Significant outcomes
We identified distinct changes in gene expression of CA1 neurons following both single and multiple administrations of cannabidiol (CBD). This highlights the molecular impact of CBD on hippocampal neurons and expands our understanding of its mechanisms of action. We revealed that repeated CBD administration led to a greater number of gene expression alterations compared to a single administration, emphasizing the importance of treatment frequency in modulating gene expression. We found that daily CBD administration for seven days resulted in the downregulation of genes related to energy metabolism and protein synthesis/degradation, while genes involved in chromatin regulation and synapse organization were upregulated. These specific gene expression changes shed light on potential cellular effects and molecular mechanisms underlying CBD actions in the hippocampus.
Limitations
One limitation of this study is its reliance on animal models, specifically C57BL/6JUnib mice, which may not fully reflect human responses to CBD. Additionally, the study primarily investigated the effects of CBD under healthy conditions and did not directly address its therapeutic effects for specific disorders or conditions. Thus, the clinical relevance and applicability of the findings to therapeutic interventions remain to be determined.
Introduction
Cannabidiol (CBD) is a natural compound found in female flowers of Cannabis sativa, an ancient plant with a long history of agriculture by humanity (Ren et al., Reference Ren, Zhang, Li, Ridout, Serrano-Serrano, Yang, Liu, Ravikanth, Nawaz, Mumtaz, Salamin and Fumagalli2021). CBD has a complex pharmacodynamic profile with targets like enzymes, membrane transporters, ionic receptors, and channels (Bih et al., Reference Ibeas Bih, Chen, Nunn, Bazelot, Dallas and Whalley2015). Regardless of being a phytocannabinoid and having a structural similarity to Δ9-THC, CBD exhibits low affinity to the CB1 receptor orthosteric site (McPartland et al., Reference McPartland, Glass and Pertwee2007). This may be a possible explanation for the lack of CBD intoxicating effects. Preclinical research has suggested that CBD may have potential therapeutic effects as an antiepileptic, antioxidant, antipsychotic, and anxiolytic agent (Elsaid et al., Reference Elsaid, Kloiber and Le Foll2019). Despite these promising findings, the underlying molecular mechanisms and gene activation associated with CBD in brain cells are still not fully understood. Until recently, almost all countries have strict drug policies and a history of drug-related violence that creates obstacles, restricting access to cannabis, hindering scientific research. These limitations prevent both researchers and patients from fully exploring the potential of cannabis as a treatment option and comprehending the intricate molecular mechanisms and gene activation associated with cannabis compounds, like CBD.
One of the brain regions most affected by a plurality of psychiatric and neurological disorders such as depression, schizophrenia, and epilepsy is the hippocampus (Anand and Dhikav, Reference Anand and Dhikav2012). The hippocampus is a structure segmented into sub-regions (CA1, CA2, CA3, and DG) that form a complex circuitry involved mainly in learning and memory processes (Fanselow and Dong, Reference Fanselow and Dong2010; Squire et al., Reference Squire, Genzel, Wixted and Morris2015). Each of these sub-regions has distinct molecular mechanisms and neuronal connections, thus selecting cellular populations precisely helps to specifically explore hippocampal functionality (Machado et al., Reference Machado, Athie, Matos, Lopes-Cendes and Vieira2022).
In the present study, we used the laser capture microdissection (LCM) in conjunction with RNA-Seq to select particular cell populations, dorsal and ventral CA1, preserving specific expression characteristics of such regions. With these strategies, we explore the effect of CBD on the transcriptome of dorsal and ventral CA1 after intraperitoneal administration for 1 or 7 days.
