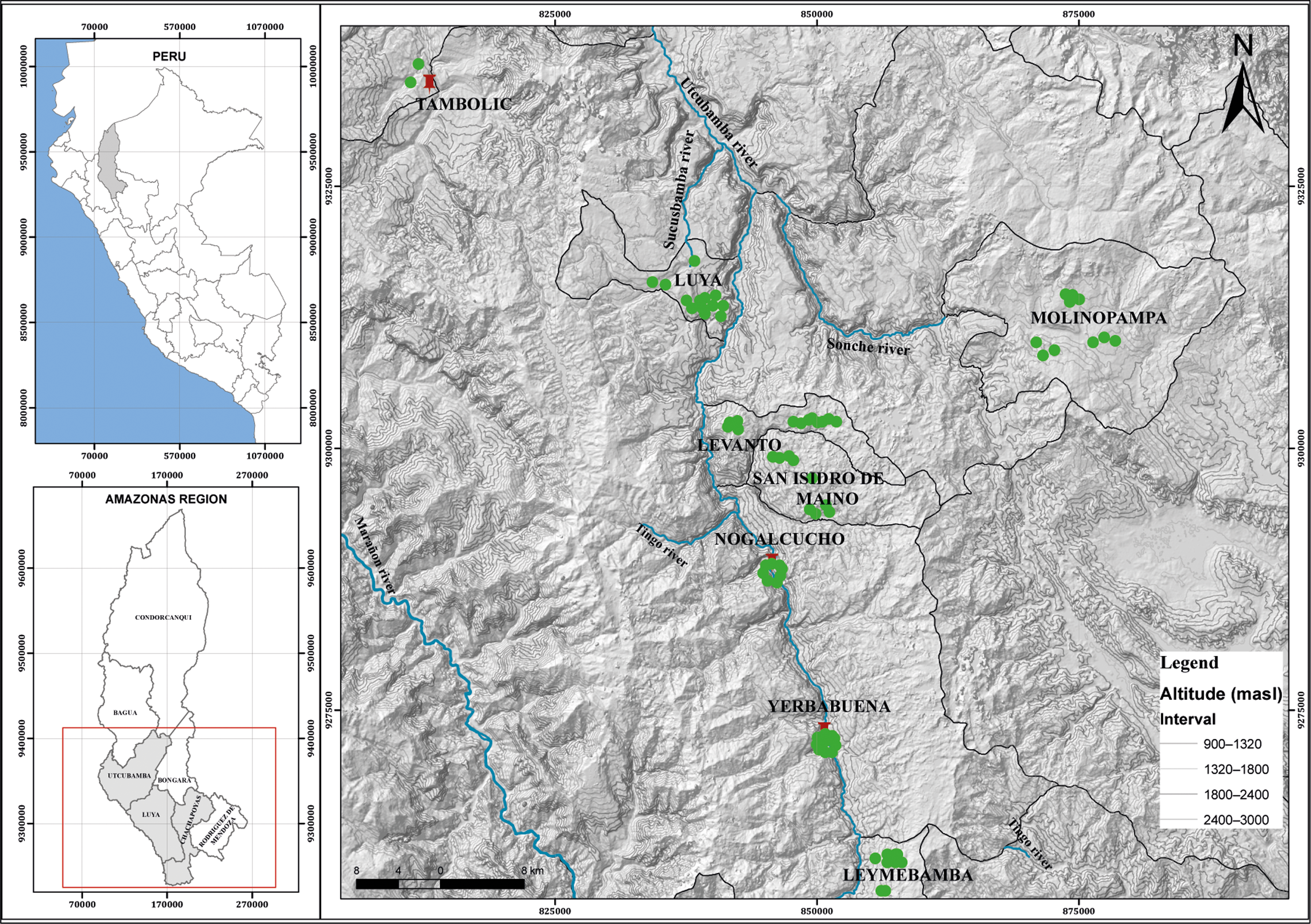
Figure 1. Collection sites for black walnut J. neotropica specimens in the Amazonas region, northern Peru.
Table 1. List of black walnut J. neotropica samples collected in the Amazonas region, northern Peru
Molecular analysis
Sample preparation and extraction
A stereoscope (Labtech, Linitron) was used to thoroughly clean the specimens of black walnut J. neotropica. Then, small 3 cm2 fragments were cut and deposited in 1.5 mL microtubes, immersed in liquid nitrogen and crushed using freezing and crushing equipment (Disruptor SK-10, Japan). Genomic DNA extraction from the specimens was performed using the NucleoSpin Plant II kit (Macherey-Nagel, Düren, Germany) following the manufacturer’s protocol. The quality of the extracted DNA was determined on a 1% agarose gel and by fluorometric quantification with Quantus™ (Promega, Madison, United States). Finally, the extracted DNA was stored in a freezer at –80 °C for preservation.
DNA amplification and purification
Preliminary analysis revealed the problematic topology observed in Juglans species when using the coding (rbcL, matK) and intergenic markers (trnT-trnF, psbA-trnH) (Figures A.1–A.5). In addition to the better phylogenetic resolution of the trnS-trnfM marker (Figure A.6), it is one of the most commonly used genes for plants in genetic analysis (Aradhya et al. Reference Aradhya, Potter, Gao and Simon2007; Minami et al. Reference Minami, Nishio, Ajioka, Kyushima, Shigeki, Kinjo, Yamada, Nagai, Satoh and Sakurai2009). Accordingly, the intergenic marker trnS-trnfM was amplified with primers 2F (5′-CGGAGCTATCAACCACTCGG-3′) and 539R (5′-ACTCGACCAACCATCAGGAG-3′) (Aradhya et al. Reference Aradhya, Potter, Gao and Simon2007). Polymerase chain reaction (PCR) was performed in a 10 μL reaction mixture containing 5.0 μL of master mix, 2.6 μL of distilled water, 0.2 μL of each primer (forwards or reverse) and 2 μL of genomic DNA (Deng et al. Reference Deng, Liu, Xie, Wei, Xie, Shi and Deng2020). The PCR protocols for the marker were as follows: 1 cycle of predenaturation at 94 °C for 5 minutes, 40 cycles of denaturation at 94 °C for 45 seconds, alignment at 62 °C for 1 minute, extension at 72 °C for 2 minutes, and a final extension cycle at 72 °C for 7 minutes. The obtained amplicons were visualized by electrophoresis in 1% agarose gels in buffer (0.045 M Tris-borate; 0.001 M EDTA) for 15 minutes at 100 V (Porth and El-Kassaby Reference Porth and El-Kassaby2014). The clear banded amplicons were purified with the Zymo Reseach DNA Clean & ConcentratorTM-5 cleaning kit (Zymo, California, United States) following the manufacturer’s instructions and were sequenced commercially by Macrogen (Seoul, South Korea).
