Introduction
Nematodes within the Tylenchidae family are one of the most abundant soil-inhabiting species worldwide in natural and agricultural environments, although they do not comprise economically important plant-parasites (Geraert Reference Geraert2008; Qing and Bert Reference Qing and Bert2019). The genus Neothada was proposed by Khan (Reference Khan1973) to include small nematodes (under 1 mm) with a thick cuticle marked by transverse and longitudinal striae or groves dividing the surface into minute squares or rectangular blocks (Siddiqi Reference Siddiqi2000). Neothada Khan, Reference Khan1973 comprises a small genus within the family Tylenchidae Thorne, Reference Thorne1935 distributed almost worldwide and associated with algae, mosses, lichens, and plant roots (Geraert Reference Geraert2008; Siddiqi Reference Siddiqi2000). Currently, Neothada comprises six valid species, namely: N. tatra (Thorne and Malek Reference Thorne and Malek1968) Khan, Reference Khan1973 (type species), N. cancellata (Thorne Reference Thorne1941) Khan, Reference Khan1973, N. costata (Geraert and Raski Reference Geraert and Raski1986) Siddiqi, Reference Siddiqi2000, N. geraerti (Andrássy Reference Andrássy1982) Siddiqi, Reference Siddiqi1986, N. hades Heyns and van den Berg, Reference Heyns and van den Berg1996, and N. major Maqbool and Shahina, Reference Maqbool and Shahina1989. The systematic position of the genera Thada and Neothada has been controversial in the nematological literature. Siddiqi (Reference Siddiqi2000) divided the family Tylenchidae in five subfamilies, including Thadinae Siddiqi, Reference Siddiqi1986, that contain exclusively the genera Thada Thorne, Reference Thorne1941 and Neothada characterised by an axial spermatheca and a short conoid to subcylindrical tail. However, Geraert (Reference Geraert2008) based on the sensilla in the lip region, the plain vulval region (without flap or membrane), and elongated tail, often rounded or clavate included these genera under the subfamily Boleodorinae Khan, Reference Khan1964 together with Boleodorus Thorne, Reference Thorne1941, Atetylenchus Khan, Reference Khan1973, Basiria Siddiqi, Reference Siddiqi1959, Neopsilenchus Thorne and Malek, Reference Thorne and Malek1968, Psilenchus de Man, Reference de Man1921, and Ridgellus Siddiqi, Reference Siddiqi2000.
Molecular phylogeny inferred from ribosomal RNA (rRNA) genes suggests that Tylenchidae is polyphyletic (Bert et al. Reference Bert, Leliaert, Vierstraete, Vanfleteren and Borgonie2008; Holterman et al. Reference Holterman, Van Der Wurff, Den Elsen, van Megen, Bongers, Holovachov, Bakker and Helder2006; Kantor et al. Reference Kantor, Handoo, Subbotin, Mowery, Hult, Rogers and Skantar2023; Subbotin et al. Reference Subbotin, Sturhan, Chizhov, Vovlas and Baldwin2006). However, currently, there is little molecular data on species of the genera Thada and Neothada which could help clarify their taxonomic and systematic position within the family Tylenchidae. These studies comprise Yaghoubi et al. (Reference Yaghoubi, Pourjam, Atighi and Pedram2015) on N. cancellata, Hosseinvand et al. (Reference Hosseinvand, Eskandari, Ghaderi and Karegar2020a, Reference Hosseinvand, Eskandari and Ghaderi2020b) on Thada populus Hosseinvand et al., 2020, N. hades, N. major, and N. cancellata, and in these studies, using partial sequences of 18S rRNA and 28S rRNA D2-D3 expansion segments, Neothada spp. clustered together with Basiria spp., concurs with the proposal raised by Geraert (Reference Geraert2008).
During nematode surveys conducted in the spring of 2023 to investigate the biodiversity of plant-parasitic nematodes in Mediterranean olive groves with different management strategies (organic and conventional) in Greece, Italy, Morocco, Portugal, and Spain, a nematode population of the genus Neothada morphologically resembling N. major was detected in an olive grove in southern Spain (Chiclana de Segura, province of Jaén, Spain). This finding prompted us to study this population using an integrative taxonomic approach based on morphological and molecular analyses, along with comprehensive scanning electron microscopy (SEM) studies. To our knowledge, no detection of these nematodes has been reported in Spain; consequently, the recent finding of a new population of Neothada sp. in olive groves in southern Spain provides an excellent chance for ribosomal and mitochondrial molecular characterisation, as well as phylogenetic studies regarded as crucial in taxonomic and systematic studies of Tylenchidae that will progress our understanding of the evolution of these nematodes.
Therefore, the main objectives of this study were: i) to characterise morphologically and morphometrically the new Spanish population of Neothada and compare with related species; ii) to characterise molecularly the new Neothada population using the D2–D3 expansion segments of 28S rRNA, ITS rRNA, partial 18S rRNA, and cytochrome oxidase c subunit 1 (COI) gene sequences; and iii) to study the phylogenetic relationships of the identified Neothada species with available sequenced species of the family Tylenchidae.
Material and methods
Nematode population and morphological characterisation
Soil samples were collected from a total of 44 olive groves (with six samples per grove) during olive flowering stage (Spring 2023), giving a total of 264 sampling sites widely distributed across the most important olive-growing regions of the Mediterranean basin (Greece, Morocco, Italy, Portugal, and Spain). Soil samples were collected within 5–30 cm soil depth, and nematodes were extracted from a 500 cm3 subsample of soil by centrifugal flotation (Coolen Reference Coolen, Lamberti and Taylor1979). Extracted specimens were heat killed, fixed in TAF, processed to glycerol by a slow evaporation method, and mounted on permanent slides (De Grisse Reference De Grisse1969; Seinhorst Reference Seinhorst1966). The light micrographs and measurements of nematode populations, including the main diagnostic characteristics (i.e., de Man indices, body length, stylet length, lip region, tail length and shape, longitudinal ridges) were performed using a Leica DM6 compound microscope with a Leica DFC7000 T digital camera. All abbreviations used are as defined in Siddiqi (Reference Siddiqi2000).
For SEM, fixed specimens were dehydrated in a gradient series of ethanol, critical-point dried, sputter-coated with gold according to the protocol by Abolafia et al. (Reference Abolafia, Liébanas and Peña-Santiago2002), and observed with a Jeol IT 800 SHL Scanning Electron Microscope (5 kV; Tokio, Japan).
Molecular characterisation and phylogenetic analyses
For molecular analyses, and to avoid mistakes in case of mixed populations in the same sample, single specimens from the sample were temporarily mounted in a drop of 1M NaCl containing glass beads (to prevent nematode specimens from crushing/damaging) to ensure specimens conformed with the target population. All necessary morphological and morphometric data were recorded. This was followed by DNA extraction from single individuals as described by Archidona-Yuste et al. (Reference Archidona-Yuste, Clavero-Camacho, Ruiz-Cuenca, Cantalapiedra-Navarrete, Liébanas, Castillo and Palomares-Rius2024). The D2–D3 expansion segments were amplified using the D2A (5’-ACAAGTACCGTGAGGGAAAGTTG-3’) and D3B (5’-TCGGAAGGAACCAGCTACTA-3’) primers (De Ley et al. Reference De Ley, Félix, Frisse, Nadler, Sternberg and Thomas1999). The ITS region was amplified using forward primer 18S (5′-TTGATTACGTCCCTGCCC TTT-3′) and reverse primer Vrain2r (5′-TTT CACTCGCCGTTACTAAGGGAATC-3′) (Vrain et al. Reference Vrain, Wakarchuk, Levesque and Hamilton1992). The partial 18S rRNA was amplified using the primers 988 (5′-CTCAAAGATTAAGCCATGC-3′), 1912R (5′-TTTACGGTCAGAACTAGGG-3′), 1813F (5´- CTGCGTGAGAGGTGAAAT -3/), and 2646R (5/- GCTACCTTGTTACGACTTTT -3/) (Holterman et al. Reference Holterman, Van Der Wurff, Den Elsen, van Megen, Bongers, Holovachov, Bakker and Helder2006). Finally, the portion of the COI gene was amplified using the primers JB3 (5´- TTTTTTGGGCATCCTGAGGTTTAT -3`) and JB5 (5’- AGCACCTAAACTTAAAACATAATGAAAATG -3´) (Derycke et al. Reference Derycke, Remerie, Vierstraete, Backeljau, Vanfleteren, Vincx and Moens2005).