Methods
Animals, drugs, and tissue processing
Three-month-old males C57BL/6JUnib were housed in a ventilated environment (12 h/12 h light cycle) with ad libitum access to standard rodent chow and water. Mice were divided into four groups (n = 5): CR1 – 1 day of vehicle (0.15 M NaCl and 2% Tween 80 ); CBD1 – 1 day of CBD (100 mg/kg); CR7 – 7 days of vehicle (0.15 M NaCl and 2% Tween 80); CBD7 – 7 days of CBD (100 mg/kg). All solutions were prepared with NaCl at 98% and Tween 80 at 2% (Sigma-Aldrich, UK) and administered i.p. in a volume of 1 mL/kg. The 100 mg/kg dosage and the treatment duration were based on previous studies demonstrating its efficacy in producing anxiolytic-like effects in rats and mice with high doses and chronic administration (Zanelati et al., Reference Zanelati, Biojone, Moreira, Guimarães and Joca2010; Long et al., Reference Long, Chesworth, Huang, Wong, Spiro, McGregor, Arnold and Karl2012; Abame et al., Reference Abame, He, Wu, Xie, Zhang, Gong, Wu and Shen2021). After 24 hours from the last i.p. CBD administration, animals were anaesthetized, brains were collected and processed for laser microdissection as previously described (Machado et al., Reference Machado, Athie, Matos, Lopes-Cendes and Vieira2022). The ventral and dorsal CA1 boundaries were located based on previous studies (Fanselow and Dong, Reference Fanselow and Dong2010) and Allen Brain Atlas in situ hybridization markers (http://portal.brain-map.org/).
Library preparation, next-generation sequencing, and data processing
RNA was extracted and libraries were prepared as previously described using TruSeq Stranded Total RNA LT (Illumina®, CA, USA) from 200 ng of RNA (Machado et al., Reference Machado, Athie, Matos, Lopes-Cendes and Vieira2022). Pooled libraries were sequenced on a HiSeq® 2500 (Illumina®, CA, USA) in high output mode, generating 100-bp paired-end sequences. On average, 20 million paired-end reads were obtained per sample, totaling 794.682.591 reads across all samples.