Phylogenetic analysis
A phylogenetic analysis was performed using the 74 specimens collected of the black walnut J. neotropica and 49 sequences downloaded from GenBank database (length of 502 nucleotides) (Figure 2). The software PartitionFinder v. 2.1.1 was used to select the evolutionary model (Coates et al. Reference Coates, Byrne and Moritz2018). The reconstruction of the tree was performed using the maximum likelihood (ML) method with RaxML GUI v. 2.0.0 beta 10 considering a bootstrap of 1000 replicates (Rosenfeld et al. Reference Rosenfeld, Mendez, Calderon, Bahamonde, Rodríguez, Ojeda, Marambio, Gorny and Mansilla2019, Tineo et al. Reference Tineo, Bustamante, Calderon, Mendoza, Huaman and Oliva2020). For this analysis, the species Juglans neotropica (AY293368, Ecuador) was not included because it was grouped outside the taxonomic genus.
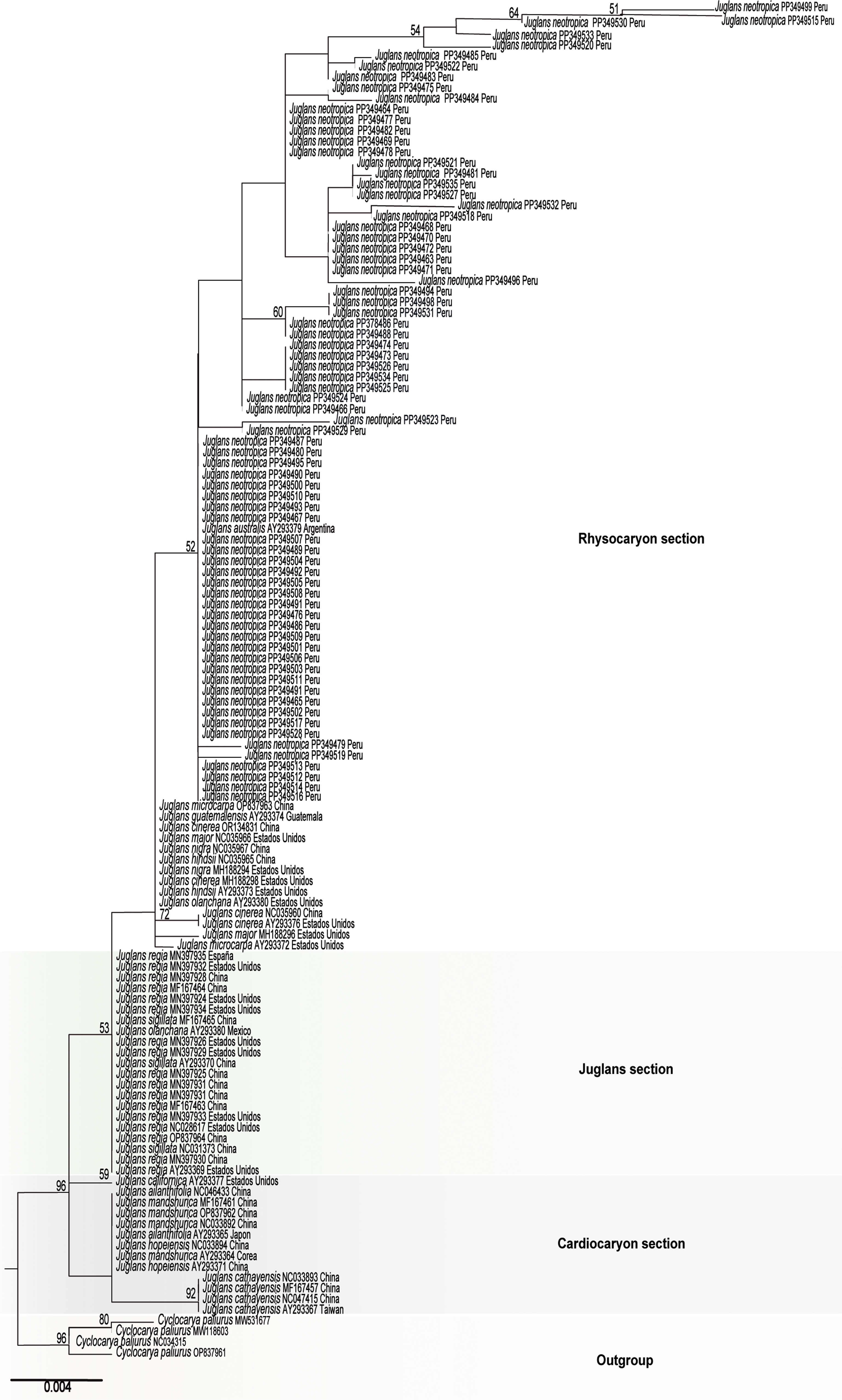
Figure 2. Phylogenetic tree based on the maximum likelihood analysis of the trnS-trnfM marker for sections of the genus Juglans. Maximum likelihood bootstrap values are indicated in the branches. The scale indicates the number of nucleotide substitutions per site. The specimens generated in this study are in bold.
Genetic diversity and haplotype network construction
Genetic diversity was obtained by calculating the frequencies of distribution via DnaSP v6 software (Librado and Rozas Reference Librado and Rozas2009), which calculated the number of haplotypes (h), the diversity of haplotypes (Hd), and the diversity of nucleotides (π). In addition, a heatmap was constructed based on pairwise distances (Magige et al. Reference Magige, Fan, Wambulwa, Milne, Wu, Luo, Khan, Wu, Qi, Zhu, Maity, Khan, Gao and Liu2022). The values of the genetic distances were obtained using the MEGAX program and then exported using the pheatmap extension to R software (R Development Core Team 3.4.2).
Connectivity analyses between the haplotypes of the populations of the Amazonas region and of the other countries were carried out by means of the median junction network (MJ) method and PopArt v1.7 software (Xu et al. Reference Xu, Zang, Wu, Wang, Wang, Huang, Zang, Ma and Zheng2021) (Table A.1).
Molecular variance analysis
The Analysis of Molecular Variance (AMOVA) was performed using Arlequin 3.11 software (Excoffier et al. Reference Excoffier, Laval and Schneider2005) to determine the distribution of variance and the level of significance among walnut populations and to determine the source of genetic variation.
