Introduction
The resistance crisis in cervical cancer
Cervical cancer continues to pose a formidable challenge to global women’s health, with epidemiological projections indicating 700,000 annual new cases by 2030 (Ref. Reference Singh, Vignat, Lorenzoni, Eslahi, Ginsburg, Lauby-Secretan, Arbyn, Basu, Bray and Vaccarella1). Current therapeutic strategies remain stratified by FIGO staging: early-stage disease (IA-IIA) is managed primarily by radical surgery, whereas locally advanced cervical cancer (LACC, IIB-IVA) necessitates definitive chemoradiotherapy (CRT) combining external beam radiation with concurrent cisplatin-based chemotherapy (Refs Reference Singh, Vignat, Lorenzoni, Eslahi, Ginsburg, Lauby-Secretan, Arbyn, Basu, Bray and Vaccarella1, Reference Xie and Xu2, Reference Ladd, Duarte, Sahin and Zarrinpar3). Despite adherence to these evidence-based protocols, therapeutic resistance manifests in 30–50% of LACC patients, evidenced by persistent or recurrent disease within 2 years of treatment completion (Ref. Reference Li, Chen, Hu, Wang, Shen, Chen, Peng, Fang, Xia, Chen, Song, Wang, Zou, Wang, Han, Feng, Yuan, Guo, Meng, Feng, Chen, Yang, Fan, Wang, Ai, Ma and Sun4). This resistance phenotype exhibits significant heterogeneity: radioresistance is driven by aberrant DNA hypermethylation and NSUN6-mediated RNA m5C methylation (an epigenetic modification regulating mRNA stability and DNA repair efficiency) overexpression, which enhance DNA double-strand break repair capacity (Ref. Reference Yu, Ni, Xu, Liu, Chen, Li, Xia, Diao, Chen, Zhu, Wu, Tang, Li and Ke5), while chemoresistance involves cancer-associated fibroblast (CAF)-driven metabolic reprogramming and drug efflux pump activation (Ref. Reference Lin, Zhou, Liu, Nie, Cao, Li, Li, Zhu, Lin, Ding, Jiang, Gu, Xu, Zhao and Cai6). Collectively, these tumour-intrinsic mechanisms explain less than 40% of the observed variance in treatment response (Ref. Reference Xie and Xu2), highlighting a critical knowledge gap in our understanding of resistance drivers. Therapeutic resistance is the core bottleneck in advanced-stage patients, leading to recurrence. Therefore, in-depth analysis of the intrinsic mechanism of therapeutic resistance and the development of new intervention strategies targeting therapeutic resistance have become key challenges that need to be urgently addressed in current clinical treatment.
The microbiome as an emerging paradigm for tumour treatment
The past decade has witnessed a paradigm shift in cancer biology, recognizing that tumour development and therapy response are co-orchestrated by host-associated microbial communities, the microbiome influences modulates tumour occurrence, progression, and treatment response of tumours through mechanisms such as metabolites, immune regulation and epigenetics (Refs Reference Dai, Tan, Qiao and Liu7, Reference Queen, Shaikh and Sears8). In cervical cancer, the vaginal microbiome – tragically viewed as a passive bystander – has emerged as an active participant in disease pathogenesis. High-throughput sequencing studies consistently demonstrate that progressive cervical intraepithelial neoplasia correlates with community state type (CST) transitions from Lactobacillus dominance to polymicrobial dysbiosis enriched in Fusobacteria, Sneathia and Prevotella (Refs Reference Liao, Chen, Ruan, Wang, Hu, Long, Li, Zhang, Yu, Ming zhang, Zhang and Liao9, Reference Wahid, Dar, Jawed, Mandal, Akhter, Khan, Khan, Jogaiah, Rai and Rattan10). Critically, microbial metabolites function as inter-kingdom signalling molecules that directly modulate therapeutic vulnerabilities. Functional studies reveal that tumour-resident Lactobacillus iners produce millimolar concentrations of l-lactate, which activate HCA1 receptors on cancer cells to enhance DNA repair capacity while simultaneously stabilizing PD-L1 expression through HIF-1α-dependent epigenetic modulation, directly fuelling radioresistance through metabolic rewiring and epigenetic modulation (Ref. Reference Colbert, El Alam, Wang, Karpinets, Lo and Lynn11). Beyond local effects, gut-derived short-chain fatty acids (SCFAs), particularly butyrate, systemically regulate CD8+ T-cell mitochondrial fitness and dendritic cell maturation, thereby influencing immune checkpoint inhibitor efficacy (Refs Reference Elkrief, Pidgeon, Maleki Vareki, Messaoudene, Castagner and Routy12, Reference Liu, Chen, Zhang and Dong13). Microbial metabolites (e.g., short-chain fatty acids, lactate) function as inter-kingdom signalling molecules, challenging the traditional tumour-centric view of resistance mechanisms (Refs Reference Jiang, Xie, Xiao, Kang, Lin, Zhang, Li, Qian, Xu, Leng, Wang, Tu, Zhong, Zhao, Chen, Wang, Liu, Hong, Chen, Chen and Fang14, Reference Liu, Lau, Cheng and Yu15). These findings establish a tripartite microbiome-immune-tumour crosstalk network, necessitating an integrated multi-omics framework spanning microbiome-metabolome-immunome analyses to decode resistance mechanisms comprehensively, providing a revolutionary strategic option for the precise treatment of cervical cancer (Refs Reference Dai, Tan, Qiao and Liu7, Reference Nobels, Van Marcke, Jordan, Van Hul and Cani16, Reference Yao, Chen, Wang, Liang, Liu, Gao and Cai17).
Microbiome-mediated mechanisms: from correlation to causation
Multiple studies have shown that the microbiome of patients with cervical cancer treatment resistance presents characteristic changes (Refs Reference Queen, Shaikh and Sears8, Reference Johnston and Bullman18, Reference Mitra, MacIntyre, Paraskevaidi, Moscicki, Mahajan, Smith, Lee, Lyons, Paraskevaidis, Marchesi, Bennett and Kyrgiou19). In addition, patients with cervical cancer often have a high burden of HPV and abnormal proliferation of Fusobacteria and Sneathia (Ref. Reference Wahid, Dar, Jawed, Mandal, Akhter, Khan, Khan, Jogaiah, Rai and Rattan10). Among patients resistant to chemotherapy and radiotherapy, both the tumour microenvironment and the intestinal microbiome have undergone significant changes. It is common for Lactobacillus iners (L. iners) to be the dominant bacteria, while the proportion of Lactobacillus crispatus (L. crispatus) has significantly decreased. The transition from observational associations to causal mechanisms has been catalysed by multi-omics integration and functional validation experiments. Multi-omics analysis revealed multiple gene mutations and abnormal signalling pathways related to drug resistance, suggesting the potential of precision medicine in the treatment of drug-resistant cervical cancer. Despite this, there are still many unsolved mysteries regarding the mechanism by which the microbiota affects drug resistance. It is urgently necessary to conduct comprehensive research combining the microbiome and the tumour genome to achieve the goal of personalized treatment (Ref. Reference Diefenbach, Greten and Efferth20). There are three core pathways now underpin microbiome-mediated resistance.
Metabolic reprogramming via lactate signalling. Tumour-colonizing L. iners generate l-lactate at rates exceeding 15 nmol/106 cells/hour, creating a self-reinforcing resistance loop (Ref. Reference Colbert, El Alam, Wang, Karpinets, Lo and Lynn11). Lactate activates the HCA1 receptor, triggering PI3K/AKT-mediated intracellular alkalinization that enhances DNA repair capacity (Refs Reference Johnston and Bullman18, Reference Ciszewski, Sobierajska, Stasiak and Wagner21). Simultaneously, lactate functions as an endogenous histone deacetylase inhibitor, promoting γH2AX hyperacetylation and accelerating double-strand break resolution (Ref. Reference Ciszewski, Sobierajska, Stasiak and Wagner21). Quantitative metabolomic analyses demonstrate that intratumoural lactate levels >12 mM correlate with a 2.3-fold increase in cancer cell survival fraction at 2 Gy (SF₂) (p < 0.001) (Ref. Reference Colbert, El Alam, Wang, Karpinets, Lo and Lynn11).
Direct drug metabolism by microbial enzymes. While β-glucuronidase (GUS) activity primarily impacts irinotecan toxicity, recent data reveal that cervical cancer biopsies exhibiting high bacterial GUS activity show 40% reduced cisplatin-DNA adduct formation (Ref. Reference Nagar and Blanchard22). This previously unrecognized mechanism suggests microbial deconjugation may limit active drug availability within the tumour microenvironment. Furthermore, tumour-enriched Gammaproteobacteria express long isoform cytidine deaminase, potentially inactivating gemcitabine and topotecan used in recurrent disease settings (Ref. Reference Geller, Barzily-Rokni, Danino, Jonas, Shental, Nejman, Gavert, Zwang, Cooper, Shee, Thaiss, Reuben, Livny, Avraham, Frederick, Ligorio, Chatman, Johnston, Mosher, Brandis, Fuks, Gurbatri, Gopalakrishnan, Kim, Hurd, Katz, Fleming, Maitra, Smith, Skalak, Bu, Michaud, Trauger, Barshack, Golan, Sandbank, Flaherty, Mandinova, Garrett, Thayer, Ferrone, Huttenhower, Bhatia, Gevers, Wargo, Golub and Straussman23).
Immune microenvironment remodelling. L. iners-derived lactate stabilizes PD-L1 nuclear translocation (a process suppressing interferon signalling via transcriptional repression of immune genes) mRNA through HIF-1α-dependent mechanisms, increasing tumour cell surface PD-L1 expression by 2.8-fold (Refs Reference Colbert, El Alam, Wang, Karpinets, Lo and Lynn11, Reference Lu, Jin, Tang, Zhou, Xu, Su, Wang, Xu, Zhao, Yin, Zhang, Jia, Peng, Zhou, Wang, Chen, Wang, Yang, Chen and Chen24). This process is compounded by microbial metabolites modulate regulatory T-cell (Treg) infiltration via butyrate-PPARα signalling, establishing an immunosuppressive niche that compromises CRT-induced anti-tumour immunity (Ref. Reference He, Fu, Li, Wang, Gong, Zhang, Dong, Huang, Wang, Mackay, Fu, Chen and Guo25). Single-cell RNA sequencing reveals that microbiome dysbiosis expands a distinct TIGIT+ CD8+ T-cell exhaustion subset, correlating with reduced progression-free survival (HR = 1.87, 95% CI 1.12–3.11) (Ref. Reference Zhu, Frank, Radka, Jeanfavre, Xu, Tse, Pacheco, Kim, Pierce, Deik, Hussain, Elsherbini, Hussain, Xulu, Khan, Pillay, Mitchell, Dong, Ndung’u, Clish, Rock, Blainey, Bloom and Kwon26).