We aligned all sequenced reads with the Mus musculus genome (GRCm38.96) using the StarAligner 2.6 program (Dobin et al., Reference Dobin, Davis, Schlesinger, Drenkow, Zaleski, Jha, Batut, Chaisson and Gingeras2013). The DESeq2 package (Love, et al., Reference Love, Huber and Anders2014) was used to normalize and statistically analyze differentially expressed genes (adjusted p-value < 0.05). Enrichment analysis was performed using clusterProfiler package (Wu et al., Reference Wu, Hu, Xu, Chen, Guo, Dai, Feng, Zhou, Tang, Zhan, Fu, Liu, Bo and Yu2021) to classify functional profiles into significant (adjusted p-value < 0.05) KEGG pathways and GO terms – biological processes (BPs).
Results
Identification of DEGs and samples visualization
A principal component analysis (PCA) was conducted for dorsal CA1 (dCBD1vsCR1 and dCBD7vsCR7) and ventral CA1 (vCBD1vsCR1 and vCBD7vsCR7) (Figs. 1A-D). Both dorsal and ventral CA1 samples revealed an overlap of the CBD1 and CR1, while the CBD7 and CR7 formed distinct groups. The number of upregulated and downregulated genes is represented in Fig. 1E. For a complete list of differentially expressed genes (DEGs) refer to Supplementary Table 1.
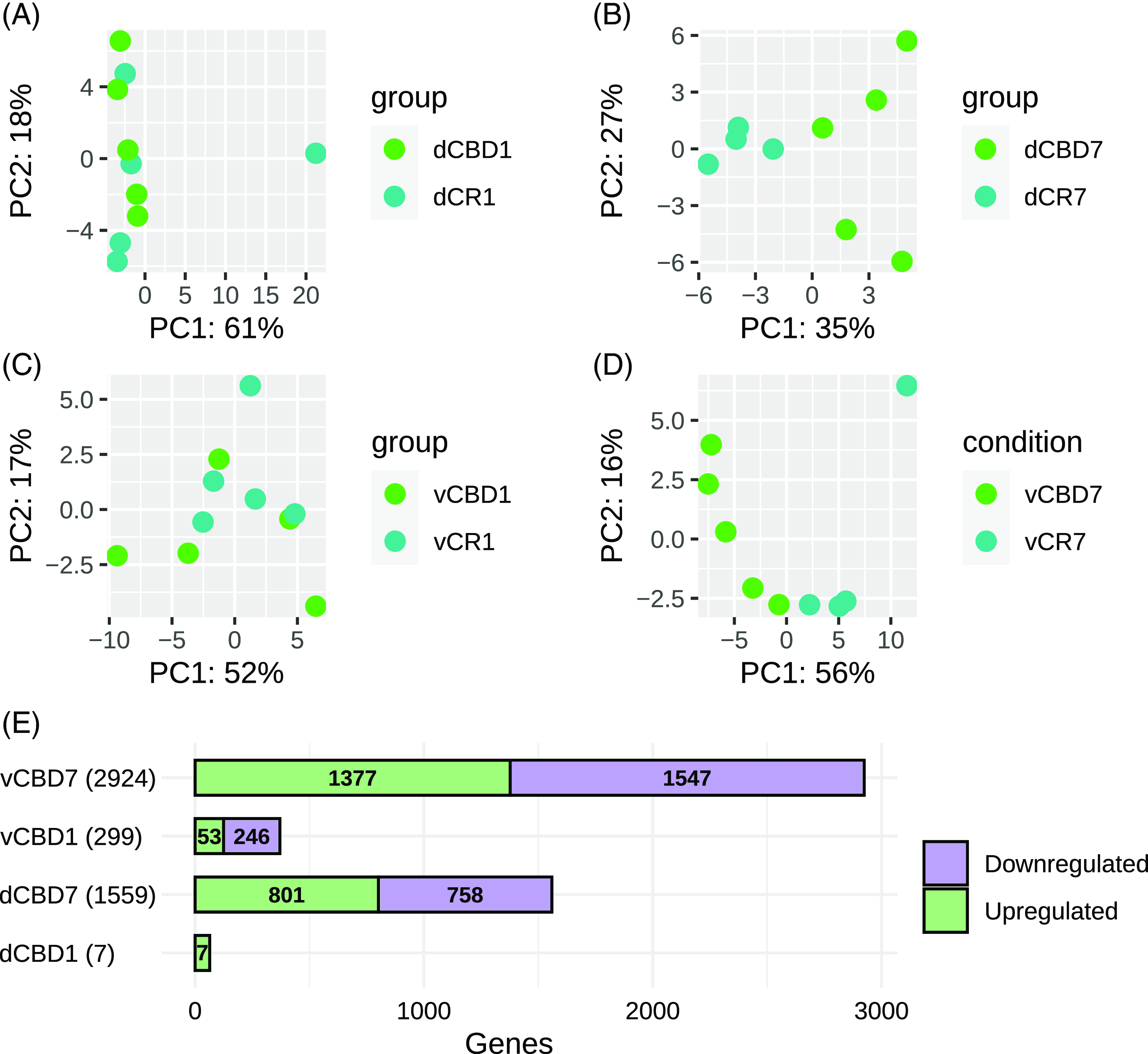
Figure 1. Sample variability and distribution of differentially expressed genes. (A) Principal component analysis of gene expression data from dorsal CBD1. (B) Principal component analysis of gene expression data from dorsal CBD7. (C) Principal component analysis of gene expression data from ventral CBD1. (D) Principal component analysis of gene expression data from ventral CBD1. (E) Barplot displaying the number of differentially expressed transcripts identified in each ventral and dorsal CBD analysis.
CBD effect on dorsal and ventral CA1
GO/KEGG analysis identified 88 significantly enriched BPs and 26 significantly enriched pathways for dCBD7, whereas vCBD7 revealed 128 enriched BPs and 24 pathways (p.adjust < 0.05) (Supplementary Table 2). GO enrichment analysis using upregulated/downregulated genes depicted transcriptional change of biological functions (Fig. 2). In the dCBD1 and vCBD1 groups, only a single significant biological process was observed (Supplementary Table 2).
