Introduction
Palmer amaranth and common waterhemp are two agronomically important weed species primarily affecting North American cropping systems (Steckel Reference Steckel2007; Van Wychen Reference Van Wychen2022); however, the recent spread of Palmer amaranth to parts of Africa and South America threatens food security and sustainable weed control on a global basis (Berger et al. Reference Berger, Madeira, Ferrell, Gettys, Morichetti, Cantero and Nuñez2016; Küpper et al. Reference Küpper, Borgato, Patterson, Netto, Nicolai, de Carvalho, Nissen, Gaines and Christoffoleti2017; Reinhardt et al. Reference Reinhardt, Vorster, Küpper, Peter, Simelane, Friis, Magson and Aradhya2022). Traits that allow Palmer amaranth and waterhemp to evade management and cause crop yield loss include rapid vegetative growth, multiple flushes of germination throughout the growing season, prolific seed production, and the ability to quickly adapt and evolve in the face of management practices or changes in environment (Costea et al. Reference Costea, Weaver and Tardif2005; Ward et al. Reference Ward, Webster and Steckel2017). This ability to quickly adapt is partially due to the large pool of genetic diversity and the reproductive system in both weed species (Kreiner et al. Reference Kreiner, Giacomini, Bemm, Waithaka, Regalado, Lanz, Hildebrandt, Sikkema, Tranel, Weigel, Stinchcombe and Wright2019; Küpper et al. Reference Küpper, Manmathan, Giacomini, Patterson, McCloskey and Gaines2018). Both Palmer amaranth and waterhemp are obligate outcrossers, which helps facilitate the combining of herbicide-resistant alleles, while wind pollination and human-mediated seed transportation allow resistant alleles to rapidly spread across long distances. Years of widespread herbicide application have resulted in the evolution of herbicide resistance to many effective sites of action (SOAs) used to control these Amaranthus weeds, up to nine SOAs for each species, leaving few effective tools for the management of these species (Heap Reference Heap2024).
Herbicides that inhibit the protoporphyrinogen oxidase (PPO) enzyme have become a commonly used tool for controlling broadleaf weeds in soybean. These include several commercial products that are registered for preemergence use on soybean (e.g., flumioxazin, saflufenacil, sulfentrazone) and postemergence use (e.g., lactofen, fomesafen, acifluorfen). Additionally, the development of transgenic crops that are resistant to a new generation of nonselective, broad-spectrum, PPO inhibitors may allow for use of more efficacious herbicides and extend the use of these herbicides to new cropping systems. Thus, Bayer CropScience is currently developing crops, including soybean [Glycine max (L.) Merr.], cotton (Gossypium hirsutum L.), and corn (Zea mays L.) that express microbial-sourced HemG PPO enzyme variants that exhibited robust tolerance to a wide range of commercial and novel PPO inhibitors (Larue et al. Reference Larue, Ream, Zhou, Moshiri, Howe, Goley, Sparks, Voss, Hall, Ellis, Weihe, Qi, Ribeiro, Wei, Guo, Evdokimov, Varagona and Roberts2020). Herbicides belonging to this SOA group were synthesized as early as 1930 (Raiford et al. Reference Raiford, Thiessen and Wernert1930) and were commercialized in the 1960s (Rhone-Poulenc Reference Rhone-Poulenc1968). The mechanism of action for these compounds eluded researchers for decades, but it was eventually found that they inhibit PPO, the last common enzyme in the heme and chlorophyll synthesis pathways (Matringe et al. Reference Matringe, Camadro, Labbe and Scalla1989). PPO isozymes are localized to the mitochondria and chloroplast. The inhibition of PPO isozymes leads to the accumulation of protoporphyrinogen, which leaks into the cytoplasm where it is converted to protoporphyrin (Lydon and Duke Reference Lydon and Duke1988). Protoporphyrin is phototoxic and creates reactive oxygen species in the presence of light, which oxidizes lipids and ultimately leads to cell death.
Target site and nontarget site resistance mechanisms are found in plants that render PPO-inhibiting herbicides ineffective. In soybean, rapid metabolism of certain PPO inhibitors leads to innate tolerance (Dayan et al. Reference Dayan, Weete, Duke and Hancock1997, Reference Dayan, Armstrong and Weete1998). Increased herbicide metabolism has been hypothesized as a mechanism for resistance in several Palmer amaranth and waterhemp accessions (Obenland et al. Reference Obenland, Ma, O’Brien, Lygin and Riechers2019; Varanasi et al. Reference Varanasi, Brabham and Norsworthy2018, Reference Varanasi, Brabham, Korres and Norsworthy2019), but no clear mechanism or genetic basis have been elucidated. The evolution of resistance to PPO inhibitors via increased expression or duplication of the target gene has not been observed, as it has been with glyphosate and glufosinate (Gaines et al. Reference Gaines, Zhang, Wang, Bukun, Chisholm, Shaner, Nissen, Patzoldt, Tranel, Culpepper, Grey, Webster, Vencill, Sammons, Jiang, Preston, Leach and Westra2010; Noguera et al. Reference Noguera, Porri, Werle, Heiser, Brändle, Lerchl, Murphy, Betz, Gatzmann, Penkert, Tuerk, Meyer and Roma-Burgos2022). The most common and widely studied mechanism of resistance to PPO inhibitors involves modification of the genes that encode PPO.
In plants, two nuclear genes designated protoporphyrinogen oxidase 1 (PPX1) and protoporphyrinogen oxidase 2 (PPX2), encode for the enzymes PPO1 and PPO2, respectively, which catalyze the oxidation of protoporphyrinogen IX. In most cases, PPO1 is targeted to the chloroplast where chlorophyll and heme are synthesized, while PPO2 is targeted to the mitochondria where heme is produced. Interestingly, PPX2 in Amaranthus species has two transcriptional start sites, leading to short (PPX2S) and long (PPX2L) isoforms of the gene. The short isoform contains the classic mitochondria localization domain only, while the long isoform also contains a chloroplast localization domain (Dayan et al. Reference Dayan, Barker and Tranel2018). It is believed that because of this dual targeting of PPO2, mutations in PPX2 that reduce affinity of herbicides are sufficient to confer resistance in Amaranthus weeds. Accordingly, variants of PPX2 have been identified to confer resistance to PPO inhibitors in Amaranthus weeds including the deletion of the 210 glycine residue (ΔG210) (Patzoldt et al. Reference Patzoldt, Hager, McCormick and Tranel2006), the substitution of the arginine residue, R128 to leucine (R128L) or glycine (R128G) (Giacomini et al. Reference Giacomini, Umphres, Nie, Mueller, Steckel, Young, Scott and Tranel2017), and the substitution of the glycine residue, G399 to alanine (G399A) (Rangani et al. Reference Rangani, Salas-Perez, Aponte, Knapp, Craig, Mietzner, Langaro, Noguera, Porri and Roma-Burgos2019). Most recently, the substitution of the valine residue, V361 to alanine (V361A), was found to confer resistance to fomesafen in Palmer amaranth (Nie et al. Reference Nie, Harre and Young2023). In addition, substitution of the alanine residue, A212 to threonine (A212T) in PPX1, was shown to cause resistance to the PPO inhibitor oxadiazon in Eleusine indica, a grass species that does not possess a dual-targeted PPO2 (Bi et al. Reference Bi, Wang, Coleman, Porri, Peppers, Patel, Betz, Lerchl and McElroy2020).
Recently gained knowledge about the biochemical basis of target site PPO-inhibitor resistance described above makes it possible to model the effect of target site mutations and predict new and existing herbicides that inhibit mutant versions of PPO (Ekins et al. Reference Ekins, Mestres and Testa2007; Wu et al. Reference Wu, Goldsmith, Pawlak, Feng, Smith, Navarro and Perez-Jones2020). While the utilization of these computational methods in herbicide design and discovery is still in its infancy, Porri et al. (Reference Porri, Betz, Seebruck, Knapp, Johnen, Witschel, Aponte, Liebl, Tranel and Lerchl2023) recently described the binding configuration of a new PPO inhibitor, trifludimoxazin, with PPO2. The authors explain from a structural and biochemical perspective why trifludimoxazin binding is less affected by known resistance-conferring target site mutations compared to other commercially available PPO inhibitors. This work complements other efforts to understand how target site variants affect the structure of PPO2 (Dayan et al. Reference Dayan, Barker and Tranel2018; Rangani et al. Reference Rangani, Salas-Perez, Aponte, Knapp, Craig, Mietzner, Langaro, Noguera, Porri and Roma-Burgos2019; Wu et al. Reference Wu, Goldsmith, Pawlak, Feng, Smith, Navarro and Perez-Jones2020). An understanding of the molecular basis of herbicide resistance will increase the power of in silico prediction tools that estimate herbicide docking to mutated targets.
As new PPO-inhibiting herbicides are introduced to the market, it is important to understand the current landscape of resistance mechanisms and how they affect the efficacy of these new molecules. In this paper, collections of Palmer amaranth and waterhemp from across the southeastern and midwestern United States were examined to understand the distribution and frequency of known PPX2 target site resistance mutations and the performance of the new active ingredient epyrifenacil (Rapidicil™), a novel PPO-inhibiting herbicide being developed by Sumitomo Chemical Company (Sada et al. Reference Sada, Jin, Hikosaka and Ido2024). Epyrifenacil is a PPO-inhibiting herbicide from the chemical class pyrimidinedione (also known as uracil) with a unique three-ring structure. It is a systemic herbicide with broad-spectrum postemergence control of grass and broadleaf weeds at lower use rates than other registered PPO-inhibiting herbicides. Results show that epyrifenacil was highly effective on nearly all collected accessions of Palmer amaranth and waterhemp, even when known resistance-causing mutations are present. However, the results also indicate a reduction in the plant injury level caused by epyrifenacil in some accessions carrying the ΔG210 allele of PPX2.
Material and Methods
Weed Seed Collection
During the 2019 growing season, 141 Palmer amaranth and 133 waterhemp accessions were collected by Bayer CropScience representatives from broadacre cropping systems (mainly soybean, cotton, and maize) in the southeastern and midwestern United States, spanning 263 different counties and 21 states (Table S1) (Singh et al. Reference Singh, Tyre, Perez-Jones, Krebel, Willis, Herrmann, Klingaman, Head and Aradhya2023). Weed seeds were collected from the inflorescence of 10 to 20 plants within the field following a zig-zag pattern and combined to form a composite sample, as outlined by Burgos et al. (Reference Burgos, Tranel, Streibig, Davis, Shaner, Norsworthy and Ritz2013). Collected seeds were air-dried in paper bags, shipped to the Bayer CropScience research facility in Chesterfield, MO, and then cleaned and stored at 4 C until assayed. This sampling was not intended to enrich for PPO-inhibitor resistant biotypes (i.e., by sampling from sites with reported low efficacy of PPO-inhibiting herbicides) like similar studies that have attempted to illustrate the landscape of resistance (Montgomery et al. Reference Montgomery, Giacomini and Tranel2021; Noguera et al. Reference Noguera, Rangani, Heiser, Bararpour, Steckel, Betz, Porri, Lerchl, Zimmermann, Nichols and Roma-Burgos2021; Salas et al. Reference Salas, Burgos, Tranel, Singh, Glasgow, Scott and Nichols2016). Instead, this collection was meant to portray a less biased picture of the frequency and distribution of resistance and sequence variants that exist in Palmer amaranth and waterhemp populations. Although a random stratified design would have further decreased bias in the experiment, these designs are difficult to execute and rarely practiced in weed science. Collectors were tasked with representing as many counties as possible from across the country and not asked to consider reports of PPO-inhibitor efficacy or weed management history when selecting sampling sites.
Plant Growth Conditions
To produce individual plants for testing, approximately 10 seeds were directly seeded into each cell of plug flats (Hummert International, Earth City, MO) containing commercial Promix BX potting soil (Hummert International) that had been saturated through subirrigation in a greenhouse at the Bayer CropScience research facility in Chesterfield, MO. Planted flats were covered with domes and subirrigated as needed until seedling emergence. Seedlings were manually thinned to one plant per cell after cotyledons were fully formed. Healthy seedlings were selected 7 to 14 d after planting and individually transplanted into 10-cm square vacuum deep (SVD) pots (Hummert International) containing commercial Promix BX potting soil. Prior to transplanting, soil in the SVD pots was thoroughly saturated via subirrigation, and transplanted plants were watered as needed by subirrigation. Plants were grown in the greenhouse at 29/26 C day/night temperature, 40% to 60% relative humidity, and 16/8 h day/night photoperiods supplemented with sodium halide lamps (560 μmol m−2 s−1).

Figure 1. Results of phenotypic and genotypic screening of Palmer amaranth (![]() ) and waterhemp (
) and waterhemp (![]() ) accessions (n = 240; ∼10 plants per accession) from across the southeastern and midwestern United States. Results include the distribution of individual accessions with % survival after epyrifenacil treatment at 20 g ha−1 21 d after treatment (A); and distribution of the ΔG210 allele of PPX2 (B).
) accessions (n = 240; ∼10 plants per accession) from across the southeastern and midwestern United States. Results include the distribution of individual accessions with % survival after epyrifenacil treatment at 20 g ha−1 21 d after treatment (A); and distribution of the ΔG210 allele of PPX2 (B).
Initial Epyrifenacil Efficacy Testing
Following the methods described by Singh et al. (Reference Singh, Tyre, Perez-Jones, Krebel, Willis, Herrmann, Klingaman, Head and Aradhya2023), a target of 10 plants from each accession were grown as described above. Each experimental run included a known PPO-inhibitor-sensitive and a resistant accession for each weed species (Table S1). The total number of plants screened per accession (Table S1) varied due to variation in seed germination, seed quality, and seed quantity. When plants reached a height of approximately 10 to 13 cm they were sprayed with epyrifenacil at 20 g ai ha−1 (Rapidicil™; Sumitomo Chemical Company Ltd., Takarazuka, Japan) along with 10 mL L−1 crop oil concentrate (Prime Oil®; WinField United, Arden Hills, MN) using a custom-built cabinet spray chamber (Bayer CropScience Technical Discovery Center, Chesterfield, MO) mounted with a 9501E flat-fan nozzle (TeeJet Technologies, Glendale Heights, IL). The nozzle was calibrated to deliver 140 L ha−1 of spray solution at 276 kPa at an approximate speed of 2.5 km h−1. Plants were evaluated for survival and visual injury approximately 21 d after treatment (DAT) on a scale from 0% (no visual injury) to 100% (complete plant mortality). Survival was determined by whether the plant would eventually produce viable flowers. Generally, plants with injury ≤90% were considered survivors. Data were plotted onto a map of the United States (Figure 1) using RStudio software (v.4.0.2; R Core Team 2023; also v.1.3.1073; RStudio Team 2023) using the ggplot2 package (v.3.3.6, Wickham Reference Wickham2016).
PPX2 Sequencing
Just prior to herbicide application, the youngest fully expanded leaf of each individual plant from the initial epyrifenacil efficacy testing was sampled into 96-well plates prefilled with metal bead balls, over dry ice and stored at −80 C until use. Plant tissues were ground in the presence of the Trizol® reagent using a tissue homogenizer and a large paint-shaker. Total RNA was isolated using Direct-zol™ RNA MiniPrep Kits (Zymo Research, Irvine, CA) following the manufacturer’s instructions.
Plant RNA was used to generate cDNA with a High-Capacity cDNA Reverse Transcription Kit (no. 4368814; Applied Biosystems, Thermo Fisher Scientific) using oligo dT primers (20 µl reactions + 200 ng total RNA). The full coding sequence of the PPX2 gene (1,580 bp) was amplified using the following primers for both weed species: forward ATGGGCAACATTTCTGAGCGG and reverse TTAYGCGGTCTTCTCATCCATCTTCAC in a 20-µl polymerase chain reaction (PCR) that contained 2 µl of oligo dT primed cDNA template, 1 µl of each primer (10 µM), 10 µl of Phusion Flash HighFidelity® PCR Master Mix (no. F-548S; Thermo Fisher Scientific), and 16 µl of nuclease-free water. The thermal cycling condition were 98 C for 10 s; 35 cycles at 98 C for 1 s, 68 C for 5 s, and 72 C for 30 s; followed by a final extension for 2 min at 72 C (Wu et al. Reference Wu, Goldsmith, Pawlak, Feng, Smith, Navarro and Perez-Jones2020). PCR products were fragmented using the Illumina DNA prep Library Preparation Kit (no. 20018705; Illumina, San Diego, CA) to build a 300-bp cDNA library. These libraries were sequenced using an Illumina NextSeq 550 instrument at the Bayer CropScience facility in Chesterfield, MO. The resulting reads from each sample were aligned to the sequence of PPX2 from Palmer amaranth using the BWA aligner (v0.7.17; Li Reference Li2013). These alignments were used to call sequence variants with GATK (v4.2.2.0; Poplin et al. Reference Poplin, Ruano-Rubio, DePristo, Fennell, Carneiro, Van der Auwera, Kling, Gauthier, Levy-Moonshine, Roazen, Shakir, Thibault, Chandran, Whelan, Lek, Gabriel, Daly, Neale, MacArthur and Banks2017; Van der Auwera and O’Connor Reference Van der Auwera and O’Connor2020). Variants were then annotated using effects of single nucleotide polymorphisms (SnpEff) to predict the effect of each sequence variant on the protein product (e.g., synonymous substitution, premature stop codon introduced, etc.) (v5.0.0; Cingolani et al. Reference Cingolani, Platts, Wang, Coon, Nguyen, Wang, Land, Lu and Ruden2012). Good-quality genotypic data were obtained for 240 out of the 274 accessions.
Confirmation of Initial Efficacy Testing
To confirm the results of the initial efficacy testing and further examine the effect of the ΔG210 allele of PPX2, seven accessions with a high estimated ΔG210 allele frequency (≥50%; six of waterhemp and one of Palmer amaranth) were grown and subjected to another round of screening. Known PPO inhibitor-sensitive and inhibitor-resistant accessions were included as controls. Both resistant control accessions used have high ΔG210 allele frequency. Growth and herbicide spraying conditions were the same as those mentioned above. Each accession was subjected to three treatments that included saflufenacil at 25 g ai ha−1 (Sharpen®; BASF Corporation, Research Triangle Park, NC), trifludimoxazin + saflufenacil at 37.5 g ai ha−1 (Voraxor™; BASF), and epyrifenacil at 20 g ha−1 (Rapidicil™; Sumitomo Chemical Company Ltd., Takarazuka, Japan). Sharpen® and Voraxor™ treatments included 10 mL L−1 methylated seed oil (Destiny® HC; WinField United, Arden Hills, MN) and Rapidicil™ treatment included 10 mL L−1 crop oil concentrate (Prime Oil®; WinField United). These treatments were intended to represent approximate field rates for each herbicide. Twenty plants from each tested accession were used for each treatment, and each accession also had two untreated control plants. Plant survival and visual plant injury were assessed 21 DAT using the rating scale described above.
Table 3. Frequencies of plant survivors in waterhemp and Palmer amaranth with or without variants of PPX2 following treatment with epyrifenacil at 20 g ha−1.

Association of Genetic Variants with Herbicide Efficacy
To understand whether any sequence variants of PPX2 were associated with a reduction in epyrifenacil efficacy, a simple linear model was fitted using R software (v.4.0.2; R Core Team 2023) to estimate the effect of each sequence variant on visual injury following epyrifenacil treatment on the same plant. Briefly, genotype information was extracted from the GATK output, separated by weed species, and paired with visual injury values for each individual. Synonymous variants (those that do not cause an amino acid change) were filtered out of the data set based on SnpEff output. For each remaining variant locus, a simple linear model described by Equation 1 was fitted:
where y is the visual injury, x is the genotype at that locus (homozygous wildtype = 0, heterozygous = 1, homozygous mutant = 2), β is the slope (expected change in control associated with each copy of the variant at that locus), and α is the y-intercept (visual injury for a plant that is homozygous for the wild-type allele at that locus). Bonferroni correction of ANOVA (Goeman and Solari Reference Goeman and Solari2014) was used to identify loci significantly associated with a change in visual injury following epyrifenacil treatment. Pearson’s correlation coefficients were calculated for significant loci to understand whether these loci explain the same variance in visual injury (R Core Team 2023).
Table 1. Frequency of survivorship following application of epyrifenacil at 20 g ha−1 and known herbicide resistance-conferring target site mutations of PPX2 in Palmer amaranth and waterhemp.a
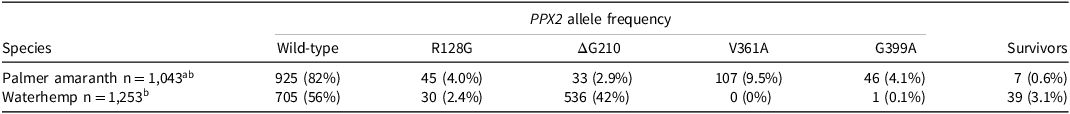
a The percent of total plants tested is listed in parentheses for each metric reported. Percentages of different genotypes may not sum to 100 because some plants possess multiple target site mutations and are represented in multiple columns.
b Total number of plants tested.
Results and Discussion
Distribution and Frequency of Accessions Surviving Epyrifenacil Treatment and Resistance Mutations in PPX2 Gene
A summary of the phenotypic results from the initial screen of 274 accessions of Palmer amaranth and waterhemp can be found in Table S1. Nearly all accessions (92%) were completely controlled by epyrifenacil. Accessions that were not completely controlled had low survivor frequency, with either one or two survivors out of 10 tested plants. PPO-inhibiting herbicides frequently show higher efficacy under optimized greenhouse settings compared to field conditions. Thus, to preserve the efficacy of epyrifenacil, it is best to monitor and control any surviving plants before seed-set production. Not allowing weeds to survive and reproduce in the field, especially highly competitive and prolific seed-producing species such as Palmer amaranth and waterhemp, is a best management practice and recommendation to reduce the risk of herbicide resistance (Norsworthy et al. Reference Norsworthy, Ward, Shaw, Lewellyn, Nichols, Webster, Bradley, Frisvold, Powles, Burgos, Witt and Barrett2012). Figure 1 shows the distribution and frequency of accessions exhibiting varied responses to epyrifenacil treatment at 21 DAT and the ΔG210 allele of PPX2 across 240 accessions where good quality genotypic data were obtained. Across these accessions, genotype information was gathered from an average of 8.7 of the 10 plants tested from each accession. Table 1 lists the frequency of known resistance-conferring mutations as well as the frequency of survivors from the initial screen. The plant survival rate observed here is lower than expected given the frequency of resistance conferring target site mutations (Table 1). In fact, of 33 Palmer amaranth plants with at least one copy of the ΔG210 allele of PPX2, only three (11%) plants survived. Of these plants, five were homozygous for the ΔG210 allele, and only one (20%) survived. Similarly, of 536 waterhemp plants with at least one copy of the ΔG210 allele of PPX2, only 36 (3%) survived. Of 215 waterhemp plants that were homozygous for the ΔG210 allele, 20 (9%) survived. All wild-type plants not carrying known resistance-conferring mutations showed 100% mortality. These results suggest that epyrifenacil can provide effective control of Amaranthus weeds that are not expected to be controlled by other PPO-inhibiting herbicides, because the ΔG210 allele of PPX2 confers high-level, broad-spectrum resistance to other commercially available PPO inhibitors such as carfentrazone, flumioxazin, fomesafen, and lactofen (Barker et al. Reference Barker, Pawlak, Duke, Beffa, Tranel, Wuerffel, Young, Porri, Liebl, Aponte, Findley, Betz, Lerchl, Culpepper, Bradley and Dayan2023, Rangani et al. Reference Rangani, Salas-Perez, Aponte, Knapp, Craig, Mietzner, Langaro, Noguera, Porri and Roma-Burgos2019; Salas-Perez et al. Reference Salas-Perez, Burgos, Rangani, Singh, Paulo Refatti, Piveta, Tranel, Mauromoustakos and Scott2017). Similarly, Sada et al. (Reference Sada, Jin, Hikosaka and Ido2024) reported that epyrifenacil applied at 20 g ha−1 controlled some biotypes of target site–based PPO-inhibitor resistant weeds, including Palmer amaranth accessions carrying the ΔG210, R128G, and G399A alleles, in the greenhouse and the field.
The frequency of resistance mutations found in this study in accessions of Palmer amaranth is considerably lower (<20%) (Table 1) than has been reported in other similar studies (Montgomery et al. Reference Montgomery, Giacomini and Tranel2021; Noguera et al. Reference Noguera, Rangani, Heiser, Bararpour, Steckel, Betz, Porri, Lerchl, Zimmermann, Nichols and Roma-Burgos2021; Salas et al. Reference Salas, Burgos, Tranel, Singh, Glasgow, Scott and Nichols2016). This is likely due to differences in the sampling methodology, which does not specifically target fields that are likely to contain PPO inhibitor-resistant weeds (see Singh et al. Reference Singh, Tyre, Perez-Jones, Krebel, Willis, Herrmann, Klingaman, Head and Aradhya2023 for more details and discussion). Nevertheless, while the sampling methodology is not completely random, we believe the results provide a better representation of the general distribution of PPX2 mutations across a larger landscape in the United States. In fact, the frequency of resistance mutations in Palmer amaranth accessions in this study is similar to the most recent characterization of the PPO-resistance landscape (Nie et al. Reference Nie, Harre and Young2023). For waterhemp, the results align with previous findings in which PPO inhibitor resistance is most frequently caused by the ΔG210 allele of PPX2 (Lee et al. Reference Lee, Hager and Tranel2008), and substitutions of R128 are relatively rare (Nie et al. Reference Nie, Mansfield, Harre, Young, Steppig and Young2019) (Table 1). Finally, it is important to note that this survey of Amaranthus species did not find any confirmed instances of multiple homozygous PPX2 resistance mutations co-occurring on the same allele as was documented by Noguera et al. (Reference Noguera, Rangani, Heiser, Bararpour, Steckel, Betz, Porri, Lerchl, Zimmermann, Nichols and Roma-Burgos2021). The plants carrying multiple PPX2 resistance mutations were all heterozygous for each mutation, meaning these resistance mutations may be on separate alleles.
Table 2. Evaluation of herbicide efficacy on accessions carrying high frequency (≥50%) of the ΔG210 mutation.a

a State abbreviations: IA, Iowa; IL, Illinois; IN, Indiana; MO, Missouri; SC, South Carolina; TN, Tennessee.
b Approximately 20 plants were used for each treatment from each accession.
c Allele frequency of the ΔG210 allele of PPX2, as determined in the genotyping from the initial screen, is represented in the %ΔG210 column.
d Sensitive controls.
e Resistant controls.
Confirmation of Epyrifenacil Efficacy on Amaranthus Accessions Carrying PPX2 Resistance Mutations
Follow-up testing of seven accessions with high frequency (≥50%) of the ΔG210 allele of PPX2 showed that epyrifenacil treatment at 20 g ha−1 provided the best control among the PPO inhibitors tested, averaging 85% mortality across these accessions, whereas applications of saflufenacil at 25 g ha−1 and trifludimoxazin + saflufenacil at 37.5 g ha−1 averaged 52% and 69% mortality, respectively (Table 2). A rapid increase in the number of cases of resistance to PPO inhibitors in Amaranthus weeds in the past years has been attributed to an increased use of PPO-inhibiting herbicides to address widespread resistance to glyphosate and acetolactate synthase inhibitors (Barker et al. Reference Barker, Pawlak, Duke, Beffa, Tranel, Wuerffel, Young, Porri, Liebl, Aponte, Findley, Betz, Lerchl, Culpepper, Bradley and Dayan2023). Epyrifenacil is part of a new generation of PPO-inhibiting herbicides with improved efficacy compared to other products such as flumioxazin, fomesafen, and lactofen that can potentially overcome resistance due to some known target site mutations in PPX2 (Sada et al. Reference Sada, Jin, Hikosaka and Ido2024). Even though the results showed that some accessions with high frequency of known resistance-conferring mutations, such as ΔG210, can be completely controlled with epyrifenacil (Table 2), it is critical to implement proactive measures that include herbicide treatments with other effective SOAs to ensure full control across accessions carrying PPO resistance mutations and prolong the efficacy of epyrifenacil. In addition, it is important to note that new mutations not yet reported could occur in the future that could potentially reduce the efficacy of epyrifenacil.
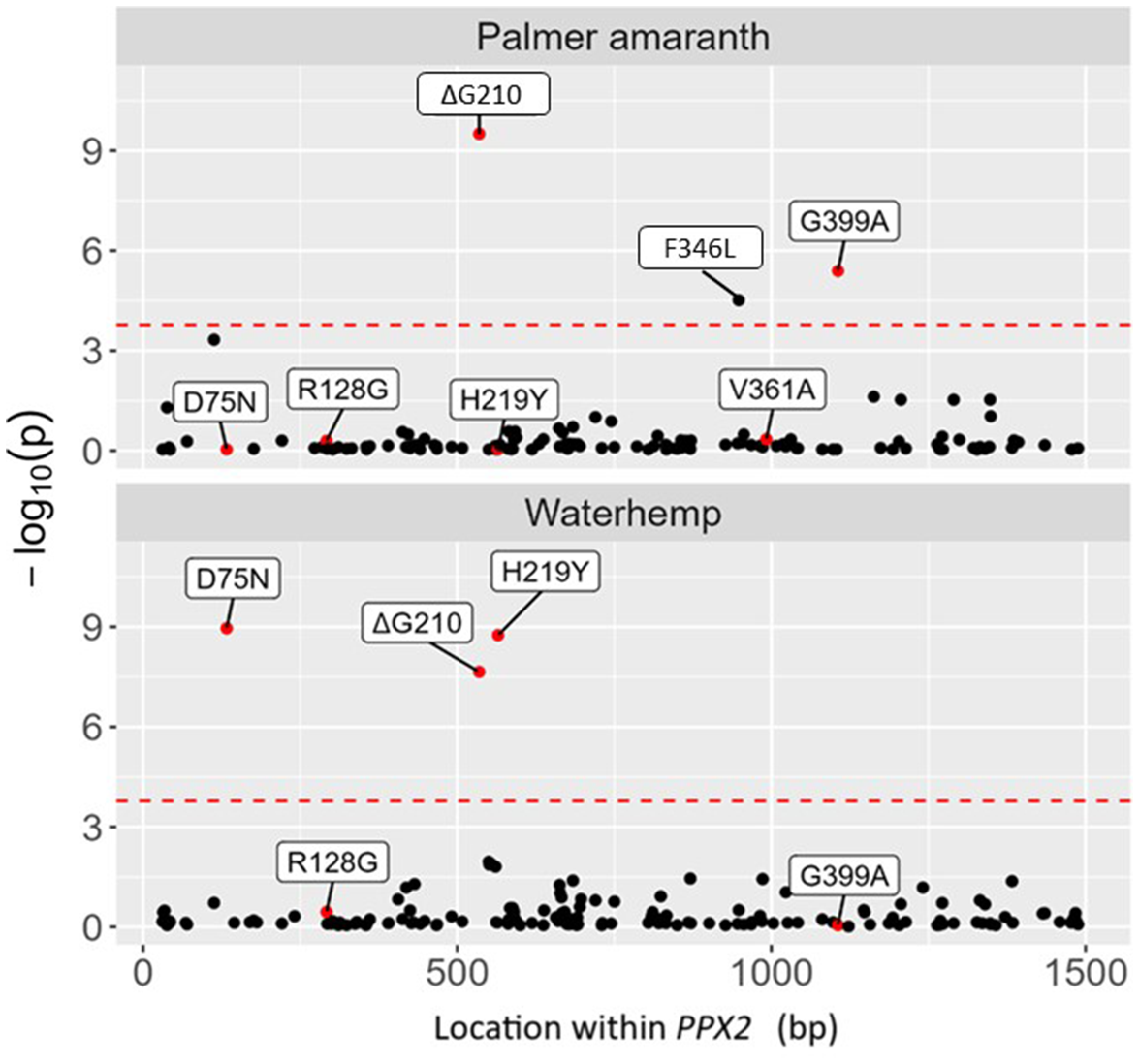
Figure 2. Association study correlating alternative allele frequency with injury following treatment with epyrifenacil at 20 g ha−1 in Palmer amaranth (top) and waterhemp (bottom). Loci of interest are colored red and labeled with predicted effect on protein sequence of PPX2 to fit with common nomenclature of known resistance mutations. The threshold for statistical significance is displayed as a dashed red line and represents a Bonferroni correction of α = 0.05.
Association of Genetic Variants with Herbicide Efficacy
To understand whether additional, uncharacterized target site mutations might be associated with reduced efficacy to epyrifenacil, an association study was conducted on plants from the initial screen to correlate alternative allele frequencies with visual plant injury following epyrifenacil treatment. Results of that analysis are shown in Figure 2 and parameter estimates for each locus are listed in Table S2. For Palmer amaranth, ΔG210 was significantly associated with a reduction in injury (Figure 2), while R128G and V361A alleles were not. Although the number of samples with these mutations was low in Palmer amaranth (Table 1), the result indicates that the efficacy of epyrifenacil is not affected by the presence of these two alleles. The analysis also showed that G399A and the substitution phenylalanine-346 to leucine (F346L) were significantly associated with reduced injury in Palmer amaranth (Figure 2). However, these mutations have smaller effects (slopes) on the efficacy of epyrifenacil compared to ΔG210 (Table S2). Finally, there was no connection between the presence of G399A and survival of plants containing the ΔG210 allele (Fisher exact test odds ratio = 0.32, df = 1, P = 0.40) (Table 3). In waterhemp, as in Palmer amaranth, the ΔG210 allele was significantly associated with a reduction in injury to epyrifenacil treatment, while the R128G was not (Figure 2). Interestingly, two additional single-nucleotide polymorphism (SNP) variants in the PPX2 gene were also significant in the analysis (Figure 2). Each of these SNPs are the consequence of a single nucleotide change and result in the amino acid substitutions of aspartate-75 to asparagine (D75N) and histidine-219 to tyrosine (H219Y), respectively (amino acid sequence of Palmer amaranth PPX2S is used for alignment and to indicate position of these SNPs to maintain consistency with other published resistance mutations). Although the presence of these two SNPs is positively correlated with the presence of ΔG210 in waterhemp (Pearson correlation coefficients of 0.48 and 0.67, respectively), they are not completely linked. Table 3 shows survivorship frequency for waterhemp plants with different genotypes at these two loci and the ΔG210 locus. There were no cases of a plant surviving epyrifenacil application with either D75N or H219Y, but without ΔG210, suggesting that these variants do not independently confer reduction of efficacy. Additionally, a chi-square test found no significant change in survivor frequency of ΔG210 plants with or without these variants (D75N χ2 = 0.19, df = 1, P = 0.66; H219Y χ2 = 0.04, df = 1, P = 0.84) (Table 3). We hypothesize that linkage of these variants with the ΔG210 allele caused them to be identified in the analysis and that they do not contribute to reduced herbicide efficacy. Although a protein modeling and in vitro enzyme inhibition assays like those performed by Rangani et al. (Reference Rangani, Salas-Perez, Aponte, Knapp, Craig, Mietzner, Langaro, Noguera, Porri and Roma-Burgos2019) and Porri et al. (Reference Porri, Betz, Seebruck, Knapp, Johnen, Witschel, Aponte, Liebl, Tranel and Lerchl2023) were not part of this study, they could help elucidate the true effect of F346L, D75N, and H219Y SNPs on the binding affinity of epyrifenacil and other PPO-inhibiting herbicides.
Practical Implications
This study represents a large-scale screening of Amaranthus weeds from across the United States and gives an updated view of the distribution and frequency of target site variants of PPX2 known to confer resistance to PPO inhibitors. This study showed that the efficacy of epyrifenacil at 20 g ha−1 on Palmer amaranth and waterhemp in a greenhouse study is not associated with several PPX2 target site variants that cause resistance to other PPO inhibitors such as lactofen, fomesafen, and saflufenacil. However, the results suggest that the ΔG210 allele of PPX2 is associated with a reduction in efficacy to epyrifenacil, although it does not confer full plant resistance. Hence, it is important to implement proactive measures that include other effective herbicide treatments with different SOAs as part of a diversified program for effective weed control to prolong the efficacy of epyrifenacil.
These results, along with those reported by Porri et al. (Reference Porri, Betz, Seebruck, Knapp, Johnen, Witschel, Aponte, Liebl, Tranel and Lerchl2023), reiterate PPO as an attractive SOA for new herbicide discovery in the fight against herbicide-resistant weeds. This study also illustrates the value of using molecular tools to predict the efficacy of new candidate PPO-inhibiting herbicides through in silico and in vitro screening (Satz et al. Reference Satz, Cai, Chen, Goodnow, Gruber, Kowalczyk, Petersen, Naderi-Oboodi, Orzechowski and Strebel2015), and demonstrates the importance of knowing specific alleles that confer resistance to PPO inhibitors, or other herbicides with different SOAs, in a weed population. Additionally, it emphasizes the significance of understanding how these alleles are associated with the efficacy of specific herbicides to help growers select more precise and targeted weed control solutions for individual farms.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/wet.2025.40
Acknowledgments
JM thanks Bayer CropScience for the opportunity to work as an intern during the summer of 2022. The authors thank the many field representatives at Bayer CropScience for collecting seeds; Jenny Krebel, Megan Gilley, and Michael Eoff for greenhouse testing and data generation; and Cheryl Johnson for tissue sample collection.
Funding
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Competing Interests
Epyrifenacil (Rapidicil™) is a new PPO-inhibiting herbicide discovered and being developed by Sumitomo Chemical Company Ltd.