Geographic and host context: the HIV modification effect
Microbiome-resistance associations exhibit profound geographic heterogeneity, necessitating region-specific validation. In sub-Saharan Africa, where cervical cancer burden is highest, HIV co-infection drives a 3-fold enrichment of inflammatory pathobionts (Prevotella bivia, Chlamydia trachomatis) and depletes protective L. crispatus (Ref. Reference Klein, Kahesa, Mwaiselage, West, Wood and Angeletti27). This dysbiotic signature, present in 68% of HIV+ patients, independently predicts CRT failure (adjusted HR = 2.34) (Refs Reference Sims, Yoshida-Court, El Alam, Ramatlho, Ketlametswe and Ning28, Reference Sims, Biegert, Ramogola-Masire, Ngoni, Solley, Ning, el Alam, Mezzari, Petrosino, Zetola, Schmeler, Colbert, Klopp and Grover29). This indicates that the high HIV infection rate in the region indirectly affects the treatment response and prognosis of cervical cancer by regulating the cervical microbiome. Latin American cohorts show CST-IV dominance (absence of Lactobacillus spp.) in 68% of high-grade squamous intraepithelial lesions (HSIL) versus 23% of controls (p < 0.001), with concurrent elevation of pro-inflammatory cytokines (IL-1β, IL-6) (Refs Reference Tosado-Rodríguez, Mendez, Espino, Dorta-Estremera, Aquino, Romaguera and Godoy-Vitorino30, Reference Lin, Huang, Zhang, Huang, Cai, Huang, Li, Zhang, Xue, Dong and Sun31). Based on the above research, it can be seen that regional microbiome differences have an important impact on the pathogenesis and treatment response of cervical cancer. The diversity of cervical microbiome structure and function among women in different regions varies due to genetic, environmental, co-infection (such as HIV) and socio-economic factors. Such differences not only affect the clearance and persistence of HPV infection but may also influence the inflammatory microenvironment and immune regulation of cervical cancer, thereby affecting treatment outcomes and patient prognosis. The high incidence of these pathogenic bacteria may lead to chronic cervical inflammation and accelerate the process of canceration, underscored that microbiome-driven resistance is modulated by host immune status and regional pathogen exposure, challenging the universality of biomarkers derived from predominantly European populations. Future research needs to integrate multi-omics techniques to deeply analyse the characteristics of the cervical microbiome in different geographical regions and its interrelationship with the host immune response, explore region-specific diagnostic biomarkers and microbiome regulatory intervention strategies, in order to achieve individualized prevention and treatment of cervical cancer (Refs Reference Pu, Wang, Wang, Gu, Zhu and Li32, Reference Li, Xiang, Liu, Chen, Zhang, Li, Kang and Wu33).
Knowledge gaps and translational imperatives
Long-term dynamic monitoring data show that the interaction between the microbiome and the host exhibits dynamic evolution characteristics. During the development of cervical cancer, the imbalance of the microbiome may lead to reproductive system inflammation and cancer occurrence. This effect is mainly achieved by activating immune responses, regulating metabolite and hormone levels (Ref. Reference Colbert, El Alam, Wang, Karpinets, Lo and Lynn11). These long-term monitoring data provide important evidence for understanding the dynamic process of microbiota–host interaction and also lay the foundation for developing microbiome-based intervention strategies (Refs Reference Liu, Chen, Zhang and Dong13, Reference Mitra, MacIntyre, Paraskevaidi, Moscicki, Mahajan, Smith, Lee, Lyons, Paraskevaidis, Marchesi, Bennett and Kyrgiou19). Despite mechanistic advances, three critical barriers impede clinical translation.
Causality versus correlation: Longitudinal studies remain observational; animal models inadequately recapitulate human vaginal microbiome complexity. Establishing causality requires microbiome depletion-reconstitution experiments in patient-derived organoid-xenograft models.
Temporal dynamics: The optimal intervention window – whether pre-treatment priming, concurrent modulation, or post-treatment restoration – remains undefined. Serial sampling studies suggest a critical ‘window of opportunity’ during the first 2 weeks of CRT when L. crispatus loss is most pronounced (Ref. Reference Dou, Ai, Zhang, Li, Jiang, Wu, Zhao, Li and Zhang34).
Standardization deficit: Heterogeneity in sampling methods (cervical swab versus vaginal aspirate), sequencing platforms (16S versus shotgun), and bioinformatic pipelines limit cross-study comparability and meta-analytic power (Ref. Reference Mitra, MacIntyre, Paraskevaidi, Moscicki, Mahajan, Smith, Lee, Lyons, Paraskevaidis, Marchesi, Bennett and Kyrgiou19).
Scope and objectives of this review
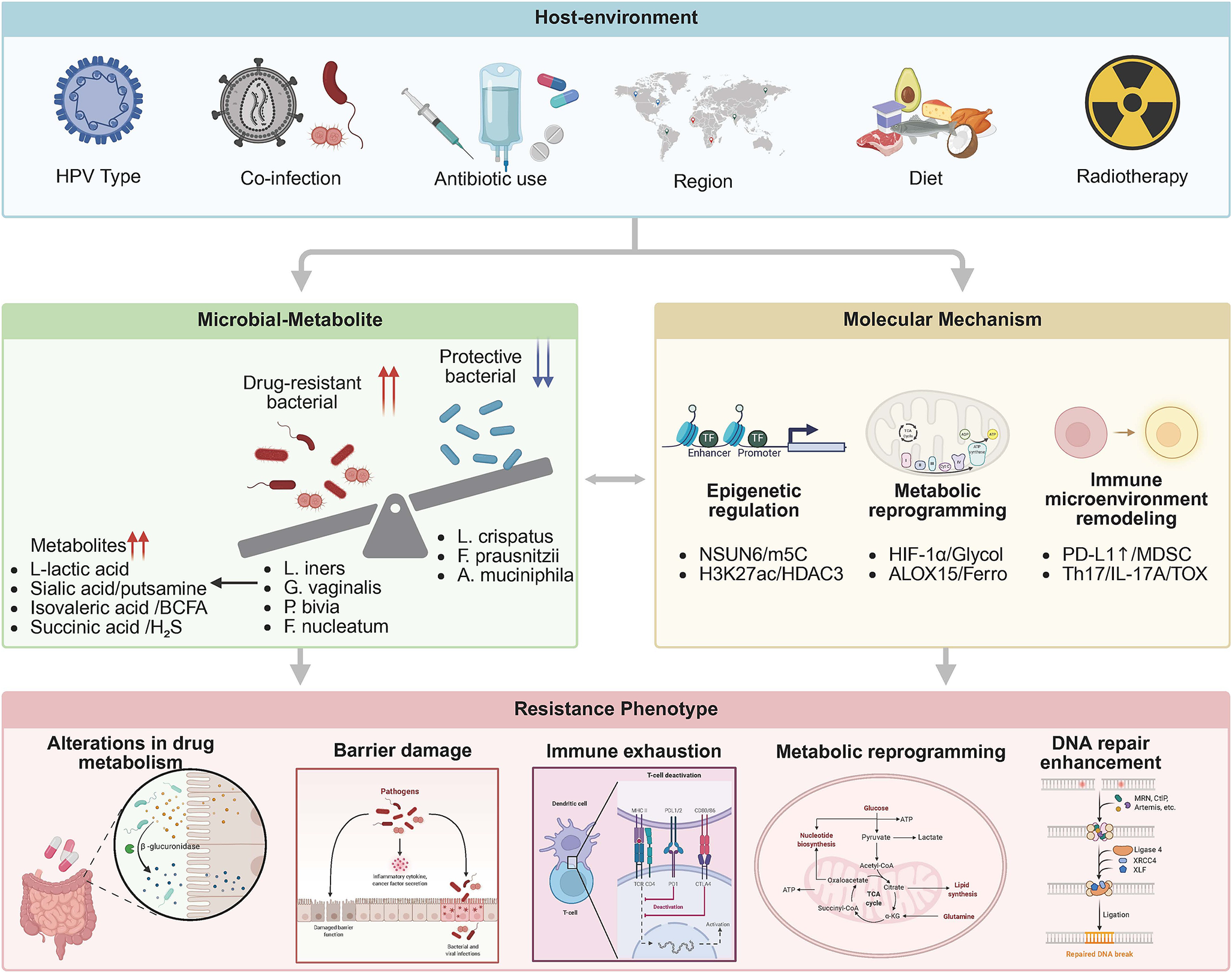
This review addresses microbiome-driven resistance in locally advanced cervical cancer (FIGO IIB-IVA) undergoing definitive CRT, aiming to bridge the systematic gap between mechanistic discovery and clinical intervention. We adopt a multi-microbial ecological network framework. This integrates the Lactobacillus paradox – L. iners’s context-dependent pathogenicity versus L. crispatus’s protective effects – with an anaerobic bacterial synergy network (Gardnerella vaginalis-P. bivia-Mobiluncus triad) to propose a three-tier regulation model: metabolic reprogramming (lactate/butyrate antagonism) – epigenetic hijacking (NSUN6-m5C/HDAC3-H3K27ac) – immune microenvironment remodelling (PD-L1 nuclear translocation/MDSC-Treg axis). By adopting a multi-omics perspective anchored in quantitative mechanistic data, we seek to redefine precision oncology in cervical cancer through microbiome-informed interventions, such as LACTIN-V with L. crispatus CTV-05 have demonstrated efficacy in colonizing dysbiotic environments and reducing genital inflammation in clinical trials (Ref. Reference Hemmerling, Mitchell, Demby, Ghebremichael, Elsherbini, Xu, Xulu, Shih, Dong, Govender, Pillay, Ismail, Casillas, Moodley, Bergerat, Brunner, Liebenberg, Ngcapu, Mbano, Lagenaur, Parks, Ndung’u, Kwon and Cohen35) (Figure 1).