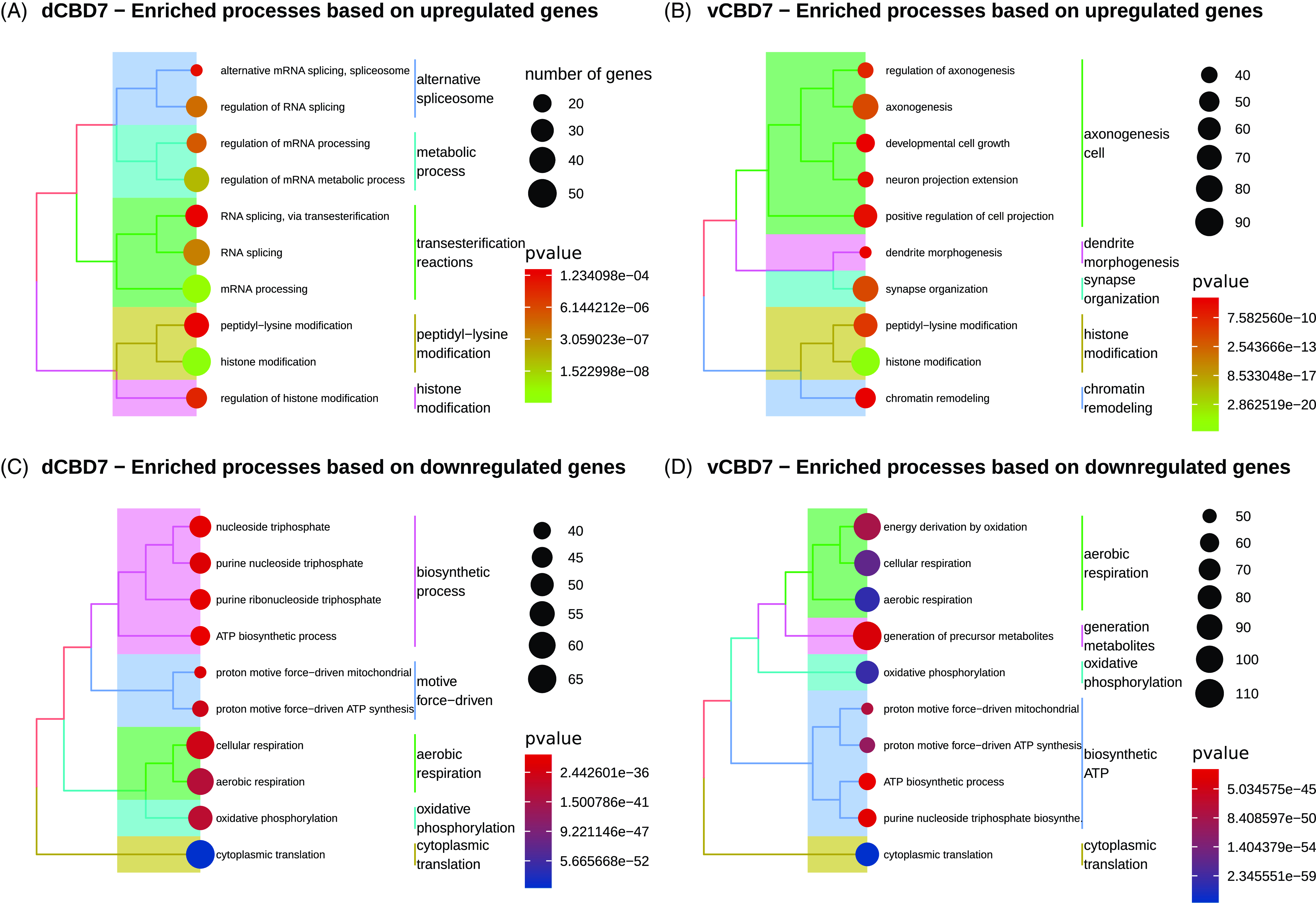
Figure 2. Enriched biological processes for differentially expressed genes in CBD7. (A) Top enriched biological processes for upregulated genes in dorsal CBD7. (B) Top enriched biological processes for upregulated genes in ventral CBD7. (C) Top enriched biological processes for downregulated genes in dorsal CBD7. (D) Top enriched biological processes for downregulated genes in ventral CBD7. All displayed biological processes reached an adj. p-value < .05.
Discussion
In this study, we used LCM/RNA-Seq to examine gene expression changes in distinct hippocampal neuron subtypes after CBD treatment. We aimed to unveil the CBD impact on transcriptome modifications and pertinent BPs/pathways associated with these changes. Additionally, we aimed to differentiate between effects within different hippocampal regions, dorsal and ventral, across two administration protocols (1-day and 7-day intervals). Our findings revealed, for the first time, a reduction in the expression of several genes linked to the respiratory chain and ribosomal subunits in mice, accompanied by an increase in the expression of genes related to histone regulation. Furthermore, vCA1 exhibited upregulation of neural projection formation genes, while dCA1 showed upregulation of RNA splicing genes. Overall, these results contribute to advancing our knowledge of the CBD effects in hippocampal neuron gene expression.
Administration frequency
Our study reveals a significant difference in the gene expression between the two treatments. Prior research has noted behavioural and molecular distinctions between single and chronic CBD administrations (50 mg/kg) in C57BL/6JArc mice (Long et al., Reference Long, Chesworth, Huang, McGregor, Arnold and Karl2010; Darweesh, et al., Reference Darweesh, Khamis and El-Elimat2020). CBD treatments for pain, psychiatric disorders, and various other conditions are often administered as a single acute dose, but effective doses may vary across disease states (Britch, et al., Reference Britch, Babalonis and Walsh2021). CBD ability to bind to multiple molecular targets may play a role in producing different physiological effects. Furthermore, binding duration is critical in determining the effectiveness and potency of pharmacological responses of several targets affected by CBD affinity and bioavailability.
While some similarities in molecular effects of dosages are observed, there are discrepancies in administration frequency (Long et al., Reference Long, Chesworth, Huang, McGregor, Arnold and Karl2010; Valvassori et al., Reference Valvassori, Bavaresco, Scaini, Varela, Streck, Chagas, Hallak, Zuardi, Crippa and Quevedo2013). CBD doses of 60 mg/kg resulted in increased mitochondrial activity in both acute and chronic administration in rats hippocampus and prefrontal cortex (Valvassori et al., Reference Valvassori, Bavaresco, Scaini, Varela, Streck, Chagas, Hallak, Zuardi, Crippa and Quevedo2013). On the other hand, reducing the dosage to 15 mg/kg led to no response in a single dose of CBD, but increased mitochondrial activity in the hippocampus and striatum of the chronic dosage group (Valvassori et al., Reference Valvassori, Bavaresco, Scaini, Varela, Streck, Chagas, Hallak, Zuardi, Crippa and Quevedo2013). Although previous studies have demonstrated similarities and differences, we are the first to report a significant number of differentially expressed genes in two administration frequencies. Therefore, our data clarify that the transcriptome of CA1 neurons undergoes more significant alteration after 7 days of CBD administration than a single dose.
Respiratory chain and TCA cycle
We observed a reduction in transcriptional levels of all mitochondrial respiratory chain complexes in CA1 after administering CBD for 7 days (Figure S1). In vitro studies have shown that CBD can reduce oxygen consumption and mitochondrial complex activity (Fišar, et al., Reference Fišar, Singh and Hroudová2014; Singh, et al., Reference Singh, Hroudová and Fišar2015). However, Valvassori et al. (Reference Valvassori, Bavaresco, Scaini, Varela, Streck, Chagas, Hallak, Zuardi, Crippa and Quevedo2013) observed an increased mitochondrial complex activity in rats hippocampus after administering CBD (60 mg/kg) for 14 consecutive days. Differences in models, doses, and administration methods account for the variability in CBD effects on mitochondria. Additionally, reduced respiratory chain complex expression in our data may not imply compromised mitochondrial function due to respiration rate limitations and mechanisms, like a compensatory increase in glycolytic rate (Brand and Nicholls, Reference Brand and Nicholls2011). Furthermore, another possibility is that CBD administration resulted in reduced energy demand by CA1 neurons, resulting in a reduction in mitochondrial demand and function. Such a possibility is reinforced by the lack of hypoxia and cell death in CA1 that would take place in case of a reduction of mitochondrial functional capacity with no change in neuronal energy demand. However, further experiments are needed to address such a possibility.