All PCR assays were done according to the conditions described by Archidona-Yuste et al. (Reference Archidona-Yuste, Clavero-Camacho, Ruiz-Cuenca, Cantalapiedra-Navarrete, Liébanas, Castillo and Palomares-Rius2024). Then, the amplified PCR products were purified using ExoSAP-IT (Affimetrix, USB products) and used for direct sequencing on a DNA multicapillary sequencer (Model 3130XL genetic analyser; Applied Biosystems, Foster City, CA, USA) using the BigDye Terminator Sequencing Kit V.3.1 (Applied Biosystems, Foster City, CA, USA) at the Stab Vida sequencing facilities (Caparica, Portugal). The newly obtained sequences were submitted to the GenBank database under the accession numbers indicated on the phylogenetic trees.
Phylogenetic analyses
D2–D3 expansion segments of 28S rRNA gene, ITS rRNA region, partial 18S rRNA gene, and COI mtDNA sequences of the unidentified Neothada species population were obtained in this study. These sequences, and other sequences from species of Tylenchidae from GenBank, were used for phylogenetic analyses. Outgroup taxa for each dataset were chosen following previously published studies (Bai et al. Reference Bai, Qing, Qiao, Ning, Xiao, Cheng and Liu2020; Munawar et al. Reference Munawar, Yevtushenko and Castillo2021; Qing and Bert, Reference Qing and Bert2019). Multiple sequence alignments of the different genes were made using the FFT-NS-2 algorithm of MAFFT V.7.450 (Katoh et al. Reference Katoh, Rozewicki and Yamada2019). The BioEdit program V.7.2.5 (Hall Reference Hall1999) was used for sequence alignment visualisation and manually edited and trimmed the poorly aligned positions using a light filtering strategy (up to 20% of alignment positions), which has little impact on tree accuracy and may save computation time, as suggested by Tan et al. (Reference Tan, Muffato, Ledergerber, Herrero, Goldman, Gil and Dessimoz2015). Phylogenetic analyses of the sequence datasets were based on Bayesian inference (BI) using MrBayes 3.1.2 (Ronquist and Huelsenbeck Reference Ronquist and Huelsenbeck2003). The best-fit model of DNA evolution was achieved using JModelTest V.2.1.7 (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012) with the Akaike information criterion (AIC). The best-fit model, the base frequency, the proportion of invariable sites, and the gamma distribution shape parameters and substitution rates in the AIC were then used in MrBayes for phylogenetic analyses. The transition models with invariable sites and a gamma-shaped distribution for the D2–D3 segments of 28S rRNA gene (TIM2 + I + G), the partial 18S rRNA gene (TIM1 + I + G), and general time-reversible model with a gamma-shaped distribution (GTR + G) for the COI gene were run with four chains for 10 × 106 generations. A combined analysis of the three ribosomal genes was not undertaken because some sequences were not available for all species. The sampling for Markov chains was conducted at intervals of 100 generations. For each analysis, two runs were conducted. After discarding burn-in samples of 30% and evaluating convergence, the remaining samples were retained for more in-depth analyses. The topologies were used to generate a 50% majority-rule consensus tree. For each appropriate clade, posterior probabilities (PP) were given. FigTree software version v.1.4.4 (Rambaut Reference Rambaut2018) was used for visualising trees from all analyses.
Results
Only one soil sample, collected from the rhizosphere of cultivated olive (cv. Picual) at Chiclana de Segura, Jaén province, Spain, contained a Neothada species at a high population density (252 nematodes/500 cm3 of soil). This contrasts with its very low overall frequency of occurrence across all samples (1/264 = 0.004). Detailed morphological, morphometrical, and molecular information about this species is provided below, confirming its identity as a new species of the genus Neothada described herein.
Systematics
Phylum: Nematoda Rudolphi, Reference Rudolphi1808
Class: Chromadorea Inglis Reference Inglis1983
Order: Rhabditida Chitwood Reference Chitwood1933
Suborder: Tylenchina Chitwood Reference Chitwood, Chitwood and Chitwood1950
Superfamily: Tylenchoidea Oerley 1880
Family: Tylenchidae Thorne, Reference Thorne1935
Genus: Neothada Khan, Reference Khan1973
Neothada olearum sp. nov
urn:lsid:zoobank.org:act: A44C2F20-D1F2-41AA-B08D-B82EBCD474D9
Description
Females. Body almost straight, slightly curved in posterior half after relaxed by gentle heat (Figures 1–2). Cuticle prominently annulated, 2.5–3.0 μm wide, total number of body annuli 214–226, 38–42 annuli from anterior end to posterior end of pharynx, 24–28 annuli from anus to tail tip. Lateral fields 6.5–8.0 μm wide with four smooth incisures, and 16 longitudinal ridges (14 longitudinal lines around the circumference of the body, outside the lateral fields) giving a tessellate body surface (Figures 1–3; Table 1). Lip region rounded, anteriorly flattened, continuous with body contour, with two annuli (Figures 1–3). Labial framework slightly sclerotised (Figure 2). Stylet delicate without distinguishable basal knobs, anterior conical part 2.5–3.5 μm long. Dorsal pharyngeal gland opening 2.0–3.5 μm posterior to stylet base. Pharynx tylenchoid; procorpus cylindroid 34.0–40.0 μm long; median bulb fusiform, weakly muscular and without valves 6.0–7.5 μm wide x 16.0-19.0 μm long; isthmus slender, basal bulb pyriform 13.0–14.0 μm wide x 25.0-27.0 μm long (Figures 1–2). Cardias rounded, 3.0–3.5 μm long. SEM observations (Figure 3) showed a quadrangular oral plate, a first annulus wider than second (2.0–2.5 μm vs 1.0–1.5 μm wide) and showing four rounded cephalic papillae, a slit-like oral aperture surrounded by six labial papillae in en face view. Amphidial aperture a conspicuous longitudinal to slightly bent slit extending as far as the second annulus (Figure 3). Excretory pore slightly sclerotised, at middle of basal bulb, 33–40 annuli from anterior end. Deirid at level of excretory pore. Hemizonid not always clear; when seen, one or two body annuli anterior to excretory pore. Nerve ring surrounding anterior part of isthmus. Reproductive system monodelphic prodelphic; vulva a transverse slit in ventral view (Figure 2), without lateral flaps 11.0–12.0 μm long. Post-vulval uterine sac 0.44 times body width. Ovary single outstretched with oocytes in a single row, quadricolumella present, spermatheca axial, oval (14.0–19.0 μm x 26.0–30.0 μm) filled with rounded sperm 1.5–2.0 μm wide. Vulva anus distance 1.4 (1.2–1.7) times as long as tail. Tail elongate-conoid, tail tip bluntly rounded.
Table 1. Morphometrics of Neothada olearum sp. nov. from Chiclana de Segura, Jaén province, Spain
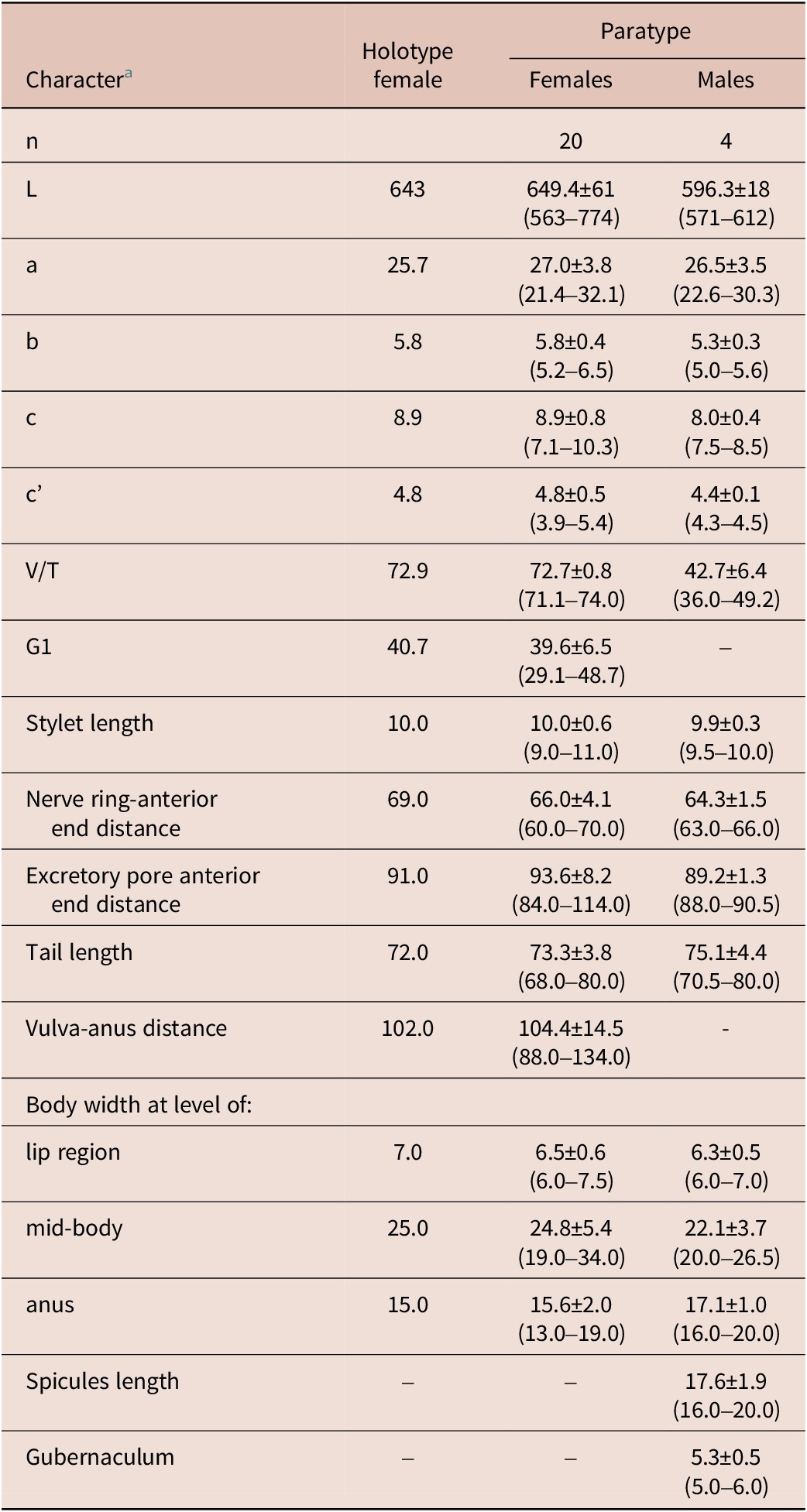
a Abbreviations are defined in Siddiqi (Reference Siddiqi2000). All measurements are in μm and in the form: mean. ± standard deviation (range) where appropriate.
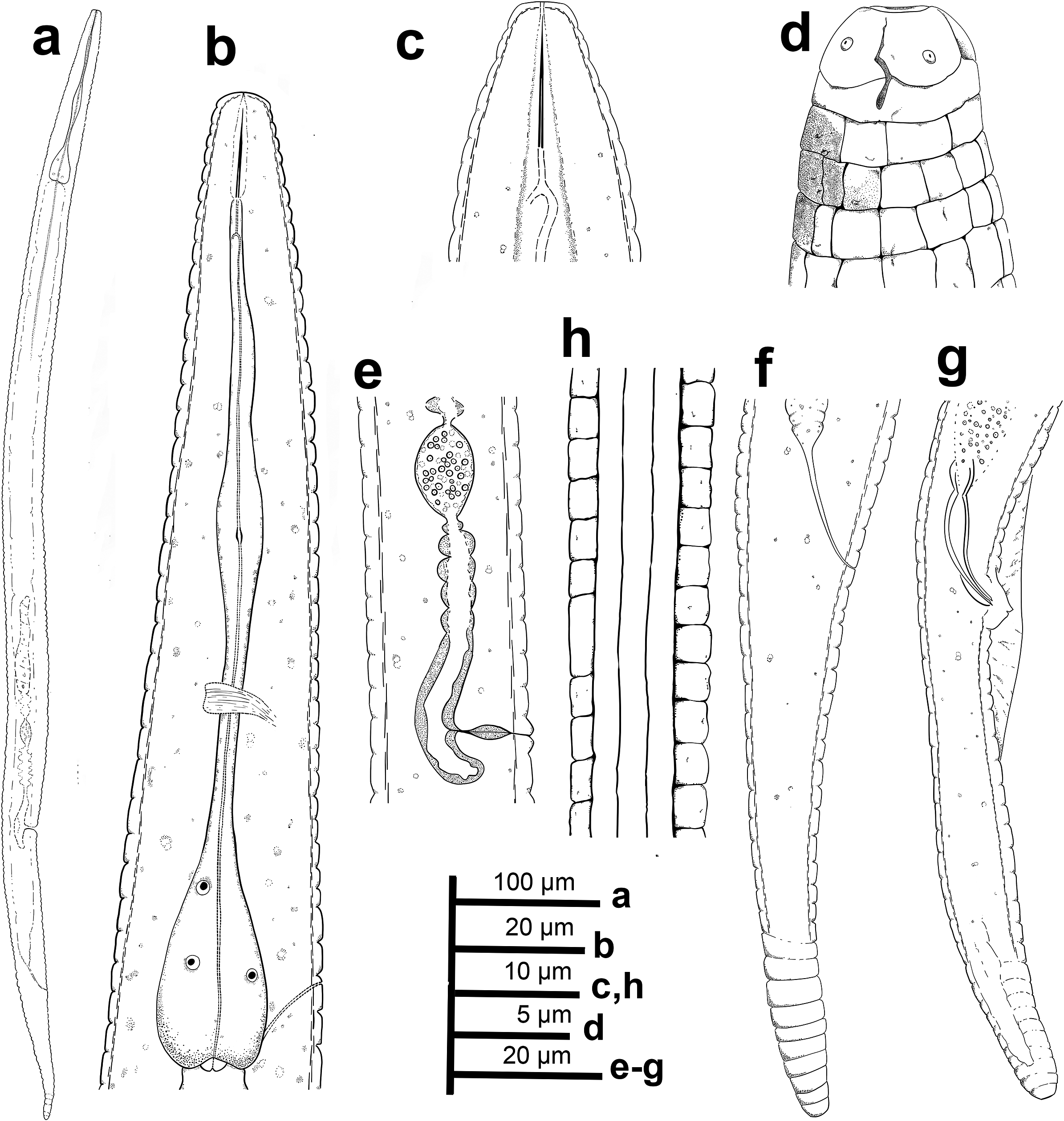
Figure 1. Neothada olearum sp. nov. (drawings). (a), entire female body; (b), female pharyngeal region; (c), detail of lip region showing stylet; (d), detail of lip region showing amphidial aperture and cervical papillae; (e), detail of vulval region showing spermatheca; (f), female tail; (g), male tail; (h), detail of lateral lines at mid of body.
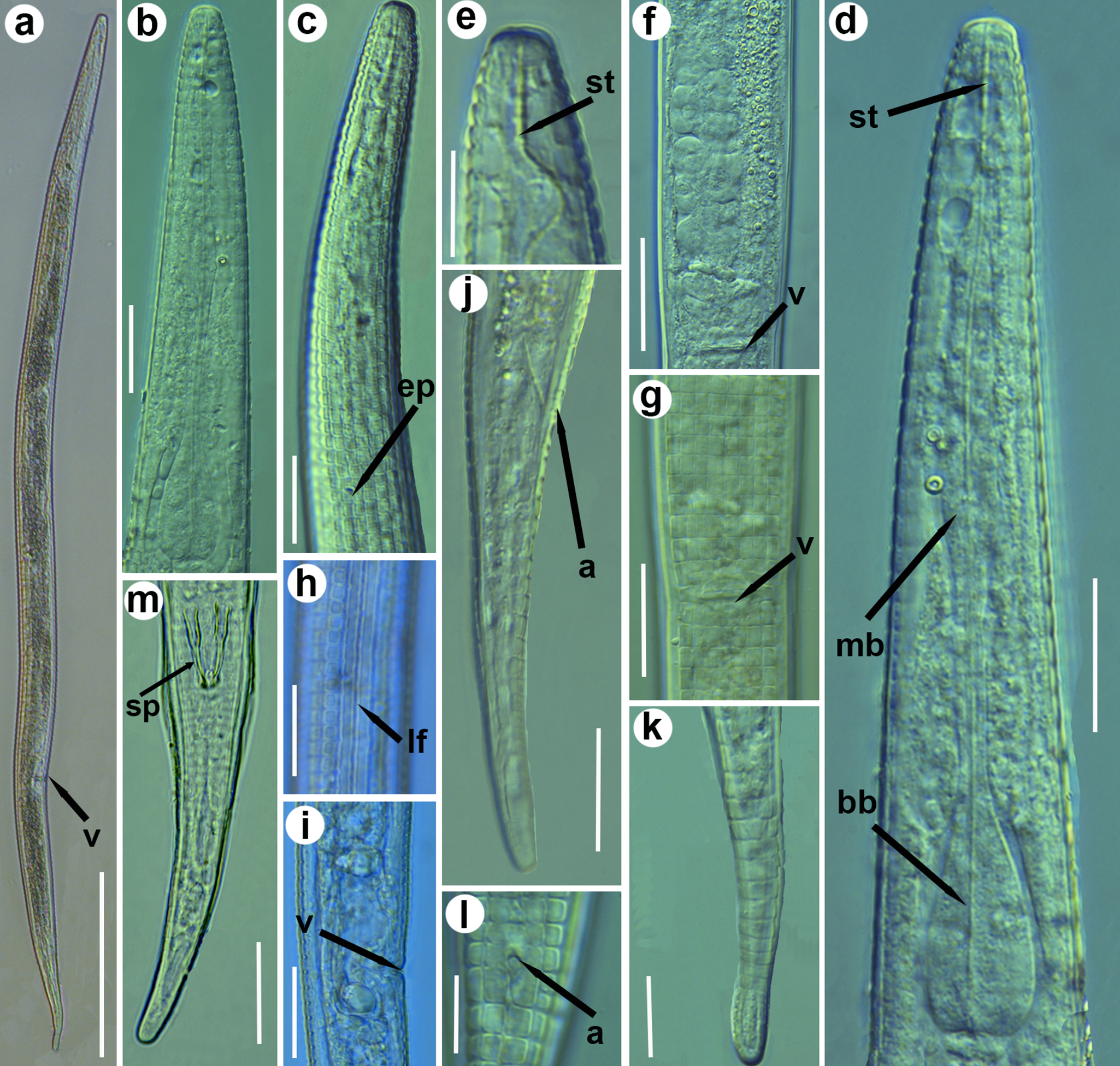
Figure 2. Light micrographs of Neothada olearum sp. nov. (a), entire female; (b–d), female pharyngeal region showing basal and median bulb, excretory pore, and stylet (arrowed); (e), detail of lip region showing stylet; (f–i), detail of vulval region showing longitudinal ridges, vulva, and crustaformeria; (j–l), female tail showing anus and rounded tip; (m), male tail showing a ventral view of spicules. a = anus; bb = basal bulb; ep = excretory pore; lf = lateral fields; mb = median bulb; sp = spicule; st = stylet; V = vulva. (Scale bars: a = 100 μm; b–d, f–k = 20 μm; e, l = 10 μm).
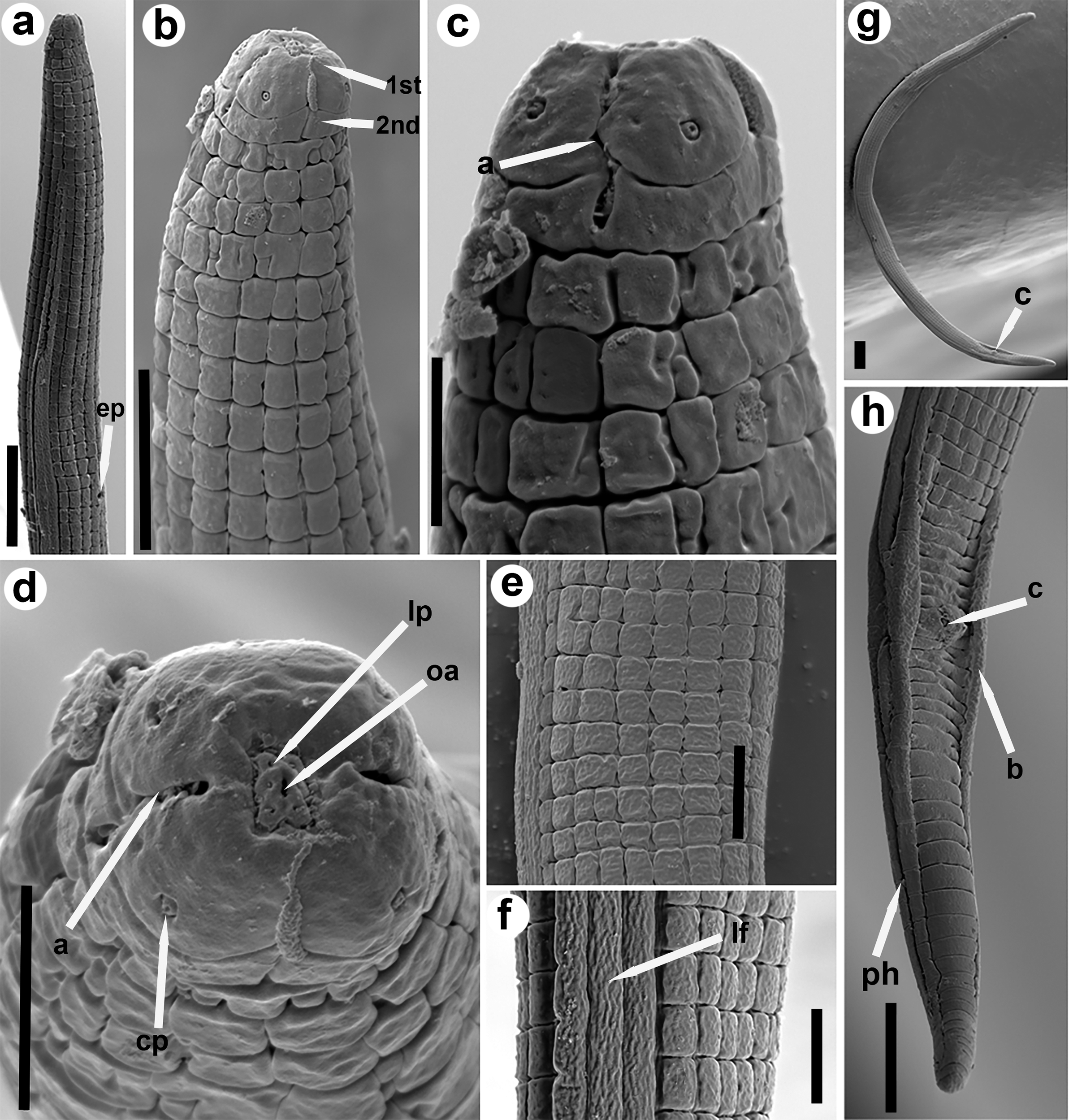
Figure 3. SEM micrographs of Neothada olearum sp. nov. a, b: female anterior region in lateral view showing excretory pore (ep), and detail of first (1st) and second (2nd) annuli; c: female lip region showing the amphidial aperture (a) arrowed; d: female lip region en face view showing the oral aperture (oa) surrounded by labial papillae (lp), cephalic papillae (cp), and amphidial aperture (a) arrowed; e, f: female mid body view showing lateral fields (lf) arrowed and longitudinal ridges; g: male whole body showing cloaca arrowed; h: male tail showing cloaca, bursa (b), and phasmid (ph) arrowed. a = amphidial aperture; b = bursa; c = cloaca; cp = cervical papillae; ep = excretory pore; lp = labial papilla; oa = oral aperture; ph = phasmid. (Scale bars: a, g, h = 20 μm; b, e, f = 10 μm; c d = 5 μm).
Males. Less frequent than female (1:5 ratio). Body habitus almost straight, slightly curved in posterior half after relaxed by gentle heat. Morphologically similar to female in body and tail shape, lip region, stylet, pharynx, and longitudinal ridges, except for genital system and secondary sexual features and posterior region more strongly curved ventrally. Testis single, outstretched, containing several rows of spermatogonia. Tail elongate-conoid, tail tip bluntly rounded. Spicules curved ventrally, gubernaculum simple and slightly ventrally curved.
Diagnosis and relationships
Neothada olearum sp. nov. is an amphimictic species characterised by a short body (563–774 μm); a widely annulated cuticle (2.5–3.0 μm); a total of 214–226 body annuli; 16 longitudinal ridges creating a tessellate body surface; a stylet without distinct basal knobs (9.0–11.0 μm); and an elongate-conoid tail with a bluntly rounded tip.
Morphological and morphometrically Neothada olearum sp. nov. is close to N. major from which can be separated in having 16 longitudinal ridges vs 20, stylet length (10.0 μm (9.0–11.0) vs 13.0 μm (12.0–14.4)), lower a ratio (27.0 (21.4–32.1) vs 37 (34–39)), and shorter spicules (17.6 μm (16.0–20.0) vs 21.5 μm (20.0–23.0)) (Maqbool and Shahina Reference Maqbool and Shahina1989). An Iranian population of N. major (Hosseinvand et al. Reference Hosseinvand, Eskandari and Ghaderi2020b) also showed morphological and morphometrical similarities with N. olearum sp. nov. but differs in longitudinal ridges (16 vs 19–20), a ratio (27.0 (21.4–32.1) vs 35.9 (29–44)), c’ ratio (4.8 (3.9–5.4) vs 6.4 (5.6–7.1)), and male presence and sperm present in spermatheca vs absence of male and spermatheca without sperm, as well as D2–D3 expansion segments of 28S rRNA (MN970002) differing in 15 bp and six indels (Figure 4).
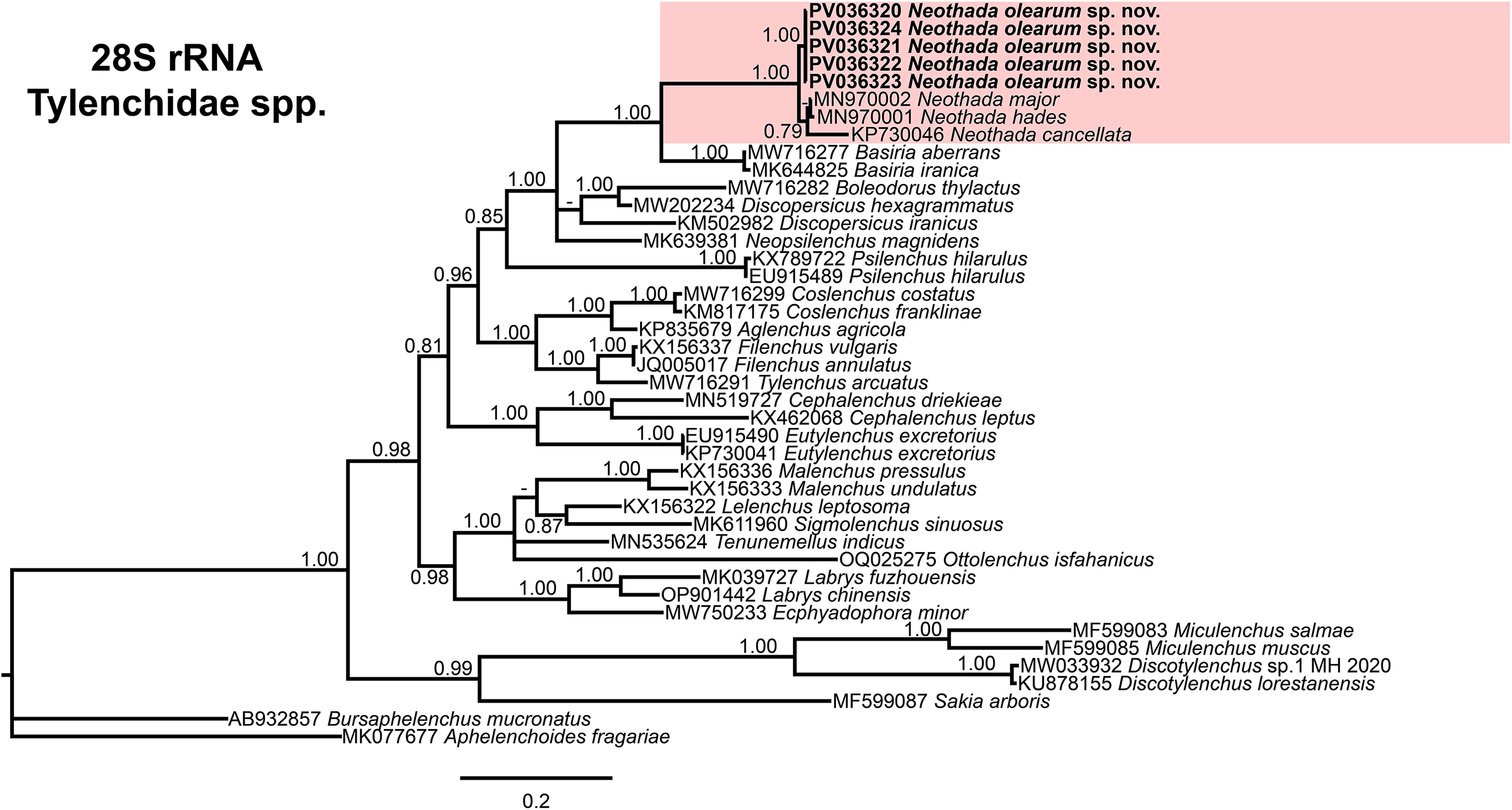
Figure 4. Phylogenetic relationships of Neothada olearum sp. nov. with species of Tylenchidae. Bayesian 50% majority rule consensus tree as inferred from D2 and D3 expansion segments of 28S rRNA sequence alignment under the TIM2 + I+ G model (−lnL = 11212.8251; AIC = 22605.650240; freqA = 0.1728; freqC = 0.2285; freqG = 0.3370; freqT = 0.2618; R(a) = 1.0546; R(b) = 2.8012; R(c) = 1.0546; R(d) = 1.0000; R(e) = 5.0928; R(f) = 1.0000; Pinva = 0.0138; and Shape = 0.6600). Posterior probabilities more than 0.70 are given for appropriate clades. Newly obtained sequences in this study are shown in bold, and coloured box indicate clade association of the new species. Scale bar = expected changes per site.
Neothada olearum sp. nov. can be separated from N. tatra by lip region (continuous with body contour vs set off), a longer body (649 μm (563–774) vs 560 μm), wider body annuli (2.5–3.0 μm vs 3.1–5.0 μm), and a longer tail (73 μm (68–80) vs 61 μm), with higher number of annuli 24–28 vs 18) (Thorne and Malek Reference Thorne and Malek1968). From N. geraerti, the new species can be separated by longitudinal ridges (16 vs 12), a longer body (649 μm (563–774) vs 490–570 μm), lower a ratio (27.0 (21.4–32.1) vs 37 (34–39)), and a longer tail (73 μm (68–80) vs 52–66 μm) (Andrássy Reference Andrássy1982). From N. cancellata, the new species can be separated by lip region shape (rounded, anteriorly flattened vs conoid), a longer body (649 μm (563–774) vs 560–580 μm), V ratio (72.7 (71–74) vs 69.5), and a longer tail (73 μm (68–80) vs 57 μm) (Geraert Reference Geraert1974; Thorne Reference Thorne1941). N. hades and N. costata mainly differ by the absence of stylet basal knobs vs presence, body length (649 μm (563–774) vs 530–620 μm, 867 μm (788–973), respectively), longitudinal ridges (16 vs 14, 14, respectively), stylet length (10.0 μm (9.0–11.0) vs 9.0–10.5 μm, 11.2 μm (11.0–13.0), respectively), V ratio (72.7 (71–74) vs 70–74, 66 (65–67), respectively), and tail length (73 μm (68–80) vs 55–66 μm, μm 122 (114–136), respectively) (Geraert and Raski Reference Geraert and Raski1986; Heyns and van den Berg Reference Heyns and van den Berg1996).
Etymology
The species epithet is the Latin term olearum = belonging to or corresponding to olives, as type material was found in an olive grove.
Type host and locality
The new species was recovered from the rhizosphere of olive (Olea europaea subsp. europaea L.) at Chiclana de Segura, Jaén province, Spain (coordinates 38°29’04.69” N, -3°05’15.43” W, 620 m a.s.l.).
Type material
Holotype female and 17 female and four male paratypes deposited at Institute for Sustainable Agriculture (IAS) of Spanish National Research Council (CSIC), Córdoba, Spain (Slide numbers ER51-02-ER51-08); and three females at the USDA Nematode Collection (T-8128p).
Molecular characterisation
Neothada olearum sp. nov. was molecularly characterised by the sequences of three ribosomal genes (D2–D3 expansion segments of 28S rRNA, ITS rRNA, and partial 18S rRNA), and the partial mitochondrial gene COI. The amplification of these regions yielded single fragments of approximately 900, 1100, 1800, and 400 bp, respectively, based on gel electrophoresis. Five D2–D3 of 28S rRNA sequences from 712 to 729 bp (PV036320–PV036324), five ITS rRNA sequences from 1126 bp (PV036325–PV036329), five 18S rRNA sequences from 1708 to 1719 bp (PV036330–PV036334), and four COI sequences of 248 to 266 bp (PV068612–PV068615) were generated for this new species without intraspecific sequence variations. D2–D3 expansion segments of 28S rRNA of N. olearum sp. nov. (PV036320–PV036324) was 98.0% similar to N. major (MN970002) from Iran, differing by 15 bp and six indels (Hosseinvand et al. Reference Hosseinvand, Eskandari and Ghaderi2020b); 95.8% similar to N. cancellata (KP730046) from Iran, differing by 28 bp and five indels (Yaghoubi et al. Reference Yaghoubi, Pourjam, Atighi and Pedram2015); 96.8% similar to N. hades (MN970001) from Iran, differing by 19 bp and five indels (Hosseinvand et al. Reference Hosseinvand, Eskandari and Ghaderi2020b); and 80.6% similar to Basiria aberrans (MW716277) from China, differing by 146 bp and 48 indels (unpublished). ITS of N. olearum sp. nov. (PV036325–PV036329) were the first sequences of this marker for Thada or Neothada genera in NCBI and no significant similarity was found in the BLAST search. In any case, our ITS sequences of N. olearum sp. nov. (PV036325–PV036329) showed an 89.1% similarity with Basiria aberrans-PP575000 (differing by 25 bp and 14 indels, but coverage = 20% only), and an 88.4% similarity with Sigmolenchus sinuosus-MK611962 (differing by 21 bp and zero indels, but coverage = 16% only). The partial 18S of N. olearum sp. nov. (PV036330–PV036334) was 95.1–94.5% similar to several Basiria species such as B. aberrans (KJ869353) from Iran, B. gracilis (EU130839) from California, USA, and B. graminophila (KJ869317) from Iran, differing by 85–95 bp and 18–31 indels (Helder et al. Reference Helder, Mooijman, van den Elsen, van Megen, Vervoort, Quist, Bert, Karegar, Karssen and Decraemer2014; Subbotin et al. Reference Subbotin, Ragsdale, Mullens, Roberts, Mundo-Ocampo and Baldwin2008). Also, 18S of N. olearum sp. nov. (PV036330–PV036334) was 92.1–92.5% similar to Boleodorus thylactus (KJ869348) from Iran and B. volutus (FJ969117) from The Netherlands, differing by 129–137 bp and 32–42 indels (Helder et al. Reference Helder, Mooijman, van den Elsen, van Megen, Vervoort, Quist, Bert, Karegar, Karssen and Decraemer2014). It also differs from Thada populus (MN557353) from Iran by 139 bp and 27 gaps (91.4% similarity) (Hosseinvand et al. Reference Hosseinvand, Eskandari, Ghaderi and Karegar2020a). Finally, COI of N. olearum sp. nov. (PV068612–PV068615) were the first sequences of this marker for the genus Neothada. COI of N. olearum sp. nov. was 83.6–84.1% similar to B. aberrans (MN577605, MN577606) from China, differing by 40–42 bp (Bai et al. Reference Bai, Qing, Qiao, Ning, Xiao, Cheng and Liu2020). It differs from Aglenchus geraerti (MN577603) or Coslenchus rafiqi (MN577602) from China by 47 bp and (79.9% similarity) (Bai et al. Reference Bai, Qing, Qiao, Ning, Xiao, Cheng and Liu2020); from B. thylactus (MN577607, MN577608) from China in 52 bp and two indels (78.4%) (Bai et al. Reference Bai, Qing, Qiao, Ning, Xiao, Cheng and Liu2020); and from Lelenchus leptosoma1 and 2 (MN577615, MN577616, MN577595, MN577596) from China in 58–70 bp (71.0–73.6%) (Bai et al. Reference Bai, Qing, Qiao, Ning, Xiao, Cheng and Liu2020).
Phylogenetic relationships of Neothada olearum sp. nov. with other Tylenchidae spp.
Phylogenetic relationships among Neothada olearum sp. nov. with other Tylenchidae spp., as inferred from analyses of D2–D3 expansion segments of 28S rRNA, the partial 18S rRNA, and the partial COI mtDNA gene sequences using BI, are shown in Figures 4, 5, and 6, respectively. The BLAST search of the newly generated ITS sequences of N. olearum sp. nov. showed a low coverage (less than 35%) with sequences of Tylenchidae spp. and as a result, no further phylogenetic analysis was performed using ITS sequences. The phylogenetic trees generated with the ribosomal and mitochondrial DNA markers included 42, 50, and 37 sequences with 738, 1690, and 441 characters in length, respectively (Figures 4–6). The D2–D3 expansion segments of 28S rRNA tree of Tylenchidae spp. showed a well-supported subclade (PP = 1.00), including N. olearum sp. nov. (PV036320–PV036324) together with a well-supported subclade (PP = 1.00), including three Neothada species (N. major-MN970002, N. hades-MN970001, and N. cancellata-KP730046). This clade clustered with a separate well-supported subclade (PP = 1.00) including B. aberrans-MW716277 and Basiria iranica-MK644825 (Figure 4).
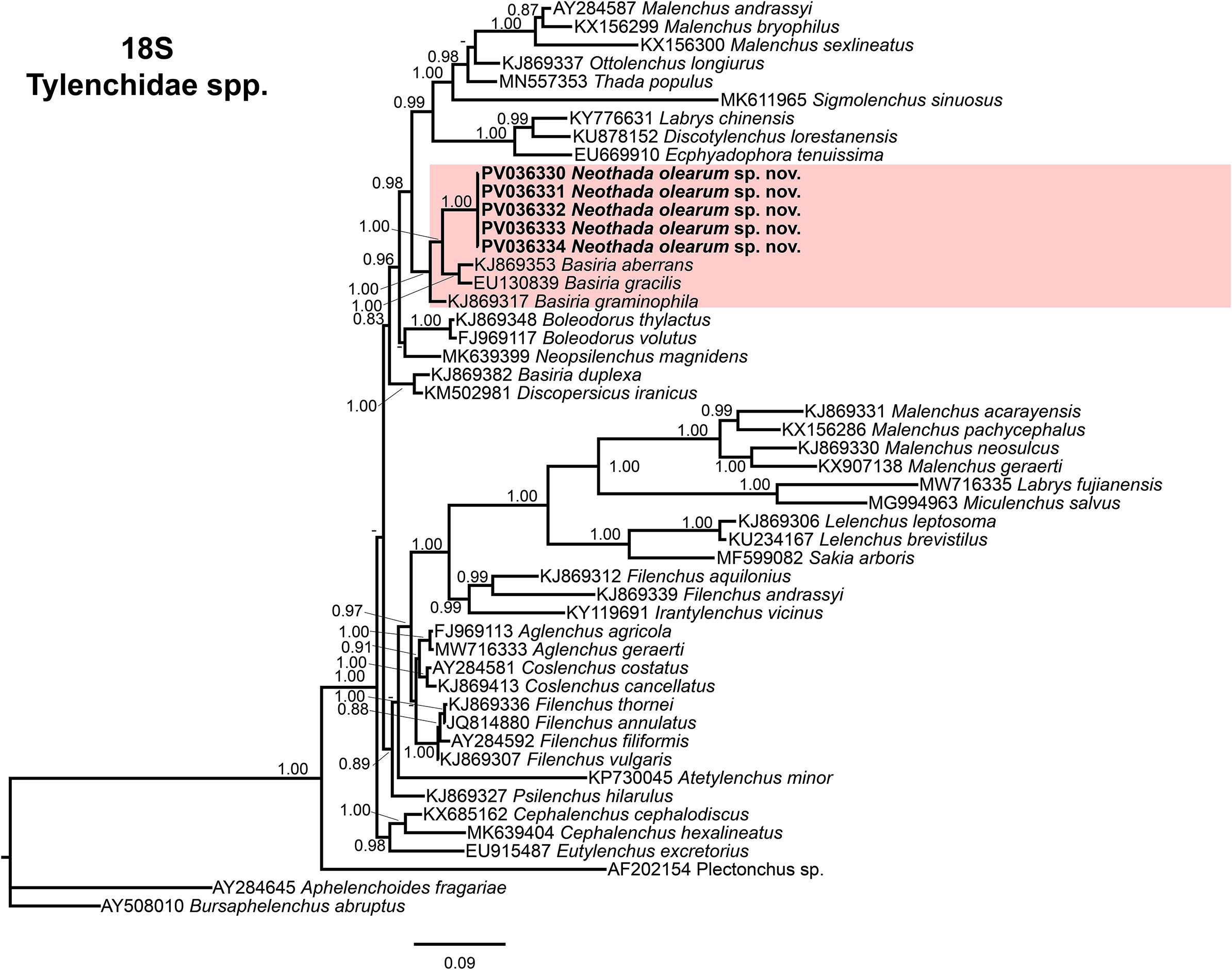
Figure 5. Phylogenetic relationships of Neothada olearum sp. nov. with species of Tylenchidae. Bayesian 50% majority rule consensus tree as inferred from 18S rRNA sequence alignment under the TIM1 + I+ G model (−lnL = 17362.9696; AIC = 34937.939200; freqA = 0.2417; freqC = 0.2246; freqG = 0.2879; freqT = 0.2458; R(a) = 1.0000; R(b) = 2.5654; R(c) = 1.1081; R(d) = 1.1081; R(e) = 4.9202; R(f) = 1.0000; Pinva = 0.1660; and Shape = 0.4570). Posterior probabilities more than 0.70 are given for appropriate clades. Newly obtained sequences in this study are shown in bold, and coloured box indicate clade association of the new species. Scale bar = expected changes per site.
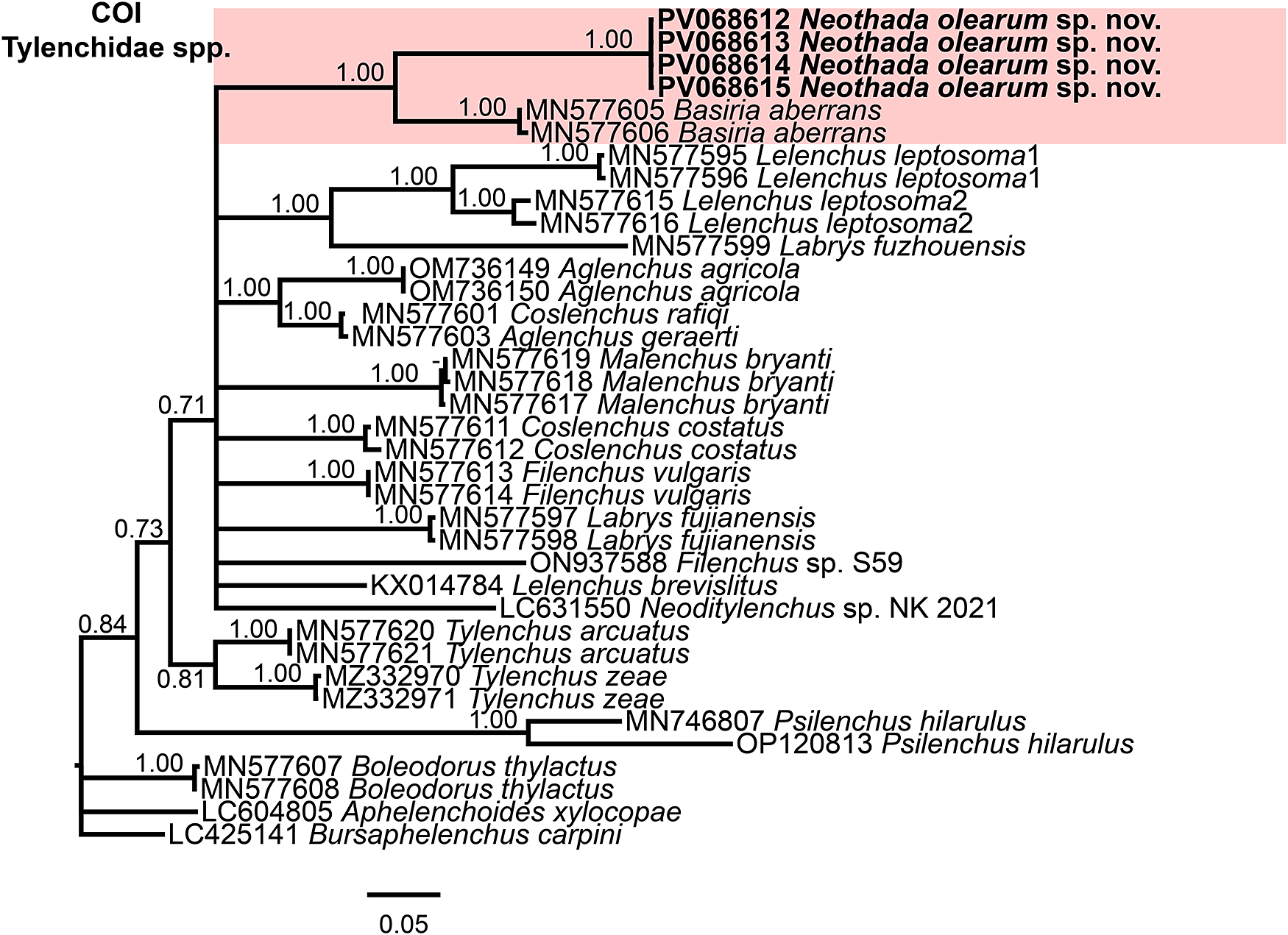
Figure 6. Phylogenetic relationships of Neothada olearum sp. nov. with species of Tylenchidae. Bayesian 50% majority-rule consensus trees as inferred from cytochrome c oxidase subunit I (COI) mtDNA gene sequence alignments under the GTR + G model (−lnL = 4051.7660; AIC = 8265.532060; freqA = 0.2622; freqC = 0.0899; freqG = 0.1832; freqT = 0.4647; R(a) = 1.2981; R(b) = 10.6042; R(c) = 4.1144; R(d) = 4.1433; R(e) = 6.3708; R(f) = 1.0000; Pinva = 0.3800; and Shape = 0.0000). Posterior probabilities more than 0.70 are given for appropriate clades. Newly obtained sequences in this study are shown in bold, and coloured box indicate clade association of the new species. Scale bar = expected changes per site.
The 50% majority rule consensus 18S rRNA gene BI tree showed also a well-supported subclade (PP = 1.00) with N. olearum sp. nov. (PV036330–PV036334) together with a well-supported subclade (PP = 1.00), including B. aberrans-KJ869353 and Basiria gracilis-EU130839, while Basiria graminophila-KJ869317 and Basiria duplexa-KJ869382 are clearly separated from this subclade (Figure 5). Interestingly, the only available 18S sequence for Thada populus (MN557353) clustered within a distinct, well-supported subclade (PP = 1.00) with other Tylenchidae genera such as Malenchus, Ottolenchus, and Sigmolenchus (Figure 5).
Finally, the 50% majority rule consensus of the partial COI gene BI tree showed also a well-supported subclade (PP = 1.00) with N. olearum sp. nov. (PV068612–PV068615) together with a well-supported subclade (PP = 1.00), including B. aberrans-MN577605-MN577606 (Figure. 6).
Discussion
The small size of Tylenchidae nematodes as well as their morphological simplicity and similarity make species identification very difficult, and integrative taxonomical approaches (morphological, including LM and SEM, and molecular) are crucial for an accurate identification (Qing and Bert Reference Qing and Bert2019). The main objective of this study was to identify and describe, morphologically and molecularly, a new population of Neothada detected in an olive grove in Chiclana de Segura, Jaén province, southern Spain, as well as clarify the phylogenetic relationships within the family Tylenchidae using ribosomal and mitochondrial markers. All the provided results from a morphological and molecular point of view confirmed that the unknown Neothada population is a new valid species of the genus herein described as N. olearum sp. nov. The present study enhances the knowledge of the biodiversity of Tylenchidae genera occurring in Spain. Our SEM results provide clear evidence that lip region structure is consistent with previous studies on N. costata, N. geraerti, and N. hades (Geraert and Raski Reference Geraert and Raski1986; Heyns & van den Berg Reference Heyns and van den Berg1996), maintaining the general structure of a quadrangular oral plate with fine oral aperture surrounded by six labial papillae, and amphidial apertures are not confined to the oral plate but continue on the lateral side as longitudinal slits, which fits with lip pattern type II-a by Qing and Bert (Reference Qing and Bert2018). The present results also supported the hypothesis that the number of longitudinal striae, which seems to be constant within each species, is an important diagnostic character, since our new species is quite close to an Iranian population of N. major (morphological and molecularly characterised) from which N. olearum sp. nov. differs mainly in the number of longitudinal ridges (16 vs 19–20) and other characters and ratio (Hosseinvand et al. Reference Hosseinvand, Eskandari and Ghaderi2020b).
Phylogenetic analyses of ribosomal and mitochondrial markers of this study confirmed that the family Tylenchidae is a heterogeneous group, which agrees with previous studies (Munawar et al. Reference Munawar, Yevtushenko and Castillo2021; Qing and Bert, Reference Qing and Bert2018, Reference Qing and Bert2019; Qing et al. Reference Qing, Decraemer, Claeys and Bert2017, Reference Qing, Pereira, Slos, Couvreur and Bert2018). Also, phylogenetic analyses are in concordance by grouping N. olearum sp. nov. with species of the genus Basiria, which are also consistent with previous studies and confirm its classical taxonomic position (Bai et al. Reference Bai, Qing, Qiao, Ning, Xiao, Cheng and Liu2020; Hosseinvand et al. Reference Hosseinvand, Eskandari and Ghaderi2020b; Munawar et al. Reference Munawar, Yevtushenko and Castillo2021; Qing et al. Reference Qing, Decraemer, Claeys and Bert2017, Reference Qing, Pereira, Slos, Couvreur and Bert2018; Qing and Bert, Reference Qing and Bert2018, Reference Qing and Bert2019; Yaghoubi et al. Reference Yaghoubi, Pourjam, Atighi and Pedram2015). Although to date, only a few Neothada species have been molecularly characterised, including N. cancellata, N. hades, N. major, and N. olearum sp. nov., based on 28S rRNA, the present results suggest that Neothada is a monophyletic genus within Tylenchidae; nevertheless further studies on other species and other gene markers are needed to confirm this hypothesis. Data on 18S rRNA gene phylogeny confirm that the genera Thada and Neothada are well-separated in two different clades, supporting that the longitudinal ridges differentiating both genera is a good trait to delimitate them as proposed by Khan (Reference Khan1973) and accepted by many other nematologists (Geraert Reference Geraert2008; Heyns and van den Berg Reference Heyns and van den Berg1996; Siddiqi Reference Siddiqi2000). Although the COI gene has only been explored for a limited number of Tylenchidae species, this molecular marker is one of the most important standard barcoding genes that has been used for many free-living and plant-parasitic nematodes (Kantor et al. Reference Kantor, Handoo, Subbotin, Mowery, Hult, Rogers and Skantar2023; Palomares-Rius et al. Reference Palomares-Rius, Cantalapiedra-Navarrete, Archidona-Yuste, Subbotin and Castillo2017; Powers Reference Powers2004). Our results on COI are congruent with the phylogenetic relationships showed by Bai et al. (Reference Bai, Qing, Qiao, Ning, Xiao, Cheng and Liu2020). We also concur with previous statements that the higher mutation rate of this marker offers a better separation of closely related species and is mainly valuable for the identification of potential cryptic species (Palomares-Rius et al. Reference Palomares-Rius, Cantalapiedra-Navarrete and Castillo2014). In addition, phylogeny of ribosomal and mitochondrial markers of N. olearum sp. nov. also confirmed that longitudinal ridges have originated several times independently within Tylenchidae (e.g., Coslenchus, Neothada, Eutylenchus) as previously proposed by Qing and Bert (Reference Qing and Bert2019).
In summary, the present study confirms the usefulness of applying integrative taxonomy in Tylenchidae to decipher the real diversity of these nematodes, which may be much higher, given that molecular diversity (rRNA and mtDNA markers) can remarkably exceed the low morphological diversity and that the majority of Tylenchidae species still remain completely undocumented (Qing and Bert Reference Qing and Bert2019). In addition, on the basis of the present results and as suggested by Heyns and van den Berg (Reference Heyns and van den Berg1996), some of the few documented reports of N. cancellata sensu lato need to be confirmed by integrative taxonomy.
Acknowledgements
The authors thank J. Martín Barbarroja, I. Casero Godoy, and G. León Ropero for their excellent technical contributions. SEM pictures were obtained with the assistance of technical staff and the equipment of ‘Centro de Instrumentación Científico-Técnica (CICT)’, University of Jaén, and CIC University of Granada, Spain. https://doi.org/10.13039/501100011033
Financial support
This research was funded by the grant Soil O-Live, from the European Union’s Horizon Europe research and innovation programme under grant agreement No. 101091255 (Soil Deal for Europe - HORIZON-MISS-2021-SOIL-02-03). R. Salazar-García’s contract is financed by Soil O-Live grant. A. Archidona-Yuste is funded by the Ramón y Cajal program (RYC2021-031108-I), funded by MCIN/AEI/10.13039/501100011033, and UE ‘Next Generation EU/PRTR’. This work was also partially supported by the Consejería de Universidad, Investigación e Innovación-Junta de Andalucía, Qualifica Project (QUAL21_023 IAS).
Competing interests
Authors stated no conflict of interest.
Ethical standard
The result of this work has not been published previously and is not under consideration elsewhere.
Ethical approval
The conducted research is not either related to human or animals use.