Results
Phylogenetic analysis
Phylogenetic analysis of the Juglandaceae family, which includes Cyclocarya paliurus as external group, revealed the grouping of three taxonomic sections (Cardicaryon, Juglans, and Rhysocaryon). The Rhysocaryon section encompassed eight species including the specimens from Amazonas region (J. cinerea, J. guatemaiensis, J. hindsii, J. major, J. microcarpa, J. neotropica, J. nigra, and J. olanchana). Additionally, the Cardiocaryon (J. ailanthifolia, J. californica, J. cathayensis, J. hopeiensis and J. mandshurica) and Juglans (J. olanchana, J. regia and J. sigillata) sections were composed of five and three species, respectively.
Genetic divergence
Interspecific genetic divergences based on pairwise distances of Juglans individuals collected in the Amazonas region and in Argentina (AY293379), China (AY293370, MF167461, KX671977, NC047415), Ecuador (AY293368), Spain (MN397935), Guatemala (AY293374), Japan (AY293365), Mexico (AY293380), the United States (AY293377, AY293373, MF167460, MH188298, MH188294, AY293372) ranged from 0 to 5%.
The greatest genetic distance was detected between the Luya population and the Ecuador population (AY293368) (4.75%). On the other hand, the lowest genetic divergence (0.19%) was obtained between the populations of Molinopampa, Tambolic, and Yerbabuena and the populations of Argentina (AY293379), the United States (MH188298, MF167460), and Guatemala (AY293374), respectively. Considering only the samples from the Amazonas region (intraspecific genetic divergence), the most divergent populations were Luya and Molinopampa (1.62–2.64% of p-distance), while the most similar populations were Levanto, Molinopampa, San Isidro de Maino and Yerbabuena (0.19%) (Figure 3).
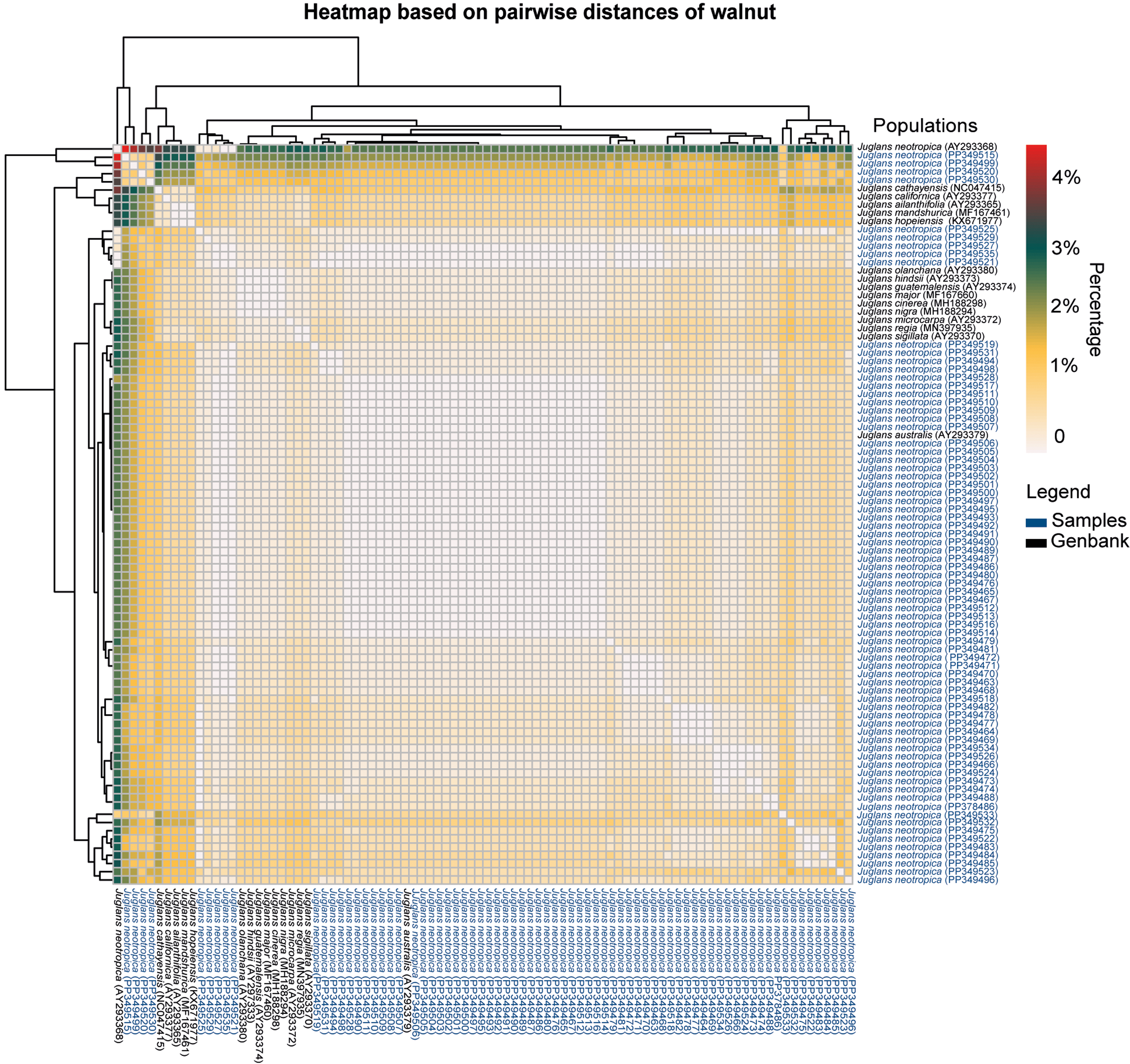
Figure 3. Heatmap based on pairwise distances (p-distance) of black walnut J. neotropica populations from the Amazonas region and foreign populations.
Genetic diversity
The nucleotide compositions of the specimens from the Amazonas region were A = 30.88%, G = 17.33%, C = 14.54%, and T = 37.25%. The data from the Amazonas region showed a mean haplotype diversity (Hd) of 0.57 and a relatively low nucleotide diversity (Pi) of 0.0035 (Table 2). Luya stood out among all populations since it was i) the population with the greatest number of haplotypes (h = 7), ii) the population with the greatest number of polymorphic sites (s = 11), iii) the population with the greatest number of diverse nucleotides (Pi = 0.01), and iv) the population with the greatest average number of nucleotide differences per pair (k = 3.97). On the other hand, the population with the greatest diversity of haplotypes was Tambolic (H = 1.00) (Table 2).