The previously mentioned studies show a biphasic CBD effect on mitochondrial respiration. Evidence suggests that CBD enhances mitochondrial respiration at lower or moderate dosages and inhibits it at higher concentrations. This observation highlights CBD dose-dependent impact on mitochondrial function, emphasising the necessity for additional functional studies on underlying mechanisms. Endogenous and exogenous CB1 ligands induced biphasic in vitro changes in mitochondrial complexes (Athanasiou et al., Reference Athanasiou, Clarke, Turner, Kumaran, Vakilpour, Smith, Bagiokou, Bradshaw, Westwell, Fang, Lobo, Constantinescu, Calabrese, Loesch, Alexander, Clothier, Kendall and Bates2007). Nevertheless, comparing respiratory chain gene activity and expression levels with different CBD dosages in animal models is crucial to comprehend its mitochondrial effects
We are the first to report a reduced expression of tricarboxylic acid cycle (TCA) genes in CA1 after CBD. It is already known that synthetic cannabinoids inhibit TCA enzymes in pancreatic cancer cells (Dando et al., Reference Dando, Donadelli, Costanzo, Dalla Pozza, D’Alessandro, Zolla and Palmieri2013). However, we only observed a reduction in the genes of isocitrate, succinyl-CoA, succinate, and malate dehydrogenase enzymes. Moreover, TCA cycle metabolites were found reduced in CBD-treated cells (Guard et al., Reference Guard, Chapnick, Poss, Ebmeier, Jacobsen, Nemkov, Ball, Webb, Simpson, Coleman, Bunker, Ramirez, Reisz, Sievers, Stowell, D’Alessandro, Liu and Old2022). Therefore, significant TCA enzyme gene expression changes suggest CBD probably influences mitochondrial activity.
CBD inhibits endocannabinoid metabolism enzymes and enhances receptor activation despite its low CB1 receptor affinity (McPartland, et al., Reference McPartland, Glass and Pertwee2007). CB1 receptor activation by endocannabinoids reduces gene expression related to mitochondrial biogenesis and oxygen consumption in mice adipocytes (Tedesco et al., Reference Tedesco, Valerio, Dossena, Cardile, Ragni, Pagano, Pagotto, Carruba, Vettor and Nisoli2010). Furthermore, our analysis found enriched BPs related to axon and dendrite formation following a 7-day CBD administration in ventral CA1. Inhibiting the respiratory chain via the endocannabinoid system and reducing ATP production may efficiently regulate high energy synaptic processes such as neurotransmitter release and neuroplasticity (Bénard et al., Reference Bénard, Massa, Puente, Lourenço, Bellocchio, Soria-Gómez, Matias, Delamarre, Metna-Laurent, Cannich, Hebert-Chatelain, Mulle, Ortega-Gutiérrez, Martín-Fontecha, Klugmann, Guggenhuber, Lutz, Gertsch, Chaouloff, López-Rodríguez, Grandes, Rossignol and Marsicano2012).
Thus, the reduction in crucial mitochondrial gene expression and increase in synaptic modulation genes might be linked to endocannabinoid signalling and may play a role in CBD ability to reduce seizure frequency in both humans and animal models (Cheung et al., Reference Kwan Cheung, Peiris, Wallace, Holland and Mitchell2019; von Wrede, et al., Reference von Wrede, Helmstaedter and Surges2021; Madireddy and Madireddy, Reference Madireddy and Madireddy2023). A reduction in energy demand by neurons, possibly linked to a increase in inhibition through enhanced gabaergic activity, could consist in a mechanism responsible for seizure reduction in epilepsy following CBD administration. Furthermore, changes in energy metabolism mediated by CBD administration could take place in other brain regions, such as the medial prefrontal cortex, possibly consisting of a mechanism responsible for CBD effects in anxiety, depression, and schizophrenia. Nevertheless, further investigation is required to uncover the underlying molecular mechanisms and confirm this hypothesis.
Ribosome and proteasome
Cell regulates the rate of protein synthesis by maintaining a balance between ATP consumption and ribosomal transcription (Jakubovic and McGeer, Reference Jakubovic and McGeer1972; Murray, et al., Reference Murray, Schneider and Gourse2003). Early studies have shown that Δ9-THC application causes a reduction of ribosome number (Hattori, et al., Reference Hattori, Jakubovic and McGeer1972). We are the first to report a significant reduction in gene expression of several ribosomal subunit constituents in CA1 after 7 days of CBD administration (Figure S1). Furthermore, our results also show several proteasome subunits downregulated after 7 days of CBD administration. Reduction in genes involved in the ubiquitin/proteasome system may reflect a lower protein synthesis, as fewer proteins are degraded or modified (Xolalpa et al., Reference Xolalpa, Perez-Galan, S. Rodríguez and Roue2013). Moreover, genes differentially expressed after a single CBD application (Supplementary Table 2) may explain the observed proteasome effect with repeated administration, considering the link between ubiquitin and proteasome in protein degradation. However, further investigation is required to validate the underlying cellular mechanisms involved in protein synthesis and degradation following CBD administration.