Figure 1. Tri-level regulatory network of microbiome-mediated therapeutic resistance in cervical cancer. External factors (e.g., HPV infection, HIV co-infection) drive baseline dysbiosis, characterized by expansion of pathobionts (L. iners, G. vaginalis, P. bivia) and depletion of commensals (L. crispatus). This alters metabolite profiles (e.g., lactate accumulation), subsequently inducing epigenetic reprogramming (NSUN6-m⁵C, HDAC3-H3K27ac) and immune remodelling (PD-L1 nuclear translocation, MDSC infiltration). Red arrows: activating interactions; blue dashed lines: inhibitory effect.
Clinical evidence: microbial signatures of treatment failure
Biphasic pattern of microbial dysbiosis
A consistent microbial signature of treatment resistance has emerged from multi-cohort analyses encompassing over 1,200 cervical cancer patients undergoing definitive CRT. Meta-analytic synthesis reveals a biphasic pattern: depletion of protective commensals and pathobiont enrichment. Five independent studies demonstrate that L. crispatus dominance (relative abundance >50%) confers a 68% reduction in recurrence risk (Refs Reference Liao, Chen, Ruan, Wang, Hu, Long, Li, Zhang, Yu, Ming zhang, Zhang and Liao9, Reference Mitra, MacIntyre, Paraskevaidi, Moscicki, Mahajan, Smith, Lee, Lyons, Paraskevaidis, Marchesi, Bennett and Kyrgiou19, Reference Wang, Wang, Yan, Zhao, Wang, Liu, Fan and Xu36, Reference Armstrong and Kaul37, Reference Zheng, Hu, Liu, Zhao, Li, Wang, Zhang, Zhang, Song, Lyu, Cui, Ding and Wang38). A prospective longitudinal study tracking 234 patients throughout CRT demonstrated that individuals maintaining L. crispatus populations achieved a 3-year progression-free survival of 71%, compared to 39% in those experiencing early loss of this species (hazard ratio of 0.43, p = 0.003) (Ref. Reference Dou, Ai, Zhang, Li, Jiang, Wu, Zhao, Li and Zhang34). This protective effect appears mediated by L. crispatus’s ability to generate a low pH environment (<4.5) through d-lactate production and to preserve local IgA-mediated immune surveillance (Refs Reference Zhu, Frank, Radka, Jeanfavre, Xu, Tse, Pacheco, Kim, Pierce, Deik, Hussain, Elsherbini, Hussain, Xulu, Khan, Pillay, Mitchell, Dong, Ndung’u, Clish, Rock, Blainey, Bloom and Kwon26, Reference Breedveld, Schuster, Van Houdt, Painter, Mebius and Van Der Veer39).
Pathobiont enrichment and lactate production
In stark contrast, L. iners abundance exceeding 30% at baseline independently predicts treatment failure, with a hazard ratio of 2.14 (95% CI 1.45–3.16) for recurrence (Refs Reference Dou, Ai, Zhang, Li, Jiang, Wu, Zhao, Li and Zhang34, Reference Wei, Chen, Lu, Cao, Tang and Yang40). The mechanism linking L. iners to resistance involves its prodigious lactate production; this species converts glucose to l-lactate at rates of 15.3 ± 2.7 nmol/106 cells/hour under hypoxic conditions typical of the tumour microenvironment, leading to intratumoural lactate accumulation from baseline 8.4 to 18.7 mM within 10 days (p < 0.001) (Refs Reference Colbert, El Alam, Wang, Karpinets, Lo and Lynn11, Reference Johnston and Bullman18). This concentration saturates monocarboxylate transporters and activates the HCA1 receptor, triggering downstream resistance pathways that reduce radiation-induced DNA damage (Refs Reference Wu, Wang, Ying, Jin, Li and Hu41, Reference Garcia-Flores, Sollome, Thavathiru, Bower and Vaillancourt42). Genomic analyses reveal that L. iners strains isolated from cancer patients uniquely encode a cytolysin operon that disrupts epithelial barrier integrity at bacterial loads exceeding 107 CFU/ml, while downregulating hydrogen peroxide production in HIV-positive individuals, thereby eliminating its protective capacity (Ref. Reference Klein, Kahesa, Mwaiselage, West, Wood and Angeletti27).
Complex pathobiont signatures beyond Lactobacilli
Beyond Lactobacilli, a pathobiont index combining Fusobacterium nucleatum, Sneathia sanguinegens and Atopobium vaginae demonstrates superior predictive accuracy (area under curve of 0.81) compared to single-taxon markers (Refs Reference Sims, El Alam, Karpinets, Dorta-Estremera, Hegde and Nookala43, Reference Tortelli, Contreras, Markovina, Ding, Wylie and Schwarz44). Fusobacterium enrichment exceeding 1% correlates with elevated interleukin-6 and interleukin-8 levels, creating a pro-inflammatory niche that enhances tumour cell survival through NF-κB activation (Refs Reference Yu, Ni, Xu, Liu, Chen, Li, Xia, Diao, Chen, Zhu, Wu, Tang, Li and Ke5, Reference Lin, Zhou, Liu, Nie, Cao, Li, Li, Zhu, Lin, Ding, Jiang, Gu, Xu, Zhao and Cai6).
Temporal dynamics during therapy
Temporal dynamics during therapy reveal distinct trajectories in responders versus non-responders. The acute phase of CRT triggers immediate L. crispatus loss (log2-fold change −3.2 ± 0.8), while responders show parallel expansion of radiation-tolerant Lactobacillus gasseri that preserves lactic acid production and low pH (Ref. Reference Dou, Ai, Zhang, Li, Jiang, Wu, Zhao, Li and Zhang34). Non-responders, however, experience rapid L. iners proliferation (log2-fold change +4.1), correlating with lactate accumulation from 8 to 18 mM (Ref. Reference Colbert, El Alam, Wang, Karpinets, Lo and Lynn11). The subacute phase witnesses radiation-tolerant Enterobacteriaceae proliferation in non-responders, facilitated by lactate-induced epithelial barrier disruption, while microbial diversity declines by 45% in non-responders but remains stable in responders (p < 0.001) (Refs Reference Garcia-Flores, Sollome, Thavathiru, Bower and Vaillancourt42, Reference Jiang, Li, Zhang, Ma, Liu, Liang, Wei, Huang and Wang45). Six months post-treatment, microbial diversity recovery correlates with reduced recurrence risk (r = 0.61, p = 0.03), and patients achieving L. crispatus reconstitution exhibit 5-year overall survival of 78% versus 51% in persistently dysbiotic individuals (p = 0.01) (Refs Reference Wu, Wang, Ying, Jin, Li and Hu41, Reference Garcia-Flores, Sollome, Thavathiru, Bower and Vaillancourt42).
Geographic heterogeneity and host factors
In sub-Saharan Africa, where cervical cancer burden is highest, HIV co-infection drives a 3-fold enrichment of inflammatory pathobionts (P. bivia, G. vaginalis) and depletes protective L. crispatus. This dysbiosis is mechanistically linked to HIV-induced CD4+ T-cell depletion (reducing counts by 50–70%), which impairs mucosal IgA secretion – a critical factor for maintaining L. crispatus adhesion to vaginal epithelium. Concurrently, diminished CD4+ T-cell surveillance permits expansion of pro-inflammatory anaerobes that elevate local IL-1β and IL-6, further disrupting the mucosal barrier and activating HIF-1α/NF-κB pathways to promote therapy resistance. This dysbiotic signature, present in 68% of HIV+ patients, independently predicts CRT failure (adjusted HR = 2.34) (Refs Reference Klein, Kahesa, Mwaiselage, West, Wood and Angeletti27, Reference Sims, Yoshida-Court, El Alam, Ramatlho, Ketlametswe and Ning28, Reference Gosmann, Anahtar, Handley, Farcasanu, Abu-Ali, Bowman, Padavattan, Desai, Droit, Moodley, Dong, Chen, Ismail, Ndung’u, Ghebremichael, Wesemann, Mitchell, Dong, Huttenhower, Walker, Virgin and Kwon46, Reference Anahtar, Byrne, Doherty, Bowman, Yamamoto, Soumillon, Padavattan, Ismail, Moodley, Sabatini, Ghebremichael, Nusbaum, Huttenhower, Virgin, Ndung’u, Dong, Walker, Fichorova and Kwon47). Recent clinical trials demonstrate that restoring L. crispatus colonization via live biotherapeutics (e.g., LACTIN-V) reduces endocervical CD4+ T-cell activation-a key HIV target cell population-and may mitigate dysbiosis-driven treatment resistance (Ref. Reference Hemmerling, Mitchell, Demby, Ghebremichael, Elsherbini, Xu, Xulu, Shih, Dong, Govender, Pillay, Ismail, Casillas, Moodley, Bergerat, Brunner, Liebenberg, Ngcapu, Mbano, Lagenaur, Parks, Ndung’u, Kwon and Cohen35). Latin American cohorts show CST-IV dominance in 68% of high-grade lesions versus 23% of controls, with concurrent elevation of sialidase-producing G. vaginalis (Refs Reference Tortelli, Contreras, Markovina, Ding, Wylie and Schwarz44, Reference Tosado-Rodríguez, Alvarado-Vélez, Romaguera and Godoy-Vitorino48, Reference Nieves-Ramírez, Partida-Rodríguez, Moran, Serrano-Vázquez, Pérez-Juárez, Pérez-Rodríguez, Arrieta, Ximénez-García and Finlay49). These regional variations necessitate population-specific biomarker validation before global implementation.
Establishing causality beyond association
Establishing causality requires experimental validation beyond observational associations. Antibiotic-mediated microbiome depletion followed by L. iners reconstitution in patient-derived xenograft models demonstrates that L. iners colonization alone reduces CRT efficacy (tumour growth inhibition: 42% versus 68% in controls, p = 0.009), with lactate inhibition rescuing this effect, thereby fulfilling modified Koch’s postulates for microbiome-mediated resistance (Ref. Reference Colbert, El Alam, Wang, Karpinets, Lo and Lynn11). Such functional studies bridge the gap between correlation and causation, strengthening the rationale for microbiome-targeted interventions.
Molecular mechanisms: the lactate-centred axis of resistance
Lactate: the master metabolite driving resistance
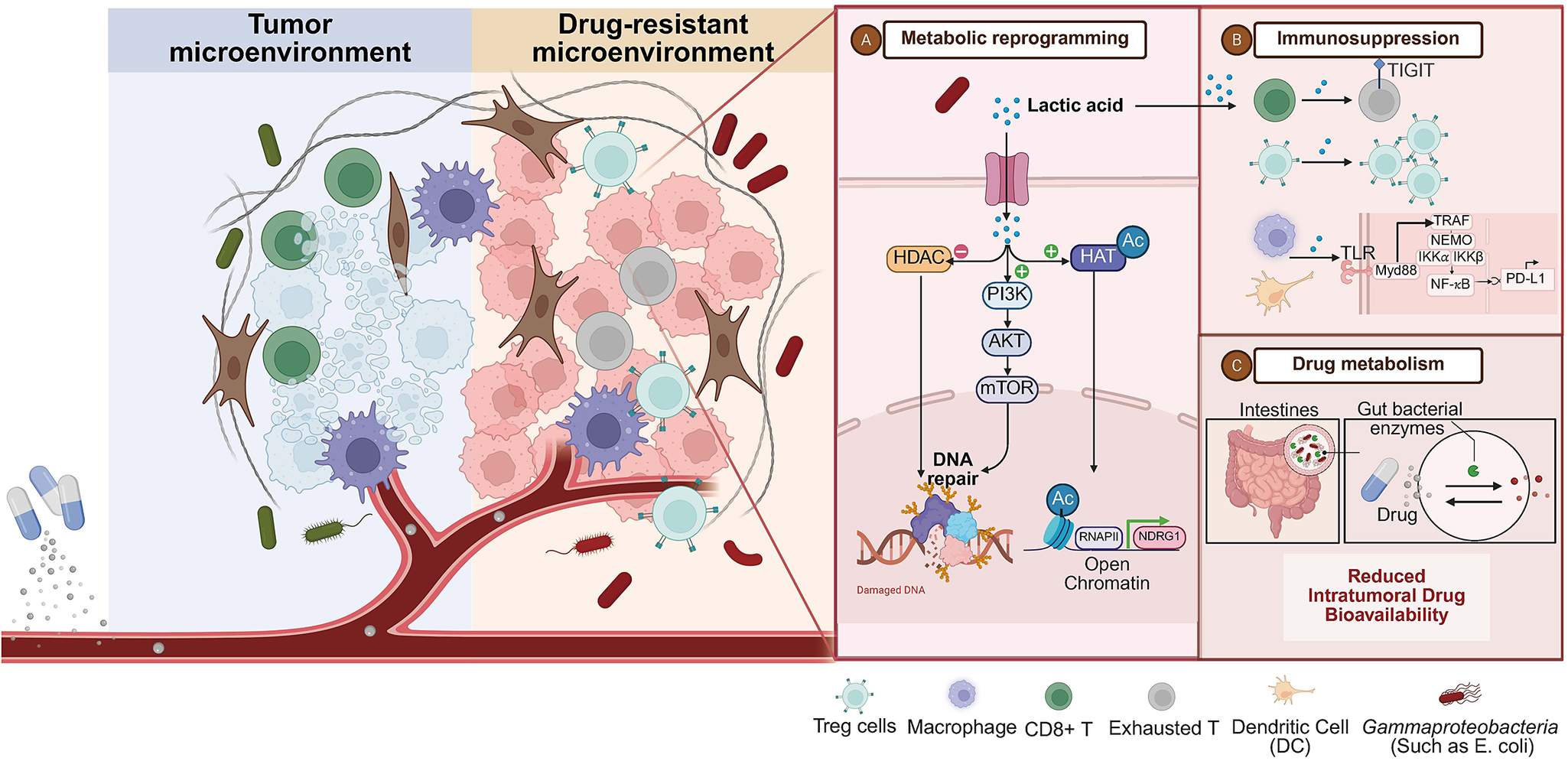
The molecular machinery underlying microbiome-mediated resistance centres on lactate as a master metabolite that reprograms tumour cells, remodels the stromal microenvironment, and suppresses anti-tumour immunity (Figure 2). Studies have shown that Lactobacillus colonized in tumours can secrete a large amount of l-lactic acid in the tumour microenvironment, and induce metabolic reprogramming of tumour cells through mechanisms such as enhancing glycolysis, tricarboxylic acid cycle activity and REDOX balance. And it activates the HIF-1α signalling pathway, p53/ P73-dependent apoptotic pathway, FGFR signal transduction, etc., accelerating the repair of DNA double-strand breaks caused by radiotherapy, ultimately leading to CRT resistance. It is worth noting that lactic acid is not only a metabolic product but also participates in the metabolic coupling between tumour cells as a ‘metabolic fuel’, further enhancing treatment resistance (Refs Reference Colbert, El Alam, Wang, Karpinets, Lo and Lynn11, Reference Wu, Wang, Ying, Jin, Li and Hu41, Reference Faubert, Li, Cai, Hensley, Kim, Zacharias, Yang, do, Doucette, Burguete, Li, Huet, Yuan, Wigal, Butt, Ni, Torrealba, Oliver, Lenkinski, Malloy, Wachsmann, Young, Kernstine and DeBerardinis50, Reference Liu, Chen, Liu, Liu, Cai, You, Ke, Lai, Huang, Gao, Zhao, Pelicano, Huang, McKeehan, Wu, Wang, Zhong and Wang51, Reference Kantarci, Gou and Riley52, Reference Hsu, Yeh, Tsai, Chen and Tsou53, Reference Rozenberg, Zvereva, Dalina, Blatov, Zubarev, Luppov, Bessmertnyi, Romanishin, Alsoulaiman, Kumeiko, Kagansky, Melino, Ganini and Barlev54, Reference Sedelnikova, Redon, Dickey, Nakamura, Georgakilas and Bonner55, Reference Ziech, Franco, Pappa and Panayiotidis56, Reference Bartesaghi, Graziano, Galavotti, Henriquez, Betts, Saxena, Minieri, A, Karlsson, Martins, Capasso, Nicotera, Brandner, de Laurenzi and Salomoni57). These findings collectively indicate that the metabolites of the microbiome influence the treatment response to cervical cancer through multi-level metabolic reprogramming. It provides a new perspective for enhancing therapeutic effects by targeting the microbiota-metabolic axis.

Figure 2. The core mechanisms of microbiome-driven resistance in cervical cancer. The tumour microenvironment (TME) exhibits dysbiosis with L. iners enrichment (red) and L. crispatus depletion (fading green). L. iners-derived lactate drives resistance via: (1) Metabolic-epigenetic axis: Lactate influx activates HCA1-PI3K/AKT signalling, enhancing DNA repair and promoting histone hyperacetylation/lactylation. (2) Immune suppression: Lactate inhibits CD8⁺ T cells and expands Tregs; L. iners lipoteichoic acid activates TLR2/NF-κB to upregulate PD-L1. (3) Direct drug metabolism: Tumour-resident Gammaproteobacteria express β-glucuronidase (GUS), inactivating chemotherapy via enzymatic modification.
At the cellular level, lactate uptake via monocarboxylate transporter 1 induces a pseudohypoxic state through stabilization of HIF-1α, even under normoxic conditions. In cervical cancer cell lines, 15 mM lactate increases HIF-1α protein half-life from 8 to 35 min (p = 0.002), leading to transcriptional upregulation of glycolytic enzymes and downregulation of oxidative phosphorylation genes (Ref. Reference Lu, Jin, Tang, Zhou, Xu, Su, Wang, Xu, Zhao, Yin, Zhang, Jia, Peng, Zhou, Wang, Chen, Wang, Yang, Chen and Chen24). Metabolic flux analysis using [13C₆]-glucose reveals that lactate-exposed cells redirect 62% of glucose carbon towards ribose synthesis, facilitating DNA repair and cell cycle progression post-irradiation (Ref. Reference Bartesaghi, Graziano, Galavotti, Henriquez, Betts, Saxena, Minieri, A, Karlsson, Martins, Capasso, Nicotera, Brandner, de Laurenzi and Salomoni57). This metabolic shift establishes a bioenergetic state that confers survival advantage during CRT. Additionally, Lactate-mediated cancer-associated fibroblast (CAF) transformation represents a critical resistance mechanism. Cervical CAFs exposed to 15 mM lactate upregulate α-SMA expression by 3.2-fold and secrete elevated levels of HGF, FGF2 and TGF-β (Ref. Reference Lin, Zhou, Liu, Nie, Cao, Li, Li, Zhu, Lin, Ding, Jiang, Gu, Xu, Zhao and Cai6). This creates a fibrotic, immunosuppressive extracellular matrix that physically impedes drug penetration and reduces oxygen diffusion, increasing hypoxia-induced radioresistance (Ref. Reference Gopalakrishnan, Spencer, Nezi, Reuben, Andrews, Karpinets, Prieto, Vicente, Hoffman, Wei, Cogdill, Zhao, Hudgens, Hutchinson, Manzo, Petaccia de Macedo, Cotechini, Kumar, Chen, Reddy, Szczepaniak Sloane, Galloway-Pena, Jiang, Chen, Shpall, Rezvani, Alousi, Chemaly, Shelburne, Vence, Okhuysen, Jensen, Swennes, McAllister, Marcelo Riquelme Sanchez, Zhang, le Chatelier, Zitvogel, Pons, Austin-Breneman, Haydu, Burton, Gardner, Sirmans, Hu, Lazar, Tsujikawa, Diab, Tawbi, Glitza, Hwu, Patel, Woodman, Amaria, Davies, Gershenwald, Hwu, Lee, Zhang, Coussens, Cooper, Futreal, Daniel, Ajami, Petrosino, Tetzlaff, Sharma, Allison, Jenq and Wargo58).
Epigenetic modulation represents another layer of lactate-mediated resistance. Lactate functions as an endogenous histone deacetylase inhibitor (Ki = 4.2 mM), competing with acetyl-lysine substrates and promoting global histone H4 hyperacetylation that correlates with intratumoural lactate levels (r = 0.68, p < 0.001) (Refs Reference Yu, Ni, Xu, Liu, Chen, Li, Xia, Diao, Chen, Zhu, Wu, Tang, Li and Ke5, Reference Ciszewski, Sobierajska, Stasiak and Wagner21). This hyperacetylation accelerates recruitment of DNA repair proteins including 53BP1 and RAD51, reducing radiation-induced γH2AX foci persistence by 45% at 24 h (p = 0.003) (Ref. Reference Sedelnikova, Redon, Dickey, Nakamura, Georgakilas and Bonner55). Lactate-derived histone lactylation, a novel post-translational modification, directly activates NDRG1 transcription, a known radioresistance driver (Ref. Reference Zhang, Fu, Leiliang, Qu, Wu, Wen, Huang, He, Cheng, Liu and Cheng59). These epigenetic changes create a durable resistance phenotype that persists beyond the initial microbial stimulus.
Microbial drug metabolism
Microbial expression of drug-metabolizing enzymes directly alters chemotherapeutic pharmacokinetics within the tumour microenvironment, which affects the bioavailability and toxicity of chemotherapy drugs. In patients with cervical cancer, the correlation between tumour and gut microbiome characteristics and the response to radiotherapy and chemotherapy has been observed. The gut microbiota can produce GUS, thereby affecting drug metabolism. Taking irinotegan as an example, its active metabolite SN-38 is detoxified by hepatic dinucleotide phosphoglucoside transferase into SN-38G and excreted into the intestine. However, GUS can re-dissociate SN-38G into the toxic SN-38, causing severe diarrhoea, which is dose-limiting toxicity (Refs Reference Nagar and Blanchard22, Reference Cheng, Tseng, Tzeng, Leu, Cheng, Wang, Chang, Lu, Cheng, Chen, Cheng, Chen and Cheng60, Reference Chen, Xu, Piao, Chang, Liu and Kong61, Reference Lin, Chen, Lin, Yeh, Hsieh, Gao, Burnouf, Chen, Hsieh, Dashnyam, Kuo, Tu, Roffler and Lin62, Reference Ting, Lau and Yu63). While GUS activity primarily impacts irinotecan toxicity, cervical cancer biopsies with high enzyme activity show reduced cisplatin-DNA adduct formation (r = −0.52, p = 0.008) and decreased apoptotic index (caspase-3+ cells: 12% versus 28% in low-activity tumours, p = 0.004) (Ref. Reference Nagar and Blanchard22). Metagenomic analysis identifies E. coli strains harbouring the gus operon in 34% of treatment-resistant tumours versus 9% of sensitive tumours (p < 0.001) (Ref. Reference Cheng, Tseng, Tzeng, Leu, Cheng, Wang, Chang, Lu, Cheng, Chen, Cheng, Chen and Cheng60). Co-administration of the specific GUS inhibitor TCH-3562 (10 mg/kg) restores cisplatin sensitivity in patient-derived xenografts, reducing tumour volume by 58% versus 23% with cisplatin alone (p = 0.009) (Ref. Reference Chen, Xu, Piao, Chang, Liu and Kong61). Tumour-enriched Gammaproteobacteria similarly express cytidine deaminase that metabolizes topotecan, reducing its cytotoxicity in vitro (IC₅₀ increase from 2.1 to 8.7 μM, p < 0.001), an effect reversible by tetrahydro-uridine (Ref. Reference Geller, Barzily-Rokni, Danino, Jonas, Shental, Nejman, Gavert, Zwang, Cooper, Shee, Thaiss, Reuben, Livny, Avraham, Frederick, Ligorio, Chatman, Johnston, Mosher, Brandis, Fuks, Gurbatri, Gopalakrishnan, Kim, Hurd, Katz, Fleming, Maitra, Smith, Skalak, Bu, Michaud, Trauger, Barshack, Golan, Sandbank, Flaherty, Mandinova, Garrett, Thayer, Ferrone, Huttenhower, Bhatia, Gevers, Wargo, Golub and Straussman23). Studies have shown that the microbial drug metabolism enzyme system is of universal importance in tumour treatment resistance (Ref. Reference Song, Yang, Ma, Wang and Leung64). These enzyme systems may alter the metabolokinetics of drugs under specific tumour microenvironmental conditions through spatiotemporal specific activation patterns.
Surgical outcomes and the microbiome
Studies have found that microbiome dysbiosis can affect postoperative recovery and treatment outcomes through multiple mechanisms. Emerging data from colorectal surgery suggests microbiome composition critically influences postoperative outcomes, with direct implications for cervical cancer surgery. In colorectal cancer, faecal microbiota transplantation (FMT) from anastomotic leak patients impairs wound healing in germ-free mice, associated with increased matrix metalloproteinase-9 activity and reduced collagen deposition (Ref. Reference Hajjar, Gonzalez, Fragoso, Oliero, Alaoui, Calvé, Vennin Rendos, Djediai, Cuisiniere, Laplante, Gerkins, Ajayi, Diop, Taleb, Thérien, Schampaert, Alratrout, Dagbert, Loungnarath, Sebajang, Schwenter, Wassef, Ratelle, Debroux, Cailhier, Routy, Annabi, Brereton, Richard and Santos65). Alistipes onderdonkii directly degrades extracellular matrix components, increasing leak risk (RR = 3.1, p = 0.01) (Refs Reference Hajjar, Gonzalez, Fragoso, Oliero, Alaoui, Calvé, Vennin Rendos, Djediai, Cuisiniere, Laplante, Gerkins, Ajayi, Diop, Taleb, Thérien, Schampaert, Alratrout, Dagbert, Loungnarath, Sebajang, Schwenter, Wassef, Ratelle, Debroux, Cailhier, Routy, Annabi, Brereton, Richard and Santos65, Reference Li, Zhang, Fu, Jiang, Zhang, Zhang, Chen, Tao, Chen and Zeng66). Translating to cervical cancer, radical hysterectomy patients with preoperative vaginal dysbiosis (Nugent score > 7) demonstrate increased postoperative cuff cellulitis (18% versus 6%, p = 0.02) and delayed wound healing (median 21 versus 14 days, p = 0.004) (Ref. Reference Ohigashi, Sudo, Kobayashi, Takahashi, Nomoto and Onodera67). These findings are not only applicable to colorectal cancer surgery but also provide important implications for the surgical treatment of cervical cancer. Microbial environment imbalance may also lead to local immune environment imbalance by disrupting mucosal barrier function, promoting chronic inflammatory responses and regulating immune cell function, increasing the risk of postoperative infection and delaying wound healing (Refs Reference Hajjar, Gonzalez, Fragoso, Oliero, Alaoui, Calvé, Vennin Rendos, Djediai, Cuisiniere, Laplante, Gerkins, Ajayi, Diop, Taleb, Thérien, Schampaert, Alratrout, Dagbert, Loungnarath, Sebajang, Schwenter, Wassef, Ratelle, Debroux, Cailhier, Routy, Annabi, Brereton, Richard and Santos65, Reference Li, Zhang, Fu, Jiang, Zhang, Zhang, Chen, Tao, Chen and Zeng66, Reference Ohigashi, Sudo, Kobayashi, Takahashi, Nomoto and Onodera67). Perioperative microbiome regulation for high-risk patients may become a new strategy to improve surgical outcomes.
Surgical site infections are similarly microbiome-dependent. Preoperative vaginal Staphylococcus aureus colonization increases post-hysterectomy infection risk 4.7-fold, with antibiotic prophylaxis efficacy reduced in the presence of biofilm-forming Enterococcus faecalis (Ref. Reference Di Modica, Gargari, Regondi, Bonizzi, Arioli and Belmonte68). Metagenomic analysis of infected surgical sites reveals enrichment of bacterial genes encoding antibiotic resistance and virulence factors, likely transferred from the vaginal reservoir (Ref. Reference Chen, Wei, Zhao, Zhou, Wang, Zhang, Zuo, Dong, Zhao, Hao, He and Bian69). Broad-spectrum prophylaxis reduces surgical site infections but causes long-term vaginal dysbiosis, with L. crispatus recovery taking >6 months in 68% of patients (Ref. Reference Zhang, Fu, Leiliang, Qu, Wu, Wen, Huang, He, Cheng, Liu and Cheng59). A pilot study implementing preoperative vaginal probiotic supplementation (L. crispatus suppositories, 109 CFU twice daily for 7 days) reduced postoperative infections from 14 to 4% (p = 0.03) and accelerated microbiome normalization, suggesting perioperative microbiome optimization could improve both infectious and oncologic outcomes (Ref. Reference Mitra, MacIntyre, Paraskevaidi, Moscicki, Mahajan, Smith, Lee, Lyons, Paraskevaidis, Marchesi, Bennett and Kyrgiou19).
Radiotherapy outcomes and microbiome
Pelvic intensity-modulated radiation therapy combined with cisplatin-based chemotherapy represents the standard of care for locally advanced cervical cancer; however, radioresistance remains a major constraint on therapeutic efficacy. Longitudinal 16S rRNA sequencing analyses reveal significant dynamic shifts in the microbial community structure of both the vagina and local tumour sites during radiotherapy. Notably, the relative abundance of taxa such as Gammaproteobacteria and Acinetobacter increases progressively over the course of treatment (Ref. Reference Jiang, Li, Zhang, Ma, Liu, Liang, Wei, Huang and Wang45). Concurrently, reduced microbial diversity within cervical tumours is observed, with specific taxa like Bifidobacteriaceae showing a positive correlation with radiosensitivity. A higher abundance of these beneficial bacteria is found in patients exhibiting favourable responses to radiotherapy, suggesting their potential utility as predictive biomarkers for treatment outcome (Ref. Reference Dou, Ai, Zhang, Li, Jiang, Wu, Zhao, Li and Zhang34).
These microbial dynamics are closely linked to the modulation of the local immune microenvironment. A proposed mechanistic hypothesis suggests that intratumoural microbiota influence radiosensitivity by regulating immune cell infiltration, inflammatory cytokine production and the DNA damage repair capacity of tumour cells. For instance, certain bacterial species may enhance radiation-induced apoptosis by activating TLR/NF-κB signalling pathways, thereby augmenting tumour response to radiotherapy (Ref. Reference Zhou, Li, Ren, Wang and Wang70). Furthermore, microbial metabolites, including SCFAs, are implicated in modulating tumour cell responses to radiation by influencing metabolic reprogramming and immune regulation (Ref. Reference Miya, Marima, Damane, Ledet and Dlamini71).
Critically, the composition of the tumour-associated microbiome not only affects immediate radiotherapy response but is also associated with patient prognosis and recurrence risk. Overall, high diversity in both the gut and tumour microbiome is generally correlated with better treatment response and improved survival rates. Conversely, an increased abundance of specific pathobionts, such as Fusobacterium, may promote therapeutic resistance and tumour recurrence (Refs Reference Sims, El Alam, Karpinets, Dorta-Estremera, Hegde and Nookala43, Reference Tortelli, Contreras, Markovina, Ding, Wylie and Schwarz44).
In summary, microbial shifts are not merely epiphenomena of radiotherapy but likely function as active modulators of treatment efficacy. Future research should prioritize elucidating the precise molecular mechanisms orchestrated by key microbial species and their metabolites. This knowledge is essential for developing microbiome-based predictive models and targeted interventions aimed at enhancing the efficacy of CRT and improving patient outcomes in cervical cancer.
Therapeutic interventions: evidence, controversies and translational reality
Microbiome-based interventions occupy a precarious space between mechanistic promise and clinical uncertainty. While preclinical models demonstrate profound effects of microbial modulation on treatment efficacy, human evidence remains fragmented, underpowered and frequently contradictory.
The L. iners paradox: a Janus-faced commensal
L. iners, as an important strain in the vaginal microecology, shows a complex duality in its role in cervical cancer and related diseases. Studies have shown that it has a certain protective effect in maintaining the balance of vaginal microecology. Under vaginal health conditions, L. iners are common and abundant. They can regulate the pH of the local environment by producing metabolic products such as lactic acid, and inhibit the colonization and growth of pathogenic bacteria. L. iners exemplifies the challenge of binary classification, as this species contributes to ecological stability through hydrogen peroxide production and NOD2-dependent antimicrobial peptide secretion in homeostatic environments (Refs Reference Zhu, Frank, Radka, Jeanfavre, Xu, Tse, Pacheco, Kim, Pierce, Deik, Hussain, Elsherbini, Hussain, Xulu, Khan, Pillay, Mitchell, Dong, Ndung’u, Clish, Rock, Blainey, Bloom and Kwon26, Reference Armstrong and Kaul37). In HPV-infected women, L. iners co-dominance correlates with increased viral clearance (OR = 1.73, 95% CI 1.12–2.68) (Ref. Reference Fan, Wu, Li, An, Yao, Wang, Wang, Yuan, Jiang, Li and Li72). However, in oncogenic contexts, the species transforms into a pathobiont, with cancer-derived strains showing 3.5-fold higher lactate production and upregulated cytolysin expression that disrupts epithelial barriers (Ref. Reference Colbert, El Alam, Wang, Karpinets, Lo and Lynn11). Studies have found through 16S sequencing analysis that the abundance of L. iners in the vaginal microbiota of patients with recurrent cervical cancer is significantly increased. As the most important microbial marker, it can be used to predict the risk of cancer recurrence before chemotherapy and radiotherapy (Ref. Reference Wang, Wang, Yan, Zhao, Wang, Liu, Fan and Xu36). This context-dependent pathogenicity precludes universal recommendations and demands personalized risk stratification integrating immune status and microbial load.
L. crispatus probiotics: bridging the efficacy gap
The microbiota structure mainly composed of L. crispatus in the vaginal microbiota is closely related to the cervical health status. Compared with the state of vaginal microbiota imbalance, the vaginal environment dominated by L. crispatus exhibits stronger immunomodulatory characteristics and can maintain a more favorable reproductive tract health state (Ref. Reference Armstrong and Kaul37). L. crispatus probiotics represent the most intuitive intervention, yet clinical validation remains incomplete. Preclinical data show that L. crispatus supernatant reduces L. iners-mediated lactate production by 45% through competitive glucose consumption and enhances epithelial barrier integrity via Wnt signalling activation (Refs Reference Dou, Ai, Zhang, Li, Jiang, Wu, Zhao, Li and Zhang34, Reference Fan, Wu, Li, An, Yao, Wang, Wang, Yuan, Jiang, Li and Li72). A phase I trial in 45 patients with CIN3 demonstrated achievable colonization in 73% of participants receiving vaginal suppositories (109 CFU twice daily), with a non-significant trend towards reduced treatment-related adverse events (Grade 3+ toxicity: 31% versus 44% in historical controls, p = 0.19) (Ref. Reference Zeng, An, Li and Zhang73). The 2-year recurrence rate of 18% versus 29% in matched controls failed to reach statistical significance (p = 0.09), reflecting inadequate statistical power (Ref. Reference Zeng, An, Li and Zhang73). Dose-escalation studies reveal a narrow therapeutic window, with 109 CFU superior to 107 CFU for colonization (68% versus 28%), but 1010 CFU provoking inflammatory cytokines without added benefit (Refs Reference Zeng, An, Li and Zhang73, Reference Wang, Wang, Yu, Zhang, Hu, Xu, Zhang, Tian, Zheng, Lu, Hu, Guo, Cai, Geng, Zhang, Xia, Zhang, Li, Liu and Zhang74). Probiotics can enhance anti-cancer immune function (Refs Reference Gopalakrishnan, Spencer, Nezi, Reuben, Andrews, Karpinets, Prieto, Vicente, Hoffman, Wei, Cogdill, Zhao, Hudgens, Hutchinson, Manzo, Petaccia de Macedo, Cotechini, Kumar, Chen, Reddy, Szczepaniak Sloane, Galloway-Pena, Jiang, Chen, Shpall, Rezvani, Alousi, Chemaly, Shelburne, Vence, Okhuysen, Jensen, Swennes, McAllister, Marcelo Riquelme Sanchez, Zhang, le Chatelier, Zitvogel, Pons, Austin-Breneman, Haydu, Burton, Gardner, Sirmans, Hu, Lazar, Tsujikawa, Diab, Tawbi, Glitza, Hwu, Patel, Woodman, Amaria, Davies, Gershenwald, Hwu, Lee, Zhang, Coussens, Cooper, Futreal, Daniel, Ajami, Petrosino, Tetzlaff, Sharma, Allison, Jenq and Wargo58, Reference Ting, Lau and Yu63, Reference Matson, Fessler, Bao, Chongsuwat, Zha, Alegre, Luke and Gajewski75) by reducing Treg levels, promoting the activation of CD8 + T cells, the differentiation of CD4 + T cells and the infiltration of NK cells within tumours. L. crispatus supplementation can restore vaginal microecology, enhance local immunity, and improve the response to radiotherapy and immunotherapy. Applying Grading of Recommendations, Assessment, Development and Evaluation (GRADE) criteria, the certainty of evidence for L. crispatus probiotic efficacy is low, derived from small pilot studies with indirect endpoints, warranting only conditional recommendations within clinical trial settings.
Faecal microbiota transplantation: compartmental failure and safety risks
FMT has shown potential value in regulating the response to tumour treatment. Animal experiments have confirmed that FMT treated with antibiotics or from donors receiving antibiotic treatment can significantly weaken the anti-tumour activity of trastuzumab, manifested as impaired recruitment of CD4 T cells and granzyme B positive cells within the tumour (Ref. Reference Di Modica, Gargari, Regondi, Bonizzi, Arioli and Belmonte68). In the stress-induced colorectal cancer model, both FMT and antibiotic treatment can eliminate the stimulating effect of chronic stress on tumour progression, and the abundance of Lactobacillus johnsonii in stressed mice is significantly reduced (Ref. Reference Cao, Zhao, Su, Liu, Lin and Da76). Studies on liver injury induced by PFOS have shown that FMT can verify the causal role of the gut microbiota in mediating liver injury. After transplantation, the recipient mice exhibited similar characteristics of microbiota imbalance and amino acid metabolism disorders to the donors (Ref. Reference Song, Yang, Ma, Wang and Leung64). In terms of metabolic improvement, FMT from bariatric surgery mice can significantly improve the metabolic parameters of recipient mice, but this effect is strain-specific (Ref. Reference Liu, Tu, Shi, Fang, Fan, Zhang, Ding, Chen, Wang, Zhang, Xu, Sharma, Gillece, Reining, Jin and Huang77). FMT enhances the response rate of radiotherapy, chemotherapy and immunotherapy for cervical cancer by reshaping the intestinal/tumour microbiota, regulating the function and metabolic pathways of immune cells, and provides new ideas for overcoming drug resistance.
However, FMT faces unique barriers in cervical cancer due to anatomic compartmentalization and safety concerns. A pilot study administering oral FMT capsules to 12 platinum-resistant patients increased gut microbiome diversity but showed only transient vaginal L. crispatus elevation in 3 patients, with no objective tumour responses and one serious adverse event of donor-derived Enterococcus faecium bacteraemia (Ref. Reference Di Modica, Gargari, Regondi, Bonizzi, Arioli and Belmonte68). The mechanistic disconnect between gut and vaginal compartments, with faecal-vaginal microbial transmission efficiency below 0.1%, undermines FMT rationale (Ref. Reference Chen, Wei, Zhao, Zhou, Wang, Zhang, Zuo, Dong, Zhao, Hao, He and Bian69). Direct vaginal FMT carries unacceptable transmission risks for HPV and HIV, with no safety data in immunocompromised patients (Ref. Reference Khoruts and Sadowsky78). Current evidence supports conditional use of FMT only within clinical trials employing rigorous donor screening and direct vaginal application protocols.
Antibiotic stewardship: balancing infection control with microbiome preservation
Antibiotic stewardship presents a double-edged sword, as broad-spectrum antibiotic exposure within 30 days before CRT initiation reduces progression-free survival (pooled HR = 1.45, 95% CI 1.18–1.78) and overall survival (HR = 1.62, 95% CI 1.29–2.03) by depleting butyrate-producing taxa and impairing CD8+ T-cell function (Refs Reference Elkrief, Pidgeon, Maleki Vareki, Messaoudene, Castagner and Routy12, Reference Zhou, Li, Ren, Wang and Wang70). Conversely, targeted pathobiont eradication – such as metronidazole depletion of Fusobacterium – enhances radiation efficacy in preclinical models without compromising systemic immunity (Ref. Reference Gopalakrishnan, Spencer, Nezi, Reuben, Andrews, Karpinets, Prieto, Vicente, Hoffman, Wei, Cogdill, Zhao, Hudgens, Hutchinson, Manzo, Petaccia de Macedo, Cotechini, Kumar, Chen, Reddy, Szczepaniak Sloane, Galloway-Pena, Jiang, Chen, Shpall, Rezvani, Alousi, Chemaly, Shelburne, Vence, Okhuysen, Jensen, Swennes, McAllister, Marcelo Riquelme Sanchez, Zhang, le Chatelier, Zitvogel, Pons, Austin-Breneman, Haydu, Burton, Gardner, Sirmans, Hu, Lazar, Tsujikawa, Diab, Tawbi, Glitza, Hwu, Patel, Woodman, Amaria, Davies, Gershenwald, Hwu, Lee, Zhang, Coussens, Cooper, Futreal, Daniel, Ajami, Petrosino, Tetzlaff, Sharma, Allison, Jenq and Wargo58). Similarly, bacteriophage therapy targeting L. iners reduces tumour lactate by 55% and restores cisplatin sensitivity (Ref. Reference Zheng, Hu, Liu, Zhao, Li, Wang, Zhang, Zhang, Song, Lyu, Cui, Ding and Wang38). These findings support a nuanced approach avoiding empiric broad-spectrum antibiotics while reserving targeted eradication for pathobiont enrichment documented by microbiome sequencing.
Engineered bacteria: translational promise meets clinical reality
With the deepening understanding of the tumour microbiome (TM), the utilization of microorganisms themselves as a strategy for tumour treatment has shown great potential, among which engineered bacteria therapy is a cutting-edge direction in tumour microbiological therapy. Engineered bacteria refer to non-pathogenic bacteria that have been genetically modified and possess specific anti-tumour functions. These non-toxic and highly selective engineered microorganisms can enhance the efficacy of radiotherapy and chemotherapy. Such bacteria as Bifidobacterium, Salmonella and Clostridium can be modified to target the hypoxic and necrotic regions specific to solid tumours (Refs Reference Sims, Yoshida-Court, El Alam, Ramatlho, Ketlametswe and Ning28, Reference Tosado-Rodríguez, Mendez, Espino, Dorta-Estremera, Aquino, Romaguera and Godoy-Vitorino30, Reference Tosado-Rodríguez, Alvarado-Vélez, Romaguera and Godoy-Vitorino48). Colonization of engineered bacteria can increase the number of CD4+ and CD8+ T cells in tumours, reduce the proportion of FOXP3+ regulatory T cells, enhance the effector function of T cells (such as TNF production) and reduce the co-expression of PD-1/LAG-3. Meanwhile, the combination of engineered bacteria with PD-1/PD-L1 monoclonal antibodies and others can improve the penetration of drugs in deep tumours and overcome drug resistance, demonstrating a synergistic effect (Refs Reference Sims, Biegert, Ramogola-Masire, Ngoni, Solley, Ning, el Alam, Mezzari, Petrosino, Zetola, Schmeler, Colbert, Klopp and Grover29, Reference Nieves-Ramírez, Partida-Rodríguez, Moran, Serrano-Vázquez, Pérez-Juárez, Pérez-Rodríguez, Arrieta, Ximénez-García and Finlay49). The above findings indicate that engineered bacteria therapy can regulate the metabolism of the tumour microenvironment, thereby enhancing the efficacy of immunotherapy. E. coli Nissle 1917 engineered to express lactate oxidase reduces intratumoural lactate by 70% and enhances radiation sensitivity, while L. crispatus engineered with cytosine deaminase achieves 20-fold higher local 5-FU concentration (Refs Reference Lim, Yin, Ye, Cui, Papachristodoulou and Huang79, Reference Brevi and Zarrinpar80).
Even engineered bacteria offer theoretically precise interventions but remain confined to preclinical development. Firstly, ensuring the genetic stability and controllable reproduction of engineered bacteria in vivo is the key to preventing them from developing drug resistance or causing infections (Refs Reference Lim, Yin, Ye, Cui, Papachristodoulou and Huang79, Reference Xie, Fan, Cheng, Yin, Li, Wegner, Chen and Zeng81). Secondly, clinical transformation is restricted by multiple aspects such as safety, immune rejection reactions, and regulatory supervision. A large number of basic and clinical studies are still needed to verify its efficacy and safety (Refs Reference Brevi and Zarrinpar80, Reference Sharma, Sharma and Gogoi82). In addition, different tumour types and individual differences among patients also have a significant impact on the therapeutic effect of engineered bacteria. It is necessary to precisely screen suitable strains and design individualized treatment plans (Refs Reference Deb, Wu, Coker, Harimoto, Huang and Danino83, Reference Radford, Vrbanac, De Nys, Worthley, Wright and Hasty84). So, no engineered bacteria have entered cervical cancer trials, with genetic instability, immune clearance and regulatory hurdles posing formidable barriers (Refs Reference Xue, Wang and Liu85, Reference Chang, Liu, Wang, Ma, Liang and Li86). Technology readiness levels remain at 3–4, restricting recommendation to research settings only.
Future research framework and clinical translation roadmap
The trajectory from mechanistic insight to clinical implementation demands a structured, milestone-driven approach that acknowledges current evidence gaps while prioritizing actionable research questions. A phased roadmap spanning basic discovery to regulatory approval is essential to avoid premature clinical adoption that could jeopardize patient safety and scientific credibility.
Multi-omics integration and predictive model development
Current research has confirmed that the tumour microbiome can influence the signalling, metabolism and proliferation processes of cancer cells by generating active metabolites (Refs Reference Song, Yang, Ma, Wang and Leung64, Reference Fernandez, Wargo and Helmink87). The immediate research priority involves constructing integrated multi-omics frameworks that transcend single-layer microbiome characterization. Current studies demonstrate that tumour microbiome features alone explain approximately 30–40% of variance in CRT response, indicating that microbial signatures must be contextualized within host genetics, immune status and metabolic networks to achieve clinically actionable predictive power (Refs Reference Dai, Tan, Qiao and Liu7, Reference Wahid, Dar, Jawed, Mandal, Akhter, Khan, Khan, Jogaiah, Rai and Rattan10). Machine learning algorithms capable of integrating vaginal microbiome sequencing data, metabolomic profiling of lactate and short-chain fatty acids and immune checkpoint expression (PD-L1, MAdCAM-1) represent a critical near-term objective (Refs Reference Liu, Chen, Zhang and Dong13, Reference Boesch, Horvath, Baty, Pircher, Wolf, Spahn, Straussman, Tilg and Brutsche88).
Prospective validation of such models requires standardized data acquisition protocols across geographically diverse cohorts. The cervical cancer microbiome consortium, comprising institutions from sub-Saharan Africa, Latin America and Asia-Pacific, has initiated harmonized sampling procedures using unified collection devices, DNA extraction kits, and bioinformatic pipelines to reduce technical heterogeneity that currently confounds meta-analyses (Ref. Reference Mitra, MacIntyre, Paraskevaidi, Moscicki, Mahajan, Smith, Lee, Lyons, Paraskevaidis, Marchesi, Bennett and Kyrgiou19). This effort must incorporate host genotyping for HLA-DRB1 and other alleles that modulate microbiome resilience, as emerging data indicate host genetic factors contribute significantly to microbial community stability during therapy. Importantly, these predictive models must be trained on hard clinical endpoints – 3-year progression-free survival rather than surrogate markers like microbial diversity indices – to ensure clinical relevance.
Synthetic biology: engineering next-generation microbiome therapeutics
Synthetic biology offers theoretically precise interventions, but current designs remain confined to preclinical development requiring substantial safety engineering before human application. The most promising near-term application involves developing ‘smart probiotics’ with enhanced colonization capacity and controlled metabolite production. Chromosomal integration of lactate oxidase or butyrate synthesis operons into L. crispatus chassis strains could circumvent plasmid instability issues that cause 15–20% gene loss per generation in current designs (Ref. Reference Lim, Yin, Ye, Cui, Papachristodoulou and Huang79). However, such engineering must be coupled with robust biocontainment strategies – auxotrophic dependencies that restrict growth to the tumour microenvironment (e.g., requirement for exogenous d-alanine) have proven effective in colorectal cancer models but remain untested in vaginal applications (Ref. Reference Brevi and Zarrinpar80).
Engineered bacteriophage therapy represents a parallel strategy with potentially superior safety profiles. Lytic phages specific for L. iners (e.g., φLinq1) can selectively deplete pathogenic populations without disrupting beneficial commensals, reducing tumour lactate by 55% in preclinical xenografts (Ref. Reference Zhang, Fu, Leiliang, Qu, Wu, Wen, Huang, He, Cheng, Liu and Cheng59). Unlike bacterial therapies, phages are self-limiting and do not persist after target elimination, minimizing ecological disruption. However, phage development is hindered by narrow host ranges and rapid bacterial resistance evolution, necessitating cocktail formulations that increase manufacturing complexity. Regulatory pathways for phage products remain undefined, with current FDA guidance requiring demonstration of purity, consistency and absence of endotoxin contamination that many academic laboratories cannot achieve (Ref. Reference Lee, McClure, Weichselbaum and Mimee89).
Clinical trial standardization and adaptive design
The heterogeneity across current microbiome intervention trials – encompassing disparate patient populations, dosing regimens and outcome measures – precludes meaningful synthesis and delays progress. Establishing an international master protocol for adaptive platform trials is imperative. Such trials should stratify patients by baseline microbiome signatures (L. iners-dominant versus L. crispatus-dominant) and randomize to microbiome modulation arms (probiotic, phage, or engineered bacteria) versus standard-of-care (Ref. Reference Fernandez, Wargo and Helmink87). Primary endpoints must include progression-free survival at 2 years, with mandatory serial microbiome and metabolome sampling to capture dynamic changes. Exploratory endpoints should integrate patient-reported quality-of-life metrics, as vaginal dysbiosis correlates with dyspareunia and treatment-related discomfort that traditional oncologic outcomes overlook (Ref. Reference Liao, Chen, Ruan, Wang, Hu, Long, Li, Zhang, Yu, Ming zhang, Zhang and Liao9).
Standardization extends beyond trial design to sample processing and data analysis. The International Cancer Microbiome Consortium has proposed guidelines requiring parallel 16S rRNA and shotgun metagenomic sequencing, with mandatory reporting of absolute bacterial load via spike-in controls rather than relative abundance alone (Ref. Reference Bhattarai, Du, Zeamer, Morzfeld, Kellogg and Firat90). Bioinformatic pipelines must be benchmarked against synthetic mock communities to ensure inter-laboratory comparability. These technical standards should be enforced by regulatory agencies as prerequisites for Investigational New Drug applications for microbiome-based therapies.
Critical controversies and cautionary principles
Several foundational controversies must be addressed to prevent premature translation that could harm patients and discredit the field. First, the causality question remains incompletely resolved. While PDX models and antibiotic depletion studies support causal roles for specific taxa, these experiments cannot fully exclude host factors that may independently drive both dysbiosis and resistance. Mendelian randomization studies using host genetic variants that influence microbiome composition may help disentangle causation from confounding, but such analyses require massive sample sizes currently unavailable (Ref. Reference Javdan, Lopez, Chankhamjon, Lee, Hull, Wu, Wang, Chatterjee and Donia91).
Second, ecological complexity warnings against oversimplified ‘good bacteria/bad bacteria’ frameworks. The L. iners/L. crispatus ratio, while predictive, captures only a fraction of microbial interactions. Keystone species like A. vaginae may exert disproportionate effects through cross-feeding networks, and elimination of one pathobiont may enable replacement by another equally deleterious species (Ref. Reference Cao, Zhao, Su, Liu, Lin and Da76). This ecological unpredictability necessitates whole-community intervention approaches rather than single-species targeting.
Third, cost-effectiveness and equity considerations loom large. Microbiome sequencing and targeted probiotics add approximately $800–1,200 to treatment costs per patient, a significant burden in low-resource settings where cervical cancer mortality is highest (Ref. Reference Deb, Wu, Coker, Harimoto, Huang and Danino83). Without demonstrated cost-effectiveness compared to standard supportive care, such interventions risk exacerbating health disparities. Implementation research must prioritize affordable diagnostics, such as point-of-care lactate assays, and locally produced probiotics to ensure global applicability. To reduce costs, alternative diagnostic methods like 16S rRNA sequencing could be used to identify key microbial markers at a lower cost. Additionally, partnerships with local manufacturers could enable the production of cost-effective probiotics, ensuring broader access in underserved populations.
Finally, regulatory science lags behind basic discovery. The FDA and EMA have yet to establish quality metrics for ‘live biotherapeutic products’ specific to vaginal applications, including acceptable limits on gene transfer, environmental persistence and immunogenicity (Ref. Reference Lee, McClure, Weichselbaum and Mimee89). Without clear regulatory pathways, academic discoveries will stall in translation, and patients may turn to unproven commercial probiotics with unsubstantiated claims. Urgent dialogue between researchers, regulators and patient advocacy groups is needed to create fit-for-purpose approval frameworks.
In immunocompromised patients, such as those with HIV co-infection, microbiome interventions must be approached with caution due to the heightened risk of infection and immune dysregulation. These patients are more susceptible to opportunistic infections, and microbiome modulation may inadvertently introduce harmful pathogens or exacerbate imbalances in the microbiota. Additionally, interventions like FMT or probiotics could trigger unwanted immune responses. Thus, strict safety protocols, including comprehensive screening and monitoring, are essential when applying microbiome therapies to immunocompromised individuals. Preclinical studies and close clinical supervision are critical to ensuring the safety and efficacy of such interventions.
Conclusion and expert opinion
The field of microbiome research in cervical cancer therapy resistance is at a critical juncture. Robust mechanistic data have established lactate-mediated resistance as a key, druggable pathway, while longitudinal clinical studies confirm the L. iners/L. crispatus ratio as a predictive biomarker, with effect sizes comparable to well-established histopathological risk factors. However, the gap between correlation and causation, combined with the lack of definitive phase III trials for any microbiome-based intervention, necessitates cautious and methodical progression.
The most prudent course of action is to incorporate pre-treatment microbiome assessments into prospective observational trials of novel chemoradiation therapy (CRT) combinations. This approach will both validate the utility of microbiome biomarkers and generate valuable real-world evidence. Concurrently, efforts should be intensified in synthetic biology safety engineering and phage therapy development, with a focus on designs that ensure biocontainment and clear regulatory pathways.
Until robust clinical evidence emerges, clinicians should adopt a ‘do no harm’ principle: refrain from administering empiric antibiotics to CRT candidates, discourage the use of unproven probiotic products and direct patients to well-validated clinical trials. While the microbiome holds genuine promise for transforming cervical cancer outcomes, realizing this potential demands the same rigorous evidence standards that have revolutionized oncology with targeted therapies and immunotherapy. Premature adoption of microbiome-based interventions risks repeating past mistakes, where biologically promising concepts failed due to insufficient clinical validation, ultimately delaying breakthroughs for patients in urgent need. The model of interdisciplinary collaboration will be key to overcoming the current bottleneck in translating microbiome research from mechanistic insight to clinical application.
Abbreviations
- 5-mC
-
5-methylcytosine
- 16S rRNA
-
16S ribosomal RNA
- ABCB1
-
ATP-binding cassette subfamily B member 1
- α-SMA
-
alpha-smooth muscle actin
- AKT
-
protein kinase B
- AMPK
-
AMP-activated protein kinase
- APC
-
antigen-presenting cell
- ATM
-
ataxia telangiectasia mutated
- ATR
-
ataxia telangiectasia and Rad3-related protein
- AUC
-
area under curve
- BV
-
bacterial vaginosis
- CAF
-
cancer-associated fibroblast
- CCR
-
C-C motif chemokine receptor
- CD
-
cluster of differentiation
- cDNA
-
complementary DNA
- cGAS
-
cyclic GMP-AMP synthase
- CHK1
-
checkpoint kinase 1
- CI
-
confidence interval
- CMV
-
cytomegalovirus
- CIN
-
cervical intraepithelial neoplasia
- CR
-
complete response
- CRT
-
chemoradiotherapy
- CSF
-
colony-stimulating factor
- CST
-
community state type
- CTC
-
circulating tumor cell
- CtDNA
-
circulating tumor DNA
- CTL
-
cytotoxic T lymphocyte
- CyTOF
-
Cytometry Time-of-Flight
- DC
-
dendritic cell
- DCR
-
disease control rate
- DMSO
-
dimethyl sulfoxide
- DNA
-
deoxyribonucleic acid
- DSB
-
double-strand break
- ECOG
-
Eastern Cooperative Oncology Group
- EGFR
-
epidermal growth factor receptor
- EMT
-
epithelial-mesenchymal transition
- ERK
-
extracellular signal-regulated kinase
- FACS
-
fluorescence-activated cell sorting
- FBS
-
fetal bovine serum
- FDA
-
Food and Drug Administration
- FIGO
-
International Federation of Gynecology and Obstetrics
- FISH
-
fluorescence in situ hybridization
- FITC
-
fluorescein isothiocyanate
- FMT
-
fecal microbiota transplantation
- FOXP3
-
forkhead box P3
- GAPDH
-
glyceraldehyde-3-phosphate dehydrogenase
- GLUT1
-
glucose transporter 1
- GM-CSF
-
granulocyte-macrophage colony-stimulating factor
- GRADE
-
Grading of Recommendations, Assessment, Development and Evaluation
- GS
-
glycogen synthase
- HCA1
-
hydroxycarboxylic acid receptor 1
- HBV
-
hepatitis B virus
- HCV
-
hepatitis C virus
- HDAC
-
histone deacetylase
- HER2
-
human epidermal growth factor receptor 2
- HGF
-
hepatocyte growth factor
- HIF-1α
-
hypoxia-inducible factor 1-alpha
- HIV
-
human immunodeficiency virus
- HLA
-
human leukocyte antigen
- HRP
-
horseradish peroxidase
- HPV
-
human papillomavirus
- HR
-
hazard ratio
- HSC
-
hematopoietic stem cell
- HSIL
-
high-grade squamous intraepithelial lesion
- IC₅₀
-
half-maximal inhibitory concentration
- ICF
-
informed consent form
- IFN-β
-
interferon beta
- IgA
-
immunoglobulin A
- IgG
-
immunoglobulin G
- IL
-
interleukin
- IND
-
Investigational New Drug
- IRB
-
Institutional Review Board
- JAK
-
Janus kinase
- LAG-3
-
lymphocyte-activation gene 3
- LDH
-
lactate dehydrogenase
- LPS
-
lipopolysaccharide
- MAPK
-
mitogen-activated protein kinase
- MCT
-
monocarboxylate transporter
- MDSC
-
myeloid-derived suppressor cell
- MHC
-
major histocompatibility complex
- mRNA
-
messenger RNA
- mTOR
-
mechanistic target of rapamycin
- NAC
-
N-acetylcysteine
- NAD+
-
nicotinamide adenine dinucleotide
- NDRG1
-
N-myc downstream-regulated gene 1
- NF-κB
-
nuclear factor kappa-light-chain-enhancer of activated B cells
- NLRP3
-
NOD-like receptor family pyrin domain containing 3
- NOD2
-
nucleotide-binding oligomerization domain-containing protein 2
- NSUN6
-
NOP2/Sun RNA methyltransferase 6
- OR
-
odds ratio
- ORR
-
objective response rate
- OS
-
overall survival
- PARP
-
poly (ADP-ribose) polymerase
- PBS
-
phosphate-buffered saline
- PCR
-
polymerase chain reaction
- PD-1
-
programmed cell death protein 1
- PD-L1
-
programmed death-ligand 1
- PDX
-
patient-derived xenograft
- PE
-
phycoerythrin
- PEG
-
polyethylene glycol
- PFS
-
progression-free survival
- PI3K
-
phosphatidylinositol 3-kinase
- PPARα
-
peroxisome proliferator-activated receptor alpha
- RIPA
-
radioimmunoprecipitation assay buffer
- RNA
-
ribonucleic acid
- ROS
-
reactive oxygen species
- RR
-
relative risk
- RT-PCR
-
reverse transcription polymerase chain reaction
- SDS-PAGE
-
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SF₂
-
survival fraction at 2 Gy
- shRNA
-
short hairpin RNA
- siRNA
-
small interfering RNA
- STAT1
-
signal transducer and activator of transcription 1
- STING
-
stimulator of interferon genes
- TCR
-
T-cell receptor
- TCGA-CESC
-
The Cancer Genome Atlas - Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma
- TGF-β
-
transforming growth factor beta
- TIGIT
-
T cell immunoreceptor with Ig and ITIM domains
- TLR
-
toll-like receptor
- TNF
-
tumor necrosis factor
- Treg
-
regulatory T cell
- WHO
-
World Health Organization
- WT
-
wild type
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Authors contribution
X.Q.W., D.J.W. and X.Q.M. was involved in conceptualization, investigation, writing the original draft, review and editing, and visualization; B.H.D. and Y.W. was involved in conceptualization, reviewed the manuscript and project administration.
Funding statement
This work was supported by the National Key R&D Program of China (grant No. 2025YFC2708404); Fujian Provincial Natural Science Foundation of China (grant No. 2025J01199); Fujian Provincial Science and Technology Innovation Joint Fund (grant No. 2025Y9615); Project of Fujian Provincial Health Commission (No. 2024GGA061); Startup Fund for scientific research, Fujian Medical University (No. 2024QH2048).
Competing interests
The authors declare that they have no competing interests.