Table 2. Genetic diversity of black walnut J. neotropica trees in the Amazonas region. n: number of samples; h: number of haplotypes; s: number of polymorphic sites; Hd: diversity of haplotypes; Pi: nucleotide diversity; K: average number of nucleotide differences per pair
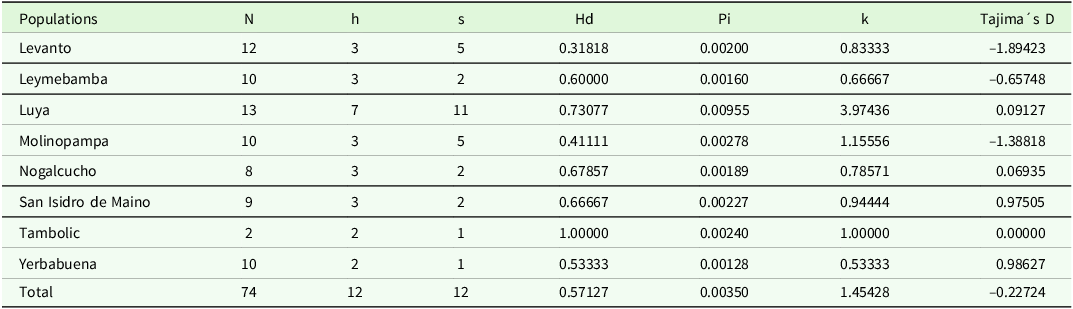
Tajima’s D test revealed negative values (P < 0.05) for the Levanto, Molinopampa, and Leymebamba populations. This would indicate that these populations recently expanded after a selective sweep (Ebrahimi et al. Reference Ebrahimi, Zarei, McKenna, Bujdoso and Woeste2017). For the other populations of Luya, Nogalcucho, San Isidro de Maino, Tambolic, and Yerbabuena, the Tajima’s D test values were positive, indicating balanced selection (Magige et al. Reference Magige, Fan, Wambulwa, Milne, Wu, Luo, Khan, Wu, Qi, Zhu, Maity, Khan, Gao and Liu2022).
Haplotype network analysis
The haplotype network based on the median-joining network analysis of the eight populations of J. neotropica from the Amazonas region (n = 74) allowed the identification of three groups. Group I was the most diverse and representative, comprising 7 haplotypes: the first haplotype (H1) included 46 individuals from the 8 populations; haplotype 2 (H2) included 16 individuals from the Leymebamba populations (3), Luya (1), Molinopampa (2), Nogalcucho (3), San Isidro de Maino (2), Tambolic (1) and Yerbabuena (4); and haplotype 3 (H3) included only 2 individuals from the populations of Levanto (1) and San Isidro de Maino (1). Additionally, group I was composed of 4 unique haplotypes from Levanto (1), Leymebamba (1), Luya (1), and Nogalcucho (1). Group II consisted of 2 unique haplotypes from Luya (1) and Molinopampa (1). Group III was composed of three unique haplotypes belonging to Luya individuals (Figure 4).
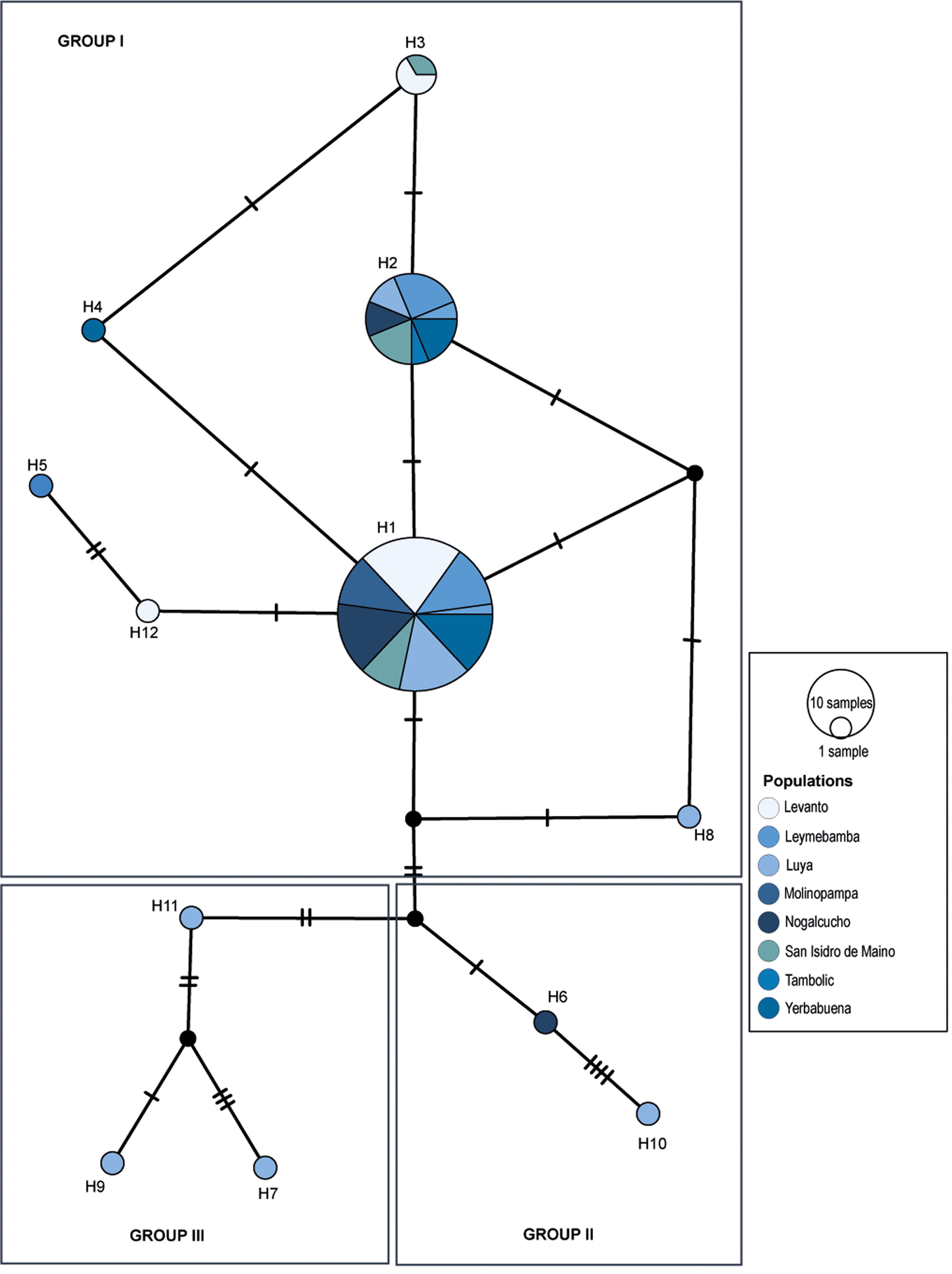
Figure 4. Haplotype network of black walnut J. neotropica populations in the Amazonas region. The populations are represented by colours. The lines perpendicular to the branches represent the mutational points. H: haplotype.
The second network was composed of a total of 124 individuals. The formation of two additional groups (Group IV and Group V) was observed. Group IV consisted of two haplotypes (H14, H18), and its individuals came from only Asian populations (China, Korea, Japan, Taiwan) (Figure 5, Table A.2). Group V included 5 haplotypes (H13, H15, H16, H17, H19), of which haplotype 19 was the most predominant and consisted of 35 individuals from populations from China (NC035960, NC035967, NC035965, OR134831, OP837963, MN397931, MN397930, MN397928, MN397925, MF167465, MF167464, MF167463, NC031373, OP837964, AY293370), Guatemala (AY293374), Mexico (AY293380), Spain (MN397935) and the United States (MH188296, MH188298, MH188294, NC035966, AY293373, MN397934, MN397933, MN397932, MN397929, MN397927, MN397926, NC028617, AY293369, MN397924, AY293372, AY293376, AY293377).
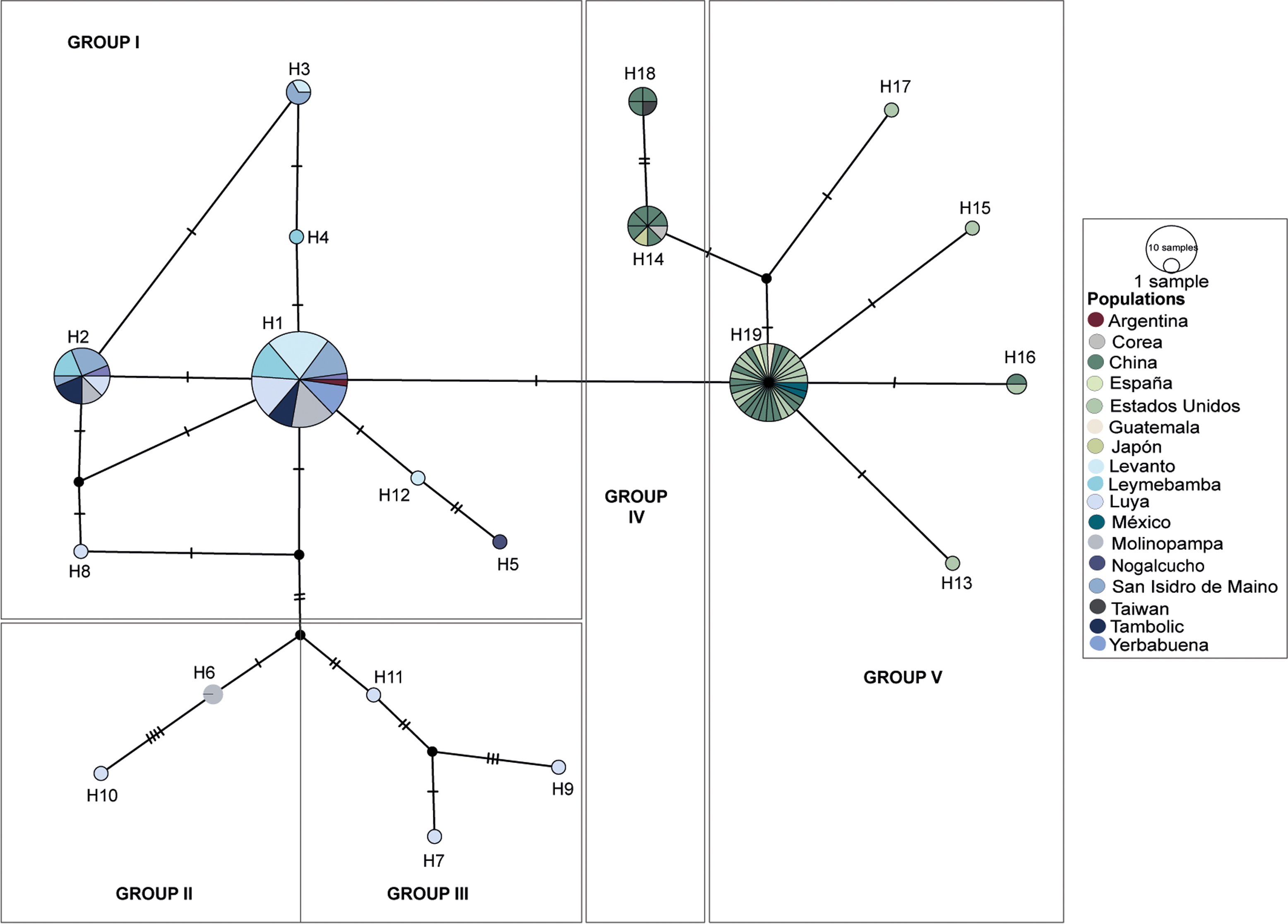
Figure 5. Haplotype network of black walnut J. neotropica populations from the Amazonas region and other countries (74 from the Amazonas region and 51 additional individuals). The lines perpendicular to the branches represent the mutational points. H: haplotype.
Genetic structure of populations
The global fixation index (Fst), based on the calculation of correlation values between genetic distances, allowed the estimation of connectivity values, and its values ranged from 0–0.05 (low connectivity), to 0.05–0.15 (moderate connectivity), and to 0.15–0.25 (high connectivity) (Aguirre-Pabon et al. Reference Aguirre-Pabon, Berdugo and Narváez2022). In the Amazonas region, a value of 0.04 was obtained, which represents a low level of genetic connectivity between populations. The populations with the highest level of genetic connectivity were Leymebamba – Levanto (Fst = 0.10052), Leymebamba – Molinopampa (Fst = 0.10077), Nogalcucho – Levanto (Fst = 0.07791) and San Isidro de Maino – Leymebamba (Fst = 0.08939). The populations with the lowest level of genetic connectivity were Yerbabuena with the following populations: Nogalcucho (Fst = –0.33397), Luya (Fst = –0.37748), Molinopampa (Fst = –0.34328), Leymebamba (Fst = 0.15559), San Isidro de Maino (Fst = –0.30178) and Tambolic (Fst = –0.22845) (Figure 6, Table A.3).
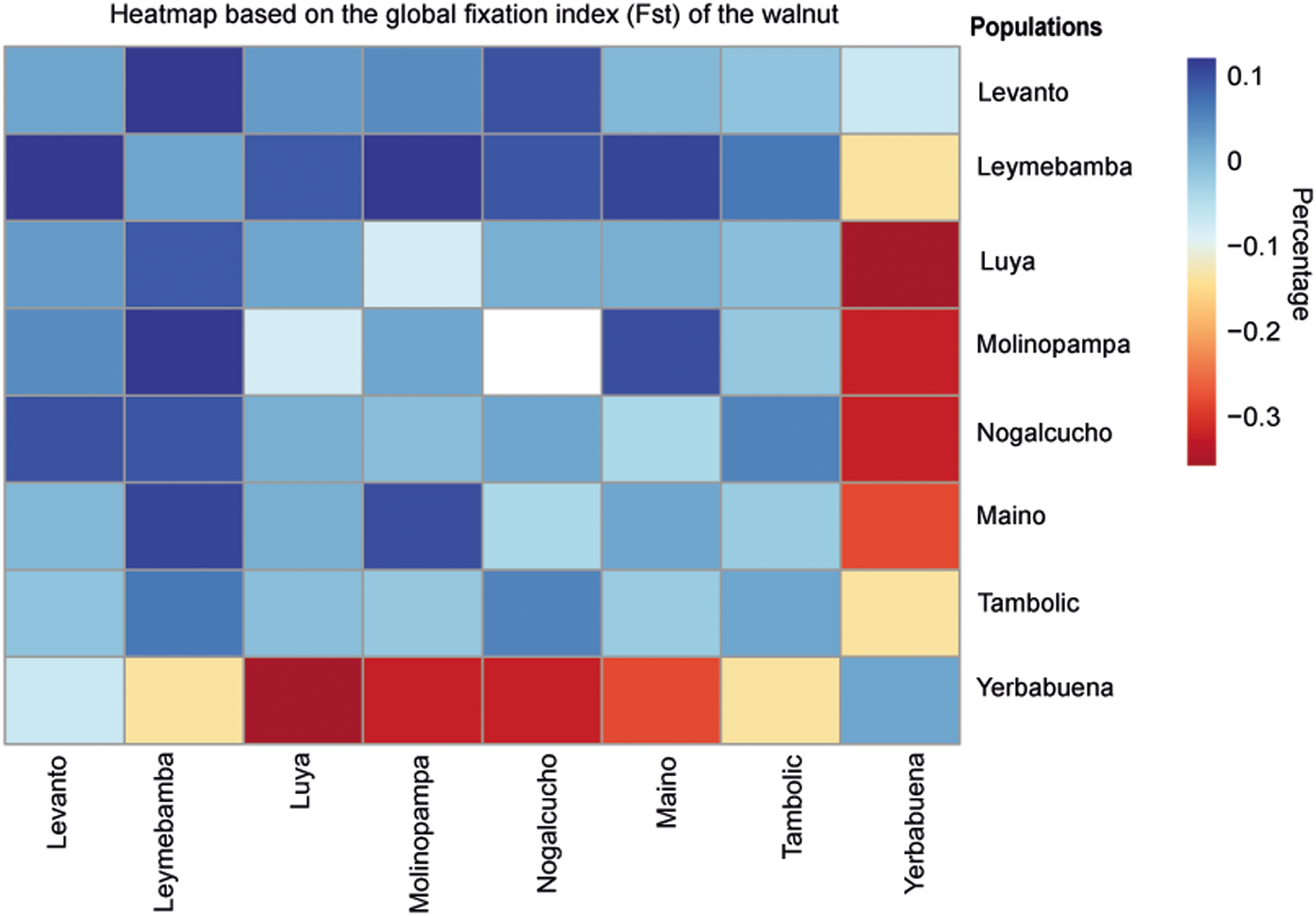
Figure 6. Heatmap based on the global fixation index (Fst) of the black walnut J. neotropica populations of the Amazonas region.
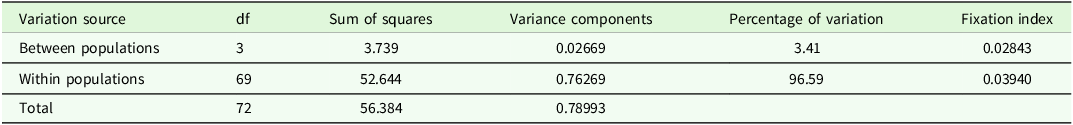
The AMOVA at the population level in the Amazonas region revealed that most of the genetic structuring and/or differentiation occurred within the populations (96.59%, P < 0.05, Fst = 0.03940), while the lowest percentage of genetic variation occurred between populations (3.41%, P < 0.05, Fst = 0.02843) (Table 3).
Table 3. Analysis of molecular variance (AMOVA) of the black walnut J. neotropica populations in the Amazonas region
Discussion
Phylogenetic analysis
In Peru, Juglans neotropica is distributed mainly in the Amazonas, Cajamarca, Cusco, Huancavelica, Junín, La Libertad, Lambayeque, and Pasco regions (Hurtado Manrique et al. Reference Hurtado Manrique, Jurado Teixeira, Ramos Llica and Calixto Cotos2015). The phylogenetic analysis of this study confirmed the presence of the black walnut J. neotropica in the Amazonas region and resolved the diversity of the family Juglandaceae in lineages that correspond to three taxonomic sections (Cardyocarion, Juglans, and Rhysocaryon). The lineage of the Rhysocaryon section, which is mainly endemic to North and Central America, included eight taxa (J. cinerea, J. guatemaiensis, J. hindsii, J. major, J. microcarpa, J. neotropica, J. nigra, and J. olanchana) (Fitz-Gibbon et al. Reference Fitz-Gibbon, Mead, O’Donnell, Li, Escalona, Beraut, Sacco, Marimuthu, Nguyen and Sork2023). The specimens of J. neotropica from the Amazonas region formed numerous small lineages that could be grouped in a sibling section to Rhysocaryon and probably different from the other two sections. However, the use of only one marker does not fully support this hypothesis. Therefore, further analysis including multiple markers and additional genomic information is strongly recommended. Additionally, the sequence of J. australis (AY293379 from Argentina) was grouped with samples of J. neotropica from the Amazonas region. This would suggest that J. australis would belong to the Rhysocaryon section. However, type materials from J. australis must be sequenced to confirm their correct phylogenetic position (Manning Reference Manning1960, GBIF 2023). The lineage in the Cardyocarion section included four species native to East Asia (J. ailanthifolia, J. cathayensis, J. hopeiensis and J. mandshurica) and one from Europe (J. californica) (Aradhya et al. Reference Aradhya, Potter, Gao and Simon2007, Dong et al. Reference Dong, Xu, Li, Xie, Lu, Liu, Jin and Suo2017). The lineage of the Juglans section, known as the Persian or cultivated English walnut, was composed of three species (J. olanchana, J. regia, and J. sigillata), which are distributed from southeastern Europe to China and the Himalayas (Ren et al. Reference Ren, Wang, Zhou, Xie, Han, Peng and Liu2023).
Genetic divergence
In the Amazonas region, the genetic divergence based on the p-distance of the black walnut J. neotropica was less than 3%, unlike that of the populations of Juglans spp. from Europe, Asia and North America, which varied by 2%–5.2% (Ebrahimi et al. Reference Ebrahimi, Zarei, McKenna, Bujdoso and Woeste2017, Magige et al. Reference Magige, Fan, Wambulwa, Milne, Wu, Luo, Khan, Wu, Qi, Zhu, Maity, Khan, Gao and Liu2022). The most divergent populations in the Amazonas region were those of Luya and Molinopampa (1.62% – 2.64% of p-distance). This could be because the flow of genes begins to change in response to genetic and geographical barriers (Vischi et al. Reference Vischi, Chiabà, Raranciuc, Poggetti, Messina, Ermacora, Cipriani, Paffetti, Vettori and Testolin2017). For example, Molinopampa is located 63 km from the Luya and contains populations of Juglans neotropica with a low level of genetic diversity and a high level of historic threat due mainly to livestock, excessive agriculture, grazing and overexploitation (Oliva et al. Reference Oliva, Collazos, Goñas, Bacalla, Vigo, Vasquez and Leyva2016). Additionally, Molinopampa and Luya are populations historically separated by two rivers (i.e., Utcubamba River and Sonche River) and a mountain range (i.e., Huancas). Consequently, we recommend integrated in situ conservation strategies for Molinopampa and Luya, including seed banking and habitat corridor establishment, to safeguard the genetic integrity of J. neotropica. On the other hand, the least divergent populations were Levanto, Molinopampa, San Isidro de Maino, and Yerbabuena (0.19%). The genetic similarity between Levanto and San Isidro de Maino is explained by their geographical proximity (8 km) (Alarcón-Méndez et al. Reference Alarcón-Méndez, Maselli, van Zonneveld, Loo, Snook, Oliva, Franco and Duminil2023). However, the localities of Yerbabuena and Molinopampa have a wider geographical distance (104 km), so genetic similarity could be explained by other factors, such as i) a common gene pool, ii) previous genetic connections of areas of distribution that were lost, or iii) possible anthropogenic or natural transport (Gaisberger et al. Reference Gaisberger, Legay, Andre, Loo, Azimov, Aaliev, Bobokalonov, Mukhsimov, Kettle and Vinceti2020).
Genetic diversity
Despite the reduced geographic space analysed in this study, the mean haplotype diversity in the Amazonas region (Hd = 0.57) was slightly lower than those in other studies conducted in Juglans. For example, Ebrahimi et al. (Reference Ebrahimi, Zarei, McKenna, Bujdoso and Woeste2017) obtained a value of Hd = 0.61 for specimens from Europe, while Dangl et al. (Reference Dangl, Woeste, Aradhya, Koehmstedt, Simon, Potter, Leslie and Mcgranahan2005) reported Hd = 0.60 for specimens from Asia, Europe, and North America. This suggests a relatively high genetic diversity within Juglans neotropica in the Amazonas region. This diversity is greater than in the genetically diverse common walnut from the Iranian Plateau (Shahi Shavvon et al. Reference Shahi Shavvon, Qi, Mafakheri, Fan, Wu, Vahdati, Al-Shmgani, Wang and Liu2023). It is also confirmed by the high nucleotide diversity (Pi = 0.004) of Juglans in the Amazonas region compared to those found in specimens from Asia (Pi = 0.002) (Hu et al. Reference Hu, Dang, Feng, Woeste and Zhao2017). For instance, in the population of Luya, nucleotide diversity (Pi = 0.009) and haplotype diversity (h = 7) were the highest in the Amazonas region. The observed high Hd and high Pi suggest either secondary contact of differentiated populations or long evolutionary history in a large stable population (Grant and Bowen Reference Grant and Bowen1998). The second scenario is supported by the haplotype network which shows star-like arrangement for H1 and H2, being a signature of prolonged demographic stability (Grant and Bowen Reference Grant and Bowen1998). In Amazonian trees, such signatures often reflect Pleistocene refugia populations (Dexter et al. Reference Dexter, Pennington, Oliveira-Filho, Bueno, Silva de Miranda and Neves2018).
The negative values of Tajima’s D for the Levanto, Leymebamba, and Molinopampa populations are caused by a sudden reduction in population size, which could be related to a possible bottleneck or a selective sweep (Ebrahimi et al. Reference Ebrahimi, Zarei, McKenna, Bujdoso and Woeste2017). These populations are constantly facing threats such as livestock, excessive agriculture, grazing and overexploitation (Casiano Reference Casiano2015, Oliva et al. Reference Oliva, Collazos, Goñas, Bacalla, Vigo, Vasquez and Leyva2016, Cachay Reference Cachay2021), which lead to the constant loss of habitat, causing fragmentation and, therefore, loss of genetic material (Vahdati et al. Reference Vahdati, Mohseni Pourtaklu, Karimi, Barzehkar, Amiri, Mozaffari and Woeste2015). On the other hand, the populations of Luya, Nogalcucho, San Isidro de Maino, and Yerbabuena presented positive values, indicating balanced selection (Muriira et al. Reference Muriira, Muchugi, Yu, Xu and Liu2018). This is because these populations do not present many threats due to their isolated locations and access difficulties and thus generate an increase in genetic diversity. This pattern may reflect ecological mechanisms such as (i) frequency-dependent pollinator foraging, where Andean bees and hummingbirds preferentially visit rare floral morphs (Eckhart et al. Reference Eckhart, Rushing, Hart and Hansen2006), and (2) rodent-mediated seed dispersal, a process previously documented in the closely related J. mandshurica (Wang et al. Reference Wang, Yan, Yan, Song, Sun and Zhu2017). These results coincide with what was found by Sun et al. (Reference Sun, Hou, Woeste, Zhang, Yue, Yuan and Zhao2019), who mentioned that there could be a greater connection of diversity in places where there is a lower incidence of human activities; therefore, these areas with fewer ecosystem disturbances lead to better adaptability with balanced selection and greater gene flow (Magige et al. Reference Magige, Fan, Wambulwa, Milne, Wu, Luo, Khan, Wu, Qi, Zhu, Maity, Khan, Gao and Liu2022).