Reduction in gene expression of several ribosomal subunit constituents does not necessarily imply lower synaptic plasticity activity (Rosenberg et al., Reference Rosenberg, Gal-Ben-Ari, Dieterich, Kreutz, Ziv, Gundelfinger and Rosenblum2014; Caccamo et al., Reference Caccamo, Branca, Talboom, Shaw, Turner, Ma, Messina, Huang, Wu and Oddo2015). Decreased protein synthesis may lead to lower levels of synaptic proteins, promoting the elimination of weaker synapses and strengthening remaining connections. This reduction can also alter proteins involved in plasticity mechanisms like long-term potentiation (LTP) or long-term depression (LTD). Interestingly, we found in our data some genes involved in the late phase of LTP (Chen et al., Reference Chen, Kawaguchi, Blum, Achiro, Coppola, O’Dell and Martin2017). Moreover, reduced expression of regulatory proteins or microRNAs can fine-tune synaptic protein levels and modulate plasticity-related signalling pathways (McClung and Nestler, Reference McClung and Nestler2008), resulting in an increase in plasticity-related pathways, which is consistent with our findings. Another point to be considered is that a reduction in overall synaptic protein levels may take place at same time as an increase in specific synapse sub-types, such as Gabaergic synapses. However, further experiments are required to verify such a possibility.
Small GTPases and neuroplasticity
Cannabinoids affect small GTPase signalling through CB1 and CB2 receptors (Kurihara et al., Reference Kurihara, Tohyama, Matsusaka, Naruse, Kinoshita, Tsujioka, Katsumata and Yamamura2006; Duman et al., Reference Duman, Mulherkar, Tu, X. Cheng and Tolias2015). We are the first to find altered expression of several genes of regulatory GTPases proteins after CBD repeated administration. Furthermore, Rho family of GTPases regulate postsynaptic density and actin cytoskeleton organization (Stankiewicz and Linseman, Reference Stankiewicz and Linseman2014). Thus, small GTPase activity influenced by CBD may also be responsible for cytoskeleton and synaptic plasticity alteration, but further research is required.
Our findings reveal that CBD increases gene expression linked to synapse organization and dendrite/axon formation solely in the ventral CA1. We found that CBD increases the gene expression of Actn1 (Alpha-actinin-1), Flna (Filamin-A), Synpo (Synaptopodin), Shank1/Shank2 (SH3 and multiple ankyrin repeat domains protein), and Itsn1 (Intersectin-1), which play a role in dendritic spine morphology and postsynaptic density organization (Muñoz-Lasso et al., Reference Muñoz-Lasso, Romá-Mateo, Pallardó and Gonzalez-Cabo2020). Thus, CBD influence on hippocampal neuronal cytoskeleton may modify dendritic morphology and promote circuit plasticity. Additionally, the data presented here demonstrate an increase in guidance molecules expression, such as Sema4d (Semaphorin-4D), Sema4f (Semaphorin-4F), Sema5a (Semaphorin-5A), Plxna2 (Plexin-A2), Plxnb2 (Plexin-B2), and Nrp2 (Neuropilin-2) in the vCA1 after CBD. Interactions like Sema5d/Plexna2 inhibits excitatory synapses formation in mouse hippocampal cells (Duan et al., Reference Duan, Wang, Song, Mironova, Ming, Kolodkin and Giger2014). In addition, Sema4d infusion into mice CA1 promotes the formation of inhibitory synapses and suppresses seizures in models generating seizures via electrical and chemical stimulation (Acker et al., Reference Acker, Wong, Kang and Paradis2018). The ability of CBD to reduce the number of seizures and increase seizure threshold, particularly in animal models, has been well established (Consroe et al., Reference Consroe, Benedito, Leite, Carlini and Mechoulam1982). Therefore, these findings may offer insights into how CBD regulates molecules responsible for inhibitory/excitatory synapse plasticity in vCA1.
Neuroplasticity defects play a role in mood disorders and diseases pathophysiology (Post, Reference Post1992; Dorszewska et al., Reference Dorszewska, Kozubski, Waleszczyk, Zabel and Ong2020). A single dose of CBