Introduction
The intra-uterine environment plays a critical role in fetal growth and development during pregnancy, and adverse maternal conditions can set the stage for lifelong offspring health problems Reference Barker, Thornburg, Osmond, Kajantie and Eriksson1 . Prior research has linked maternal pre-pregnancy obesity (ppOB; defined as body mass index [BMI] ≥30 kg/m2) to high birthweight (macrosomia) and, to a lesser degree, low birthweight, as well as higher risk for pregnancy complications including gestational diabetes, pre-eclampsia, gestational hypertension, and preterm delivery Reference Huda, Brodie and Sattar2 . The placenta functions as a key mediator of these relationships Reference Fowden, Camm and Sferruzzi-Perri3 . ppOB exerts distinct effects on the placenta, including its size, efficiency, and histopathology Reference Fowden, Camm and Sferruzzi-Perri3–Reference Kovo, Zion-Saukhanov and Schreiber5 . Some of these adverse impacts on placental growth and function may be due to alterations in expression of placental genes, including those involved in angiogenesis, or changes in their epigenetic regulators, such as long non-coding RNAs (lncRNAs) Reference Lassance, Haghiac and Leahy6–Reference Cabili, Trapnell and Goff9 .
LncRNAs are RNA transcripts longer than 200 base pairs that are not translated into proteins Reference Statello, Guo, Chen and Huarte10 . They are now understood to be new class of epigenetic biomarkers known to have transcription regulation roles through physical interactions with mRNA, DNA, proteins and miRNAs at transcriptional, post-transcriptional, translational, and post-translational stages Reference de Goede, Nachun and Ferraro11 . LncRNAs are less abundant than mRNAs and show considerable tissue-specificity and inter-individual variation Reference de Goede, Nachun and Ferraro11 . To date, around 40 placental lncRNA transcripts have been characterized to have roles in placental cell growth control, trophoblast invasion, and in cases where they appear aberrantly expressed, pregnancy complications Reference Cabili, Trapnell and Goff9,Reference Adu-Gyamfi, Cheeran, Salamah, Enabulele, Tahir and Lee12 . Further, infant-sex differences in placental lncRNAs have been reported before, mirroring other differences between male and female infants in their response to adverse perinatal exposures and maternal characteristics Reference Majewska, Lipka and Paukszto13–Reference Clark, Eaves and Gaona16 . However, to our knowledge, no prior study examined human placental lncRNA transcript levels in relation to maternal ppOB and downstream effects on infant birth weight. The current investigation sought to address these knowledge gaps by investigating overall and infant-sex specific associations of ppBMI and ppOB with placental lncRNA transcripts, as well as associations of placental lncRNAs with infant birthweight. Based upon prior evidence relating maternal obesity with pregnancy complications and birth weight phenotypes and potential roles of lncRNAs, we hypothesized that levels of placental lncRNA transcripts that regulate placenta proliferation and inflammation are associated with ppBMI and infant birth weight. Further, we hypothesized that these associations would be infant sex-specific.
Methods
Study setting and study participants
Participants in this study came from two cohorts of the ECHO PATHWAYS Consortium Reference LeWinn, Karr and Hazlehurst17 . The first was the Conditions Affecting Neurocognitive Development in Early Childhood (CANDLE) cohort, described in detail elsewhere Reference Sontag-Padilla, Burns and Shih18 . Briefly, CANDLE is a prospective pregnancy cohort study that enrolled urban, pregnant, women between 2006 and 2011 (N = 1,503). To be eligible, women were required to be Shelby County, TN residents – most were from the Memphis metro area – be 16 to 40 years of age, and plan to deliver at one of the participating study hospitals. Additionally, participants were verified at the time of enrollment to have uncomplicated singleton pregnancies. The University of Tennessee Health Sciences Center IRB approved all research protocols and CANDLE participants gave informed consent at enrollment.
The second cohort consisted of participants of a pregnancy biorepository based at Seattle Children’s Hospital in Seattle. This repository was a part of the Global Alliance to Prevent Prematurity and Stillbirth (GAPPS) Reference LeWinn, Karr and Hazlehurst17 , an initiative to investigate risk factors for adverse birth outcomes and interventions addressing them. Women were eligible to be re-recruited from this biorepository into the ECHO PATHWAYS study if they delivered in an affiliated hospital (in Seattle, WA or Yakima, WA), provided one or more pregnancy urine samples and a placenta sample at delivery, completed an enrollment questionnaire during pregnancy, and had a GAPPS child between 4–7 years of age at time of recontact. A total of N = 1,271 women were eligible at the start of re-recruitment in 2017.
A total of 794 and 392 women who provided placental samples at birth were ultimately available for inclusion in this ECHO-PATHWAYS study from CANDLE and GAPPS, respectively. Based on these records, additional exclusions were applied to women from both cohorts with multiple births or stillbirths. After final exclusions, 776 and 205 women from CANDLE and GAPPS, respectively, comprised the analytic samples. The IRBs of Seattle Children’s Hospital and University of Washington approved all GAPPS study protocols. Participants gave informed consent.
Data collection
In both cohorts, demographic covariates (e.g., maternal age, maternal education, self-reported race, family income, and smoking behavior) and self-reported maternal pre-pregnancy weight and height were collected as a part of interviewer-assisted questionnaires at enrollment. Maternal ppBMI was calculated as self-reported pre-pregnancy mass (in kg) divided by height (in meters) squared. We used both continuous ppBMI and ppOB (categorical, BMI ≥30 kg/m2 vs. no) to comprehensively characterize the relationships of pre-pregnancy maternal weight with placental lncRNA. Information on course of pregnancy and birth outcomes were abstracted from medical records.
Placental sample collection
Similar procedures were used to isolate placental samples for sequencing in CANDLE and GAPPS, as described in prior reports Reference Paquette, MacDonald and Lapehn19,Reference Enquobahrie, MacDonald and Hussey20 . For the CANDLE study, within 15 min of delivery, a piece of placental villous tissue in the shape of a rectangular prism with approximate dimensions of 2 × 0.5 × 0.5 cm was dissected from the placental parenchyma and cut into four ∼0.5-cm cubes. The tissue cubes were placed in a 50-mL tube with 20 mL of RNAlater and refrigerated at 4°C overnight (≥8h but ≤ 24h). Each tissue cube was transferred to an individual 1.8-mL cryovial containing fresh RNAlater. The cryovials were stored at − 80°C, and the fetal villous tissue was manually dissected and cleared of maternal decidua. Following dissection, the fetal samples were placed into RNAlater and stored at − 80°C.
In the GAPPS study, within 30 min of delivery, 8 mm full-thickness vertical tissue punches from the placental disc were taken and put into 5-ml tubes containing approximately 3 ml of RNAlater and stored at − 20°C before specimens were shipped to the GAPPS facility. Samples were then stored at − 80°C. Punches were thawed and inspected for identifiable fetal-side membranes and maternal sides. The fetal-side of the placental punch was cut-off from the rest of the punch, and divided into 1–3 pieces with mass ranging from 10 to 30 mg. Each sample was placed in 1 ml RNAlater and stored at − 20°C until shipped for further processing.
Sample processing and RNA sequencing
RNA was isolated for sequencing from approximately 30 mg pieces of tissue. Tissue samples were removed from storage and allowed to warm to room temperature. Next, each sample was removed from its storage tube, gently dried with a Kimwipe to remove RNAlater buffer solution, and placed in new test tube containing 600 μl Buffer RLT Plus with mercaptoethanol. A 5-mm ball bearing was then placed in each tube to aid in homogenization by a TissueLyser LT (Qiagen, Germantown, MD). We then used AllPrep DNA/RNA/miRNA Universal Kits (Qiagen, Germantown, MD) to isolate the RNA according to the manufacturer’s instructions. RNA purity was then characterized by OD260/230 and OD260/260 ratios measured using a NanoDrop 8000 Spectrophotometer (Thermo Fischer Scientific, Waltham MA), and RNA integrity was further tested using a Bioanalyzer 2100 and RNA 6000 Nanochips (Agilent, Santa Clara, CA). RNA samples that did not meet an RNA Integrity Number >7 were not included in the RNA-Seq analysis.
Transcripts were isolated from 1μg of total RNA, starting with poly-A selection and followed by cDNA synthesis. These procedures were completed with TruSeq Stranded mRNA kits (Illumina, San Diego, CA) on a Sciclone NGSx Workstation (Perkin Elmer, Waltham, MA) prior to each cDNA library being marked with a unique barcode. Libraries were then amplified over 13 PCR cycles before being quantified with Qubit Quant-it dsDNA High Sensitivity fluorometric assays (Life Technologies, Carlsbad, CA). Further, a DNA1000 assay on an Agilent 2100 Bioanalyzer was used to assess overall quality and average fragment size before final sequencing of libraries on an Illumina HiSeq 4000 to a depth of around 30 million reads per sample. Quality assessments of reads were performed using the FASTX-toolkit (v0.0.13) and FastQC (v0.11.2) Reference Brown, Pirrung and McCue21 . Sequencing operations were performed at the University of Washington Northwest Genomics Center.
Sequencing reads were aligned to the GRCh38 transcriptome (Gencode v33) with Kallisto Reference Bray, Pimentel, Melsted and Pachter22 to estimate their abundances (summarized at the transcript level), which were then scaled to the average transcript length using the R Bioconductor tximport package Reference Soneson, Love and Robinson23 . Library normalization factors were then estimated using the Bioconductor edgeR package’s Reference Robinson and Oshlack24 Trimmed Mean of M-values (TMM) function, and lncRNA transcripts with unreliably low expression (mean log counts/million counts (logCPM) ≤ 0) were excluded from the analysis. Of the ∼ 16,000 lncRNA transcripts initially sequenced, 1,077 met this threshold value in CANDLE and 1,033 in GAPPS (949 overlapping). The limma-voom pipeline Reference Law, Chen, Shi and Smyth25 of the Bioconductor limma package was used to convert the counts per transcript to logCPM values using the TMM normalization factors before computing observation-level weights based on the transcripts’ mean-variance relationships. These weights were then applied in the linear modeling functions of the same limma-voom pipeline for analysis.
Statistical analyses
We used descriptive statistics to characterize the analytic samples and the parent cohorts. As with prior transcriptomic studies in these populations Reference Enquobahrie, MacDonald and Hussey20,Reference Hussey, Enquobahrie and Loftus26 , all analyses were cohort-specific to CANDLE or GAPPS. To examine the impact of increasing BMI and BMI extremes, weighted linear regression models were fit using the limma lmFit function Reference Soneson, Love and Robinson23 , and regressed genome-wide placental expression of lncRNAs (as logCPM values, described above) on ppBMI (continuous) or ppOB (categorical, BMI ≥30 kg/m2 vs. no). Models were fit for each BMI variable separately, and both ppBMI and ppOB models adjusted for a priori-identified confounding variables and experimental variables, including RNA sequencing batch, hospital site (for GAPPS only, controlling for differences between the Seattle and Yakima, WA locations), maternal age (continuous), self-reported race (White/Black/Asian/other/multiple race, with American Indian/Alaska Native counted as a part of “other” in GAPPS), infant sex (male/female), calendar year of birth – as control for other year-linked exposures or conditions – gestational age at birth (continuous), family income in US dollars (continuous), mode of delivery (cesarean section/vaginal), labor (presence/absence), smoking (yes/no for smoking during pregnancy), maternal urine cotinine (binary variable using 200 ng/mL cutoff Reference Schick, Blount and Jacob27 ), and maternal education (less than high school/high school/college or technical school/graduate or professional degree). Prior to viewing results, the subset of models with p-value distributions deviating from the expected uniform distribution were rerun with added surrogate variable analysis (SVA) Reference Leek, Johnson, Parker, Jaffe and Storey28 to control excess unmeasured sources of variation. In all models, LncRNAs were considered to be differentially transcribed based on a false discovery rate (FDR) <0.10 Reference Benjamini and Hochberg29 . Further, we fit infant-sex stratified models with similar covariate control to examine sex-specific associations. We also refit main-effect models with an additional exposure–infant-sex interaction term to test ppBMI–infant-sex or ppOB–infant-sex interactions. An FDR <0.10 was also used to determine statistical significance of interactions terms.
We then examined the relationship between lncRNA expression and birthweight, with birthweight as the exposure and lncRNA transcripts as the outcome. This reverse model construction allowed us to take advantage of the performance properties of the limma package methodology, which require RNA levels to be the dependent variable Reference Soneson, Love and Robinson23,Reference Law, Chen, Shi and Smyth25 . These models included identical covariate sets to those described above. Models containing interaction of birthweight and infant-sex were also fit to examine infant-sex specific associations, followed by fitting of infant-sex stratified models. As in BMI-related models, all birthweight analyses used an FDR <0.10 to determine statistical significance.
All analyses were performed with complete-cases, resulting in 51 subjects being excluded from CANDLE models for missing covariate information (final N = 725). Of the original 205 subjects, 46 were removed from individual GAPPS models due to a combination of missing covariate information, resulting in a final analysis sample size of N = 159. All regression analyses were performed using R (v 4.1.2). Finally, we examined potential functions of identified lncRNA transcripts using the iPathwayGuide platform (Advaita Bioinformatics, Ann Arbor, MI).
Results
On average, study participant mothers were 27.3 (SD = 5.5) and 30.5 (SD = 5.7) years old in CANDLE and GAPPS, respectively (Table 1). Mean gestational age at birth was 39.0 (SD = 1.5) and 38.3 (SD = 3.0) weeks for CANDLE and GAPPS participants, respectively. Most CANDLE participants self-identified as Black (56%) or White (38%), while GAPPS participants were predominantly White (80%). The analytic populations of both studies were broadly representative of their source cohorts, though CANDLE mothers in the analytic sample tended to be slightly older, have slightly later births and higher birthweight, as well as higher income than the parent cohort, the latter perhaps reflecting their higher rate of college education (Supplemental Table 1). While average pre-pregnancy BMIs were similarly overweight (BMI ≥25 kg/m2) in both cohorts (27.7 kg/m2 [SD = 7.4] and 27.4 kg/m2 [SD = 7.4] for CANDLE and GAPPS, respectively), a slightly – though not significantly – larger proportion of CANDLE mothers were obese pre-pregnancy (32%) compared with mothers in GAPPS (25%) (Table 2).
Table 1. Select characteristics of the study populations
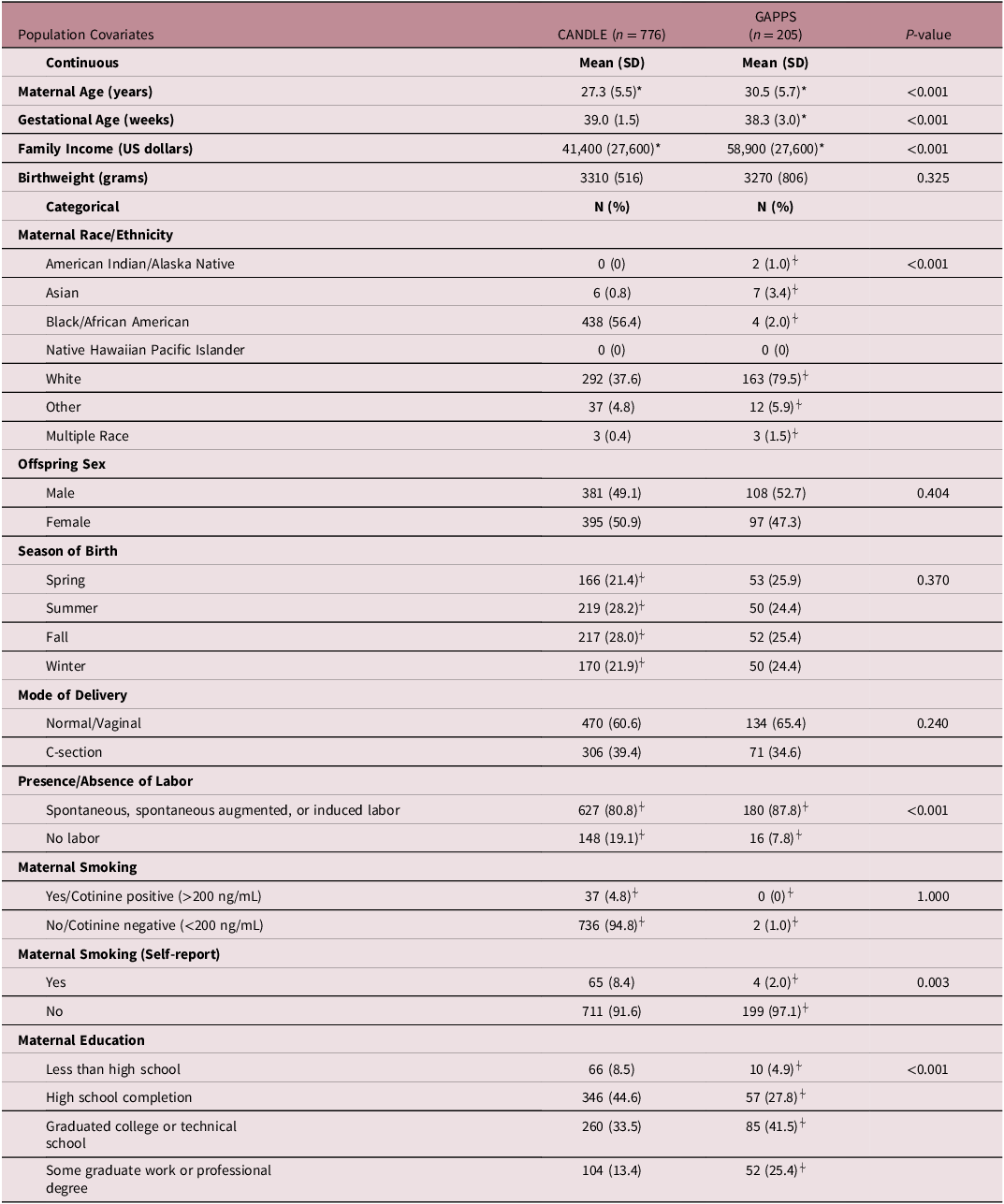
*Mean and standard deviation values calculated after removal of missing values.
⍭Percentages do not sum to 100% due to missing values of the covariates.
Table 2. Maternal pre-pregnancy body mass index (BMI) distributions
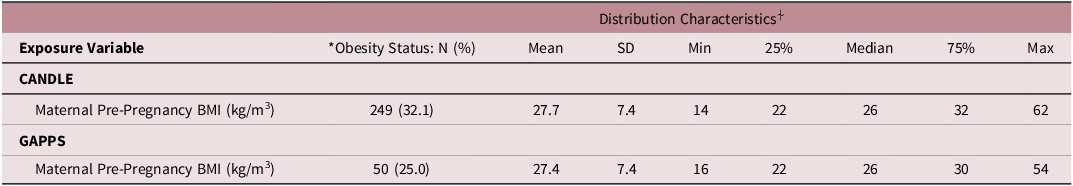
*Obese status is defined by a BMI ≥30 kg/m3.
⍭BMI was rounded to the nearest whole number upon initial calculation.
We found no overall associations of ppBMI or ppOB with lncRNA transcripts in CANDLE or GAPPS (Fig 1). In sex-stratified models, ppOB was not associated with lncRNA transcripts, while ppBMI was associated with lncRNA only among male infants (Fig 2). Among male infants in CANDLE, ppBMI was associated with the expression of three lncRNA transcripts (ERVH48-1, AC139099.1, CEBPA-DT) (Table 3). These transcripts were upregulated by approximately 0.9%, 2.0% and 1.1%, respectively, per unit increase in ppBMI among males (all FDR values = 0.085). In GAPPS, ppBMI was associated with downregulation of two lncRNA transcripts (AP000879.1 and AL365203.2) among male infants, by approximately 5.2% and 3.6%, respectively, per unit BMI (both FDR values = 0.085). Further, no ppBMI-infant sex or ppOB-infant sex interaction terms were statistically significant in either cohort.
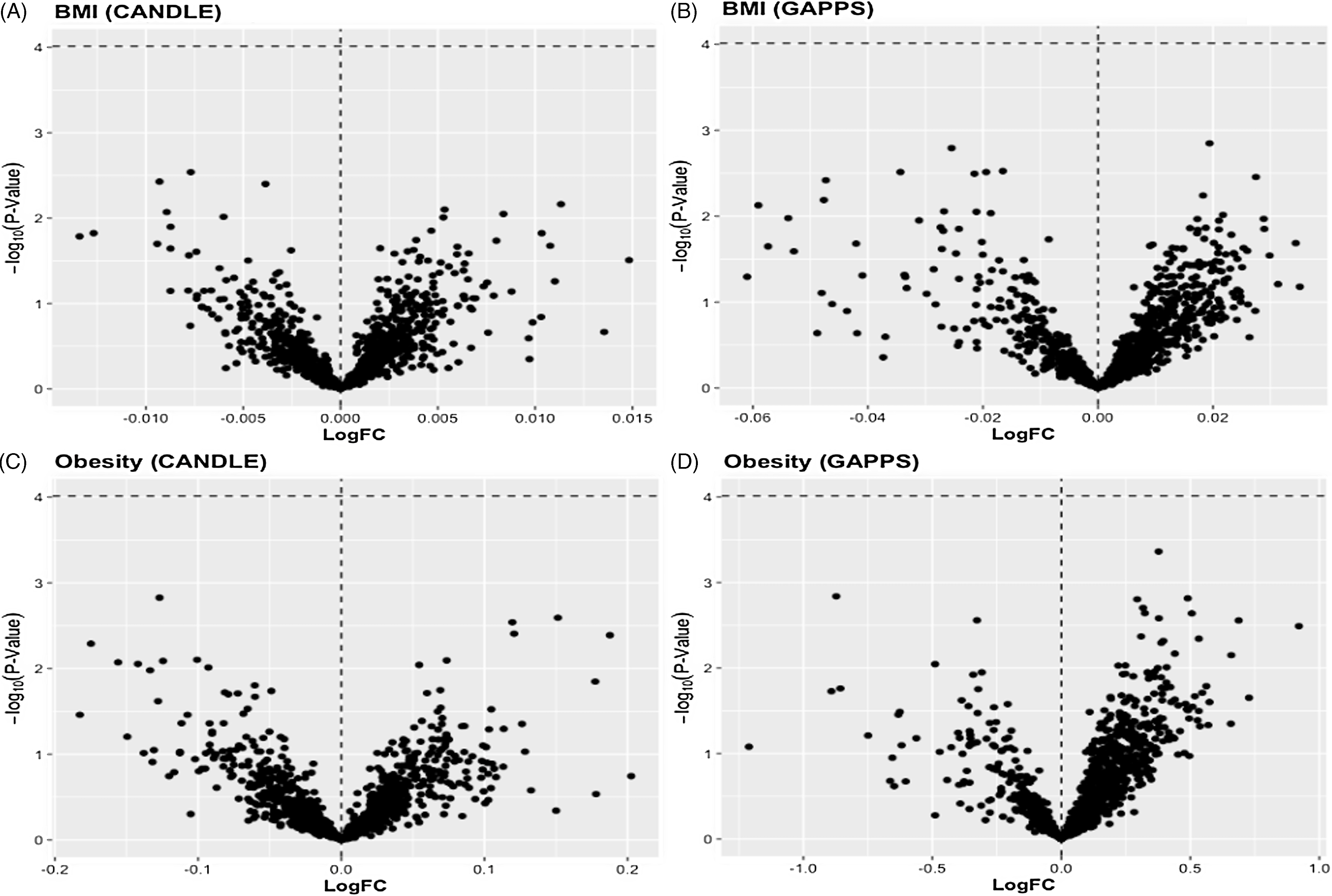
Figure 1. Volcano plots of LogFC change in placental lncRNA expression per unit change in A) maternal BMI in the CANDLE cohort, B) maternal BMI in GAPPS, or across levels of C) maternal obesity in CANDLE, and D) maternal obesity in GAPPS. Statistical significance of associations (indicated in red) was determined by a Benjamini-Hochberg false discovery rate (FDR) of 0.10.
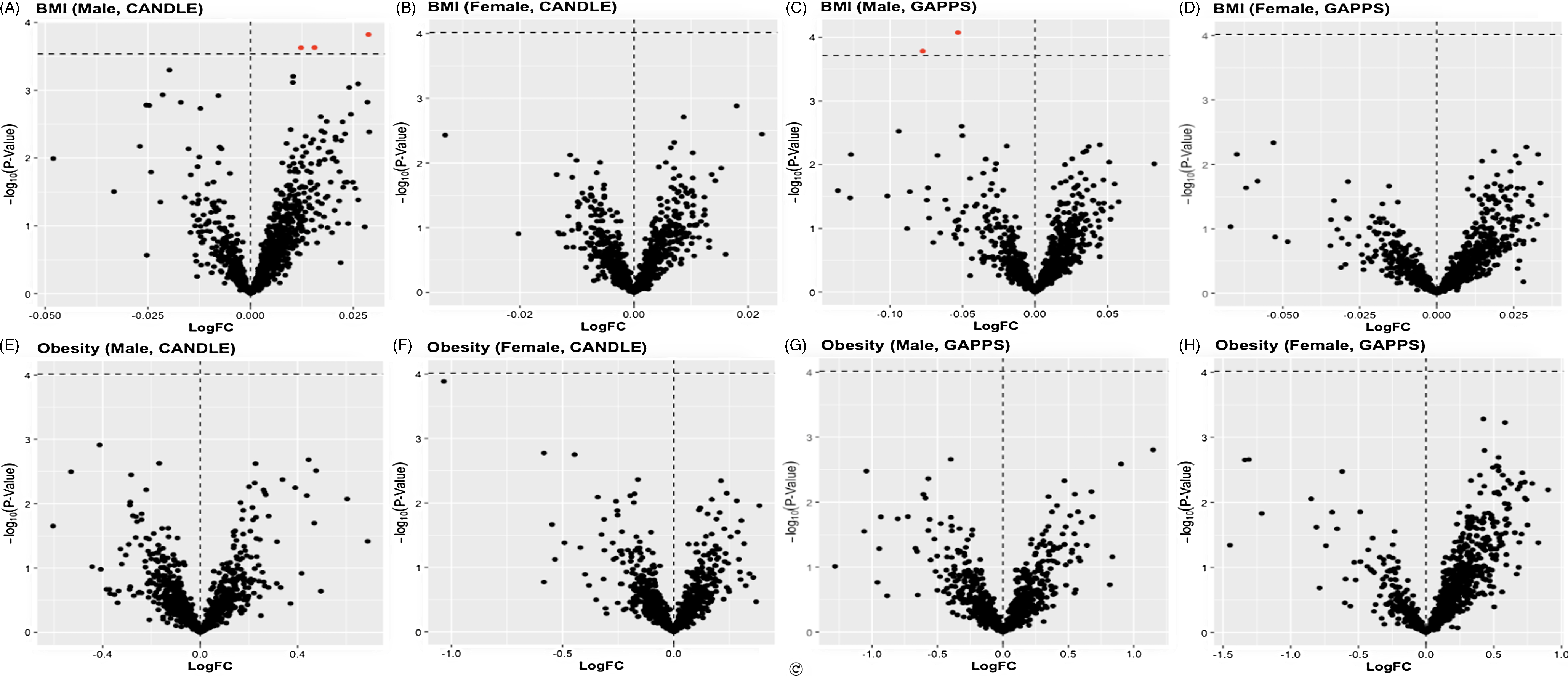
Figure 2. Volcano plots of infant-sex stratified LogFC change in placental lncRNA expression per unit change in maternal BMI A) among male CANDLE infants, B) among female CANDLE infants, C) among male GAPPS infants, and D) among female GAPPS infants, as well as the impact of maternal obesity status among E) male CANDLE infants, F) female CANDLE infants, G) male GAPPS infants, and H) female GAPPS infants. Statistical significance of associations (indicated in red) was determined by a Benjamini-Hochberg false discovery rate (FDR) of 0.10.
Table 3. Top sex-specific associations of maternal pre-pregnancy BMI with lncRNA transcripts
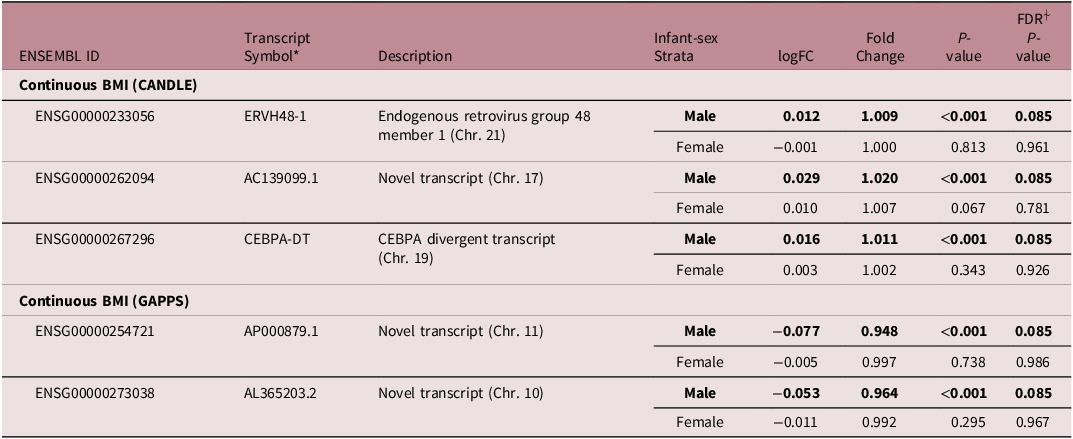
*Gene symbols and descriptions from HGNC database.
⍭The FDR cutoff for significance was <0.10.
We did not find overall associations of the expression of placental lncRNA transcripts with infant birthweight in CANDLE or GAPPS (Fig 3). In stratified models, among male infants in CANDLE we found a positive association of the expression of one lncRNA transcript (AC104083.1) with infant birthweight (logFC = 4.3e-4, FDR = 0.039) (Table 4, Fig 3). Among female infants in GAPPS, we found associations between the expression of 17 lncRNA transcripts and infant birthweight (all FDR <0.1). Seven of these lncRNAs (LINC02709, KANSL1-AS1, DANCR, EPB41L4A-AS1, GABPB1-AS1, CRIM1-DT) were previously described, while the remaining 11 lncRNAs were novel. Similar to ppBMI-related analyses, none of the infant-sex–birthweight interaction terms were statistically significant in either cohort. Attempts at pathways analysis were limited by the fact that identified lncRNA transcripts were largely not previously characterized and had unknown functions.
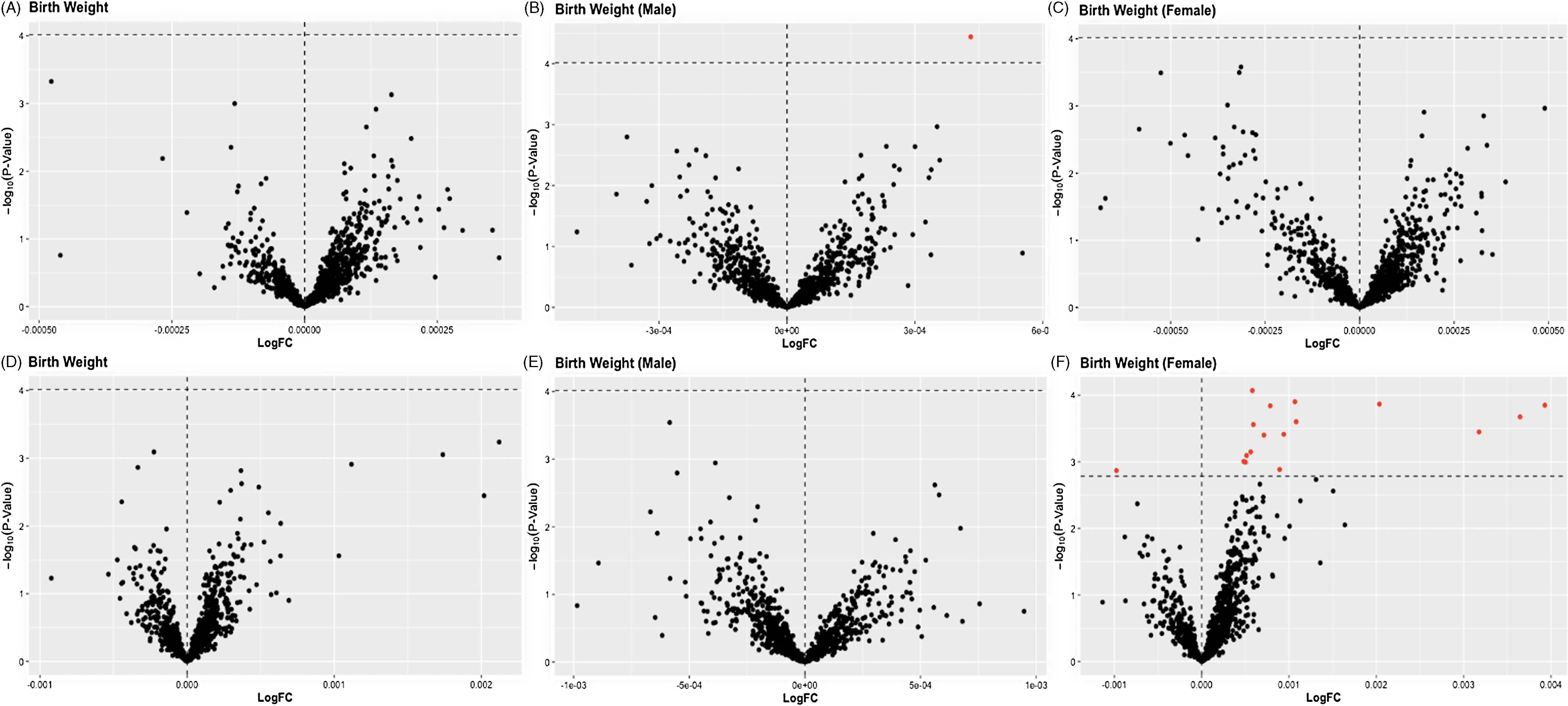
Figure 3. Volcano plots of LogFC change in placental lncRNA expression per gram change in birth weight for the A) full CANDLE cohort, B) CANDLE male infants, C) CANDLE female infants, D) the full GAPPS cohort, E) GAPPS male infants, and E) GAPPS female infants. In overall cohort and infant-sex stratified models, statistical significance of associations (indicated in red) was determined by a Benjamini-Hochberg false discovery rate (FDR) of 0.10.
Table 4. Top sex-specific associations of birthweight with LncRNA transcripts
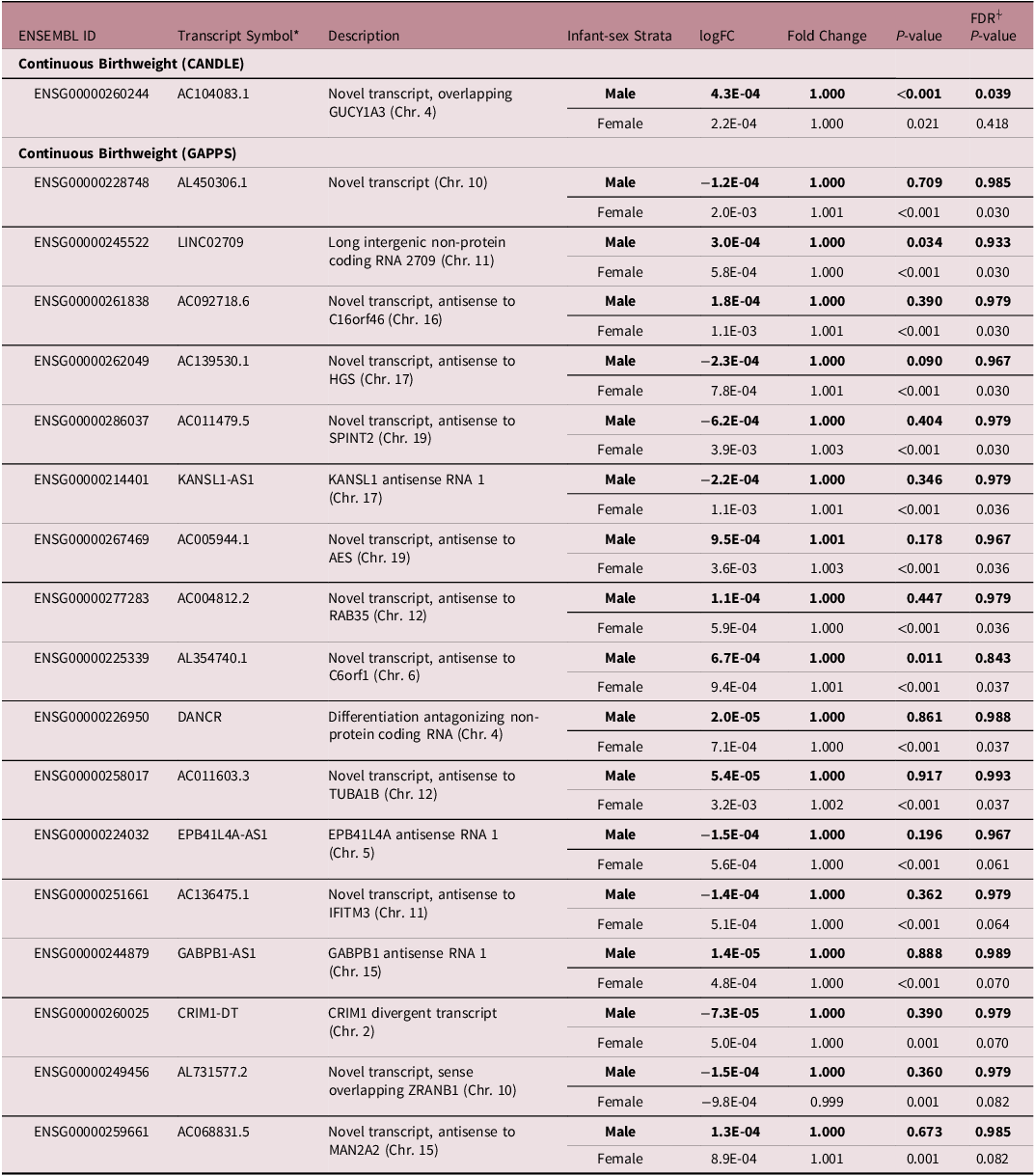
*Gene symbols and descriptions from HGNC database.
⍭The FDR cutoff for significance was <0.10.
Discussion
In the current study of CANDLE and GAPPS, we did not find overall associations of the expression of lncRNA transcripts with either ppBMI, ppOB, or infant birthweight. Among male infants only, the expression of three transcripts (ERVH48-1, AC139099.1 and CEBPA-DT) was associated with ppBMI in CANDLE and two transcripts (AP000879.1 and AL365203.2) were associated with ppBMI in GAPPS. Further, the expression of one lncRNA transcript (AC104083.1) was associated with infant birthweight among male infants in CANDLE, while the expression of 17 lncRNA transcripts (seven previously characterized and eleven novel) was associated with infant birthweight among females in GAPPS. We did not observe overlap of findings across the two cohorts and none of the infant-sex interaction terms were statistically significant.
To our knowledge, this study was the first to examine variability in the expression of human placental lncRNA transcripts in relation to maternal ppBMI or ppOB. Prior studies of human lncRNA transcripts’ expression in adipose tissue have highlighted expression of a number of transcripts related to obesity, adipogenesis, lipogenesis and metabolic dysfunctions, including TUG1, MALAT1, RP11-20G13.3, HOTAIR, GYG2P1, OLMALINC and MIR31HG Reference Ebrahimi, Toolabi and Jannat Ali Pour30–Reference Puthanveetil, Chen, Feng, Gautam and Chakrabarti34 , though we did not see these transcripts among our findings. Of note, these prior investigations did not evaluate placental lncRNA transcripts except for one study of mouse placenta and gonadal fat that compared the impacts of high-fat and low-fat diets Reference Huang, Huang and Niu35 . The study identified 52 lncRNAs that were dysregulated in placentas of high fat diet mice with roles related to inflammation, metabolism and vascular growth. Gene-gene interaction networks with placental mRNAs and lncRNAs identified prominent gene-expression changes for extra-cellular matrix interactions and metabolic pathways Reference Huang, Huang and Niu35 , though no homologs of the identified lncRNA transcripts were found in our study.
ERVH48-1 and CEBPA-DT are lncRNAs whose functions have been extensively characterized. ERVH48-1 (endogenous retrovirus group 48 member 1) – also known as HERV-Fb1 – is an endogenous retroviral sequence. The otherwise non-coding ERVH48-1 contains an exon sequence that codes for a small protein, which Sugimoto and colleagues designated suppressyn (SUPYN) after observing that knockdown of the HERV-Fb1 transcript triggered a large increase in cellular fusion of BeWo cells in vitro Reference Sugimoto, Sugimoto, Bernstein, Jinno and Schust36 . The same authors reported that SUPYN was primarily localized to villous and extravillous cytotrophoblasts, yet it was also detected in syncytiotrophoblasts and even in maternal decidua; in contrast, its cell-surface target ASCT2 was limited purely to cytotrophoblast lines Reference Sugimoto, Schust, Kinjo, Aoki, Jinno and Kudo37 . Kudaka et al. observed that ERVH48-1 expression was decreased in the placentas of preeclamptic or hypertensive women Reference Kudaka, Oda, Jinno, Yoshimi and Aoki38 , while Sugimoto et al. noticed differential expression under hyper- and hypoxia conditions – the former intended to mimic possible hyperoxia that occurs under preeclampsia Reference Sugimoto, Schust, Kinjo, Aoki, Jinno and Kudo37 . These findings tend to indicate that the downstream consequences of obesity in pregnancy, which are known to include preeclampsia, gestational diabetes and other pregnancy complications Reference Huda, Brodie and Sattar2 , can conceivably be related to the impact of ERVH48-1 expression (and SUPYN production) on aberrant placental fusion events. A recent investigation within the Rhode Island Child Health Study found higher expression of ERVH48-1 to be significantly associated with lower birthweight among infants (β = −320.9 g, 95% CI: −487.8, −154.0) Reference Hussey, Burt and Deyssenroth39 . For comparison, an ad hoc analysis of ERVH48-1 using similar linear regression models in the CANDLE and GAPPS populations (with our present covariates) estimates birthweight differences of β = −77.3 g (95% CI = −236.5, 82.1) and −184.2 g (95% CI = −555.2, 186.8), respectively, per 10x increase in ERVH48-1 expression (p > 0.05 in both cases). While the direction of birthweight associations with ERVH48-1 is similar in CANDLE and GAPPS to Hussey et al. 2020, part of the difference in scale of estimates could be contributed by the latter study’s intentional over-selection of large for gestational age and small for gestational age infants during recruitment, resulting in relatively stronger associations of ERVH48-1 with birthweight. However, associations were observed only in male infants in the current study, and the previous analyses did not examine sex-specific associations. Given prior study evidence that male and female placentas develop at different rates and respond differently to external stimulus Reference Kalisch-Smith, Simmons, Dickinson and Moritz40 , it is possible that this late-pregnancy observation of increased ERVH48-1 reflects the decrease in male placental adaptability previously seen to occur in late pregnancy. Our current observations also parallel similar sex-specific exposure-transcriptomic relationships, yet warrants further investigations Reference Majewska, Lipka and Paukszto13,Reference Martin, Smeester and Bommarito15,Reference Clark, Eaves and Gaona16 . One other study by Song et al. compared RNA expression profiles of the placentas of macrosomic and non-macrosomic infants, and found 2,929 lncRNAs upregulated and 2,127 lncRNAs downregulated in macrosomic infants Reference Song, Na, Wang and Qiao41 . However, no overlaps were observed between the small subset of these lncRNAs presented by the authors and current findings.
CEBPA-DT, previously designated ADINR for Adipogenic Differentiation Induced Noncoding RNA, is a transcript that has been linked to adipogenesis and cancer. Investigators observed that CEBPA-DT is up-regulated in oral squamous cell carcinoma (OSCC) cells, whereas silencing of CEBPA-DT resulted in reduced proliferation and induced apoptosis in OSCC cells Reference Guo, Ma, Hu, Song, Zhu and Zhong42 . These effects, as well as alterations in cell migration and invasion, were theorized to be controlled by CEBPA-DT’s influence on CEBPA and subsequent CEBPA regulatory actions on BCL2, an apoptosis promoting gene Reference Guo, Ma, Hu, Song, Zhu and Zhong42 . No studies were found that directly examined CEBPA-DT in placenta or in relation to maternal obesity or birthweight. Multiple studies have found CEBPA to be a regulator of MTOR expression: increased CEBPA was found to inhibit MTOR expression, while decreased CEBPA led to the opposite effect Reference Wang, Xiao, Zhang, Liao, Fu and Huang43–Reference Okawa, Gupta and Kahraman45 . Placental mTOR acts as a key regulator of cell growth, metabolism and adaptive response to cell stress, and in general, mTOR signaling is increased in placentas of higher birthweight infants and decreased in infants with lower birthweight Reference Dong, Shin, Chen, Lei, Burd and Wang46 . CEBPA-DT conceivably influences birthweight via control of placental cell proliferation under stress conditions, including maternal obesity.
The majority of placental transcripts related to birthweight among female infants in the current study were uncharacterized. Of the remaining transcripts, a small subset has been associated with varied processes involved in cell growth and development. Of these transcripts, DANCR, differentiation antagonizing non-protein coding RNA, is the best characterized. Previous investigators postulated that the transcript played a role in the transition from progenitor to differentiated cells. Further experiments in regenerated epidermal tissue found that depletion of DANCR resulted in expression of differentiation genes in all cell types, including epidermal basal cells that normally have no expression of such proteins Reference Kretz, Webster and Flockhart47 . This led to the conclusion that DANCR is explicitly antagonistic toward differentiation processes in cell types where it is expressed Reference Kretz, Webster and Flockhart47 . Similar to CEBPA-DT, DANCR expression has also been linked to the action of the MTOR pathway Reference Akbari, Eshghyar and Gholipour48 – though this has not been investigated specifically within the placenta. In the placenta DANCR directly binds and inhibits miR-241-5p, a microRNA that is highly expressed in placental tissues in preeclampsia and is known to impact the proliferation and invasion of trophoblasts Reference Adu-Gyamfi, Cheeran, Salamah, Enabulele, Tahir and Lee12,Reference Zhang, Wang, Cheng and Wu49 . DANCR could conceivably represent part of an adaptive response to the pro-preeclamptic pressures of maternal pre-pregnancy obesity. Consequently, DANCR’s potentially protective effect on placental pathologies provides a plausible explanation for the observed positive association between DANCR expression and birthweight in female infants.
EPB41L4A-AS1, the antisense transcript to EPB41L4A on chromosome 5, is one of a few dozen transcripts whose expression is rapidly up-regulated during the G0 phase of the cell cycle under conditions of cell cycle arrest Reference Yabuta, Onda and Watanabe50 . CRISPRi analysis of lncRNAs in seven cell types by Liu and colleagues revealed EPB41L4A-AS1 was differentially expressed in relation to anti-growth pathways for U87 (Uppsala 87 Malignant Glioma) and K562 (immortalized myelogenous leukemia) cells Reference Liu, Horlbeck and Cho51 . Further, gene ontology analysis noted positive up-regulation of genes in the K572 “response to O2 levels” pathway and slight up-regulation for U87 genes in a number of signaling pathways (p53, Integrin, VEGF), gene expression, and negative regulation of cell cycle with increasing EPB41L4A-AS1 Reference Liu, Horlbeck and Cho51 . Placental EPB41L4A-AS1 expression was also investigated in the context of recurrent miscarriage (RM), to our knowledge the only study that has specifically examined EPB41L4A-AS1 in placental pathology Reference Zhu, Liu and Liao52 . EPB41L4A-AS1 was highly upregulated in placental tissue of early RM pregnancies compared to normal controls and similarly exerted an inverse impact on levels of glycolysis in response to PCG-1 α signaling (up-regulated in early RM). Notably, the authors also found that placentas from normal pregnancies had lower EPB41L4A-AS1 expression than placentas from pregnancies with pathologies such as preeclampsia and intrauterine growth restriction (p < 0.05) Reference Zhu, Liu and Liao52 .
The GABPB1-AS1 transcript, positively associated with birthweight among female infants in GAPPS, has been noted as a marker of chemical-exposure induced cell stress Reference Tani and Torimura53 . An inverse relationship was observed between GABPB1-AS1 expression and expression of a number of genes that promote cell-cycle progression in gliomas, contrasted by high up-regulation of GABPB1-AS1 for certain auto-immune disorders Reference Chen, Cheng, Du, Liu and Wu54 . While no studies were found linking this transcript to the placenta or birthweight, these prior findings point to a potential widespread role of GABPB1-AS1 as a short-term regulator of cell response to stress conditions. Finally, KANSL1 antisense RNA 1 (KANSL1-AS1) – associated with increased birthweight among female infants in this study – is a non-coding transcript on chromosome 17 found proximal to its namesake gene, KAT8 regulatory NSL complex subunit 1 (KANSL1). Only two recent investigations have explored functional roles for KANSL1-AS1, including a TWAS-style analysis linking it to anxiety disorders Reference Su, Li and Lv55,Reference Liu, Zong and Sun56 . REACTOME and gene ontology analyses in that study indicated possible links of anxiety-associated transcripts to myogenesis, the Wnt signaling pathway/calcium modulating pathway and mRNA binding; however, the proportion of contribution KANSL1-AS1 provided to these pathways was not clear Reference Su, Li and Lv55 . The second investigation found that KANSL1-AS1 expression was associated with a decrease in osteosarcoma incidence and development risk scores Reference Liu, Zong and Sun56 . While these studies point to potential roles of KANSL1-AS1 in disease pathogenesis, its mechanistic relevance to placental tissue and infant birthweight remains unclear.
Despite a lack of prior studies on placental lncRNA modulation by ppBMI, lncRNAs have been shown to play roles in pathways underlying development of placental abnormalities and pregnancy complications, most clearly in conditions of hypoxia or improper uterine invasion contributing to preeclampsia Reference Sugimoto, Schust, Kinjo, Aoki, Jinno and Kudo37,Reference Kudaka, Oda, Jinno, Yoshimi and Aoki38 , yet also in other conditions such as gestational diabetes Reference Sugimoto, Schust, Kinjo, Aoki, Jinno and Kudo37,Reference Kudaka, Oda, Jinno, Yoshimi and Aoki38,Reference Wang, Lu and Li57,Reference Guiyu, Quan and Ruochen58 . Given lncRNAs have also been associated with some disease pathologies in a sex-specific manner, our sex-specific findings are not only plausible but conceivably relate to observed sex-specific differences in gene expression during normal placental growth or response to maternal exposures and exogenous cell stressors Reference Majewska, Lipka and Paukszto13–Reference Clark, Eaves and Gaona16 . As a part of an additional ad hoc analysis, we examined publicly available GEO DataSets submitted to the US National Library of Medicine. Out of an original 4,348 datasets available, we identified only 8 datasets with overlapping transcripts judged to be relevant to the current study (e.g., placental tissue). Only three of these studies made any comparisons by infant-sex, one finding no difference in expression profiles of ERVH48-1 between male and female placentas Reference Cvitic, Longtine and Hackl59 , a second observing no appreciable difference in expression levels of four transcripts (ERVH48-1, KANSL1-AS1, DANCR, and EPB41L4A-AS1) among mothers dosed with choline in the last trimester Reference Jiang, Bar and Yan60 , and the final observing only KANSL1-AS1 to be slightly elevated among female placentas at term among control subjects Reference Huuskonen, Storvik and Reinisalo61 . These samples tend to be quite small (6 – 42) and did not account for the impact of other covariates on transcript levels, so their findings should be interpreted with caution.
This investigation had a number of key strengths. To the best of our knowledge, this ECHO-PATHWAYS investigation is the first placental study to explicitly examine maternal BMI, obesity status and full transcriptome placental lncRNA expression in humans. It is the largest existing cohort analysis of transcriptome wide placental lncRNA expression and infant birthweight. These large sample sizes provided enough power to examine sex-specific associations with lncRNAs, something not possible in many earlier analyses of this type. Beyond its size, another strength of this investigation is its robust consideration of covariates from extensive surveys and medical record abstraction. This allowed for a priori construction of models that include a number of possible confounders and experimental variables. We hoped the relatively large number of subjects and excellent covariate control in these cohorts would allow us to explore mediation by transcripts, yet a lack of overlapping significant associations between pre-pregnancy BMI, lncRNAs, and birthweight precluded such analysis. Also, the larger of the two cohorts, CANDLE was mainly comprised of Black women and their infants (56.4%). This makes this analysis the largest of its kind to focus on placental lncRNA expression in the American Black population, a population that has rarely been a focus of this type of analysis.
Several limitations of this investigation deserve mention. While the underlying intention behind using two cohorts for this analysis was to compare these populations and provide an opportunity for result replication, it is clear that the cohorts had different distributions of population characteristics and exposures. These differences – likely linked to major underlying differences in region and demographics – led to consistently different effect sizes for various transcripts, independent of the major difference in power between the CANDLE (N = 776) and GAPPS (N = 205) analytic samples. Rather than indicating that these findings are false, however, this lack of replication may instead imply that mechanisms of placental adaptation are more population-dependent than might be previously expected, a hypothesis that deserves further study. Additionally, placental lncRNA expression was measured at birth; thus the associations identified may not reflect impacts of ppOB on lncRNAs prior to delivery. This is a key limitation, as placental transcriptome varies widely over the course of pregnancy Reference Sitras, Fenton, Paulssen, Vårtun and Acharya62 . Further, despite greater power in combined models to detect an interaction, we did not observe any statistically significant interactions for the various transcripts highlighted in stratified models. This finding – and our less stringent FDR <0.1 – increases the possibility of false positives. However, the fact that numerous transcripts that were identified are associated with controlling growth and proliferation (including under stress conditions) lends credence to our findings. Definitive confirmation of these transcripts’ importance in pregnancy would require us to address the issue of placental cell heterogeneity, and the best method to do this (i.e., single-cell RNA-seq), was not feasible in this investigation. Even though some studies have reported good validity of self-reported weight measurements, bias is anticipated self-reported weight measures related to recall or social desirability issues Reference Stommel and Schoenborn63,Reference Davies, Wellard-Cole, Rangan and Allman-Farinelli64 . Finally, generalizability of our findings likely only applies to populations with similar ppBMI, ppOB, or birthweight distributions to those in our study cohorts.
Conclusions
In conclusion, in this study we found several infant-sex specific associations of placental lncRNA with BMI, maternal pre-pregnancy obesity status, and infant birthweight. Aside from sex-specific findings, no transcripts were associated with maternal BMI or birthweight in main-effect or interaction models. Our work suggests that maternal pre-pregnancy BMI has potential infant-sex related impacts on placental growth and development via lncRNA expression, and that placental lncRNAs may also have infant-sex specific impact on birthweight. These findings warrant future confirmatory and functional studies.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S204017442500011X.
Acknowledgments
We thank Drs. J. MacIsaac and K. Ramadori from Dr Michael Kobor’s laboratory (Centre for Molecular Medicine and Therapeutics, University of British Columbia) for performing DNA and RNA extraction. We would also like to thank the participating families and study team members in the GAPPS and CANDLE cohorts.
Financial support
This study was conducted by the ECHO PATHWAYS Consortium, funded by the NIH (1UG3OD023271/4UH3OD023271). This manuscript has been reviewed by PATHWAYS for scientific content and consistency of data interpretation with previous PATHWAYS publications. This study was conducted using specimens and data collected and stored on behalf of the Global Alliance to Prevent Prematurity and Stillbirth (GAPPS) Repository. CANDLE was supported by the Urban Child Institute. Air pollution models were constructed with funding support from NIH (P01AG055367, R01ES025888, R56ES026528, and R01ES023500) and the Kresge Foundation Grant No.243365 as well as by STAR research assistance agreements RD831697 (MESA Air), RD-83830001 (MESA Air Next Stage), RD83479601 (UW Center for Clean Air Research), and R83374101 (MESA Coarse), awarded by the U.S. Environmental Protection Agency. This work has not been formally reviewed by the EPA. The views expressed in this document are solely those of the authors and the EPA does not endorse any products or commercial services mentioned in this publication. MRH was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under award number (T32ES015459). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. AGP was supported by NICHD (K99/R00HD096112). Laboratory infrastructure and hardware used for this study was supported by the University of Washington Interdisciplinary Center for Exposures, Diseases, Genomics, and Environment funded by the NIEHS (2P30ES007033). RNA extraction was conducted at the Kobor Lab of the University of British Columbia.
Competing interests
None.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national guidelines on human experimentation in the National Research Act and with the Helsinki Declaration of 1975, as revised in 2008, and has been approved by the institutional IRB committees of the University of Tennessee Health Sciences Center, the Seattle Children’s Hospital and University of Washington.