Haplotype network analysis
The haplotype network of the individuals of the populations of J. neotropica from the Amazonas region was divided into 3 groups, comprising predominantly individuals from the population of Luya, with the presence of 7 haplotypes distributed in the three groups (H1, H2, H7, H8, H9, H10, H11). This high frequency of haplotypes could be caused by adaptation to changes in habitats or by probable fragmentation or isolation over time (Shigaeva and Darr Reference Shigaeva and Darr2020, Gaisberger et al. Reference Gaisberger, Legay, Andre, Loo, Azimov, Aaliev, Bobokalonov, Mukhsimov, Kettle and Vinceti2020). However, the hypothesis of a recent separation in small, fragmented populations is discarded because the results of genetic diversity yielded a balanced selection of populations of Juglans neotropica in the population of Luya (0.09). Additionally, the prevalence of haplotypes H1 and H2, displaying structural patterns characteristic of expanding populations in Amazonas, indicates their likely ancestral origin. The negative Tajima’s D values observed in our analysis provide statistical support for this conclusion.
The inclusion of new individuals from other populations outside the Amazonas revealed the differentiation of populations present in the Amazonas region with respect to the groups formed by specimens from other countries (e.g., groups IV, V). This difference could be related to genetic groups not previously reported, in addition to the scarce molecular information on genus Juglans in the Southern Hemisphere. For instance, group IV is exclusively distributed in Asia (Hu et al. Reference Hu, Dang, Feng, Woeste and Zhao2017), while group V is more dispersed throughout the Northern Hemisphere. The individuals in group IV are impacted by geographic barriers, cultivated material or local autochthonous plants that restrict their distribution (Magige et al. Reference Magige, Fan, Wambulwa, Milne, Wu, Luo, Khan, Wu, Qi, Zhu, Maity, Khan, Gao and Liu2022), while the wide distribution of group V is a consequence of possible recent human or natural transport of propagules at long distances (Gaisberger et al. Reference Gaisberger, Legay, Andre, Loo, Azimov, Aaliev, Bobokalonov, Mukhsimov, Kettle and Vinceti2020).
Additionally, the three groups identified in Juglans neotropica from the Amazonas region could probably represent three distinct taxa based on the high genetic divergence and haplotype grouping. Nevertheless, the use of a unique maker in this study hampers the confirmation of this finding. As previously stated, a multilocus approach and plastome data are crucial for the establishment of this potentially new taxa (Mao et al. Reference Mao, Zhang, Nakamura, Guan and Qiu2014).
Genetic structure of populations
AMOVA revealed greater genetic variation in partitioning within populations (Fst = 0.04), a common scenario, particularly for species with a cross-breeding system such as walnut (Magige et al. Reference Magige, Fan, Wambulwa, Milne, Wu, Luo, Khan, Wu, Qi, Zhu, Maity, Khan, Gao and Liu2022). The AMOVA results were consistent with the differences between the populations (Fst = 0.02), which represents a low level of connectivity (Aguirre-Pabon et al. Reference Aguirre-Pabon, Berdugo and Narváez2022). This could be due to fragmentation of the habitat caused by anthropogenic activities in the region (Zhou et al. Reference Zhou, Hu, Ebrahimi, Liu, Woeste, Zhao and Zhang2021). Habitat fragmentation limits or prevents long-distance pollination events, resulting in pollination only within groups of close relatives (Shigaeva and Darr Reference Shigaeva and Darr2020).
Conclusion
The use of the intergenic marker trnS-trnfM in specimens of the black walnut J. neotropica from the Amazonas region resolved them in the Rhysocaryon section. These specimens were found in a genetic group not previously reported and further genetic information (i.e., plastid genomes, multilocus phylogeny) could probably delimit candidates for new taxa in the genus Juglans. The genetic divergences of black walnut J. neotropica in the Amazonas region were slightly lower than those reported in Europe, Asia, and North America. Within the populations of the Amazonas region, the greatest genetic diversity was found in the Luya population and high divergence was detected in the populations of Molinopampa and Luya, probably due to the presence of genetic and geographical barriers, while the populations of Levanto and San Isidro de Mayno were less divergent due to their geographic proximity and greater genetic interaction. The populations of Levanto, Leymebamba and Molinopampa showed a reduction in population size, which could be linked to a selective sweep or a possible bottleneck, while the populations of Luya, Nogalcucho, San Isidro de Maino, Yerbabuena, and Tambolic showed balanced selection. Collectively, our findings indicate pronounced genetic variability alongside restricted gene flow between populations, a pattern consistent with anthropogenic habitat fragmentation. Consequently, we propose a multi-faceted in situ conservation strategy for Molinopampa and Luya to preserve the evolutionary potential of Juglans neotropica. This should integrate (i) targeted seed banking of genetically distinct populations to capture extant diversity, (ii) establishment of habitat corridors to facilitate gene flow between fragmented subpopulations, and (iii) community-based protection initiatives to mitigate anthropogenic pressures. Such measures are critical for maintaining both the species’ adaptive capacity and the ecological functionality of these Andean forest ecosystems. Our study yields novel insights into Juglans neotropica conservation by empirically linking deforestation to genetic erosion and generating critical baseline genetic parameters for monitoring.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0266467425100102
Acknowledgements
We express our gratitude to Dr. Manuel Oliva Cruz for his logistical support.
Financial support
This research was funded by CUI 2314090 ‘Creación de los servicios de un Herbarium y centro de adaptación de especies vegetales de la Universidad Nacional Toribio Rodríguez de Mendoza’, CUI 2261386 ‘Creación de los Servicios de un laboratorio de biodiversidad y conservación de recursos genéticos de especies silvestres en la Universidad Nacional Toribio Rodríguez de Mendoza – Región Amazonas’ – BIODIVERSIDAD, and CONCYTEC under the Project MiCroResi (N° PE501079652-2022-PROCIENCIA).
Competing interests
The authors declare none.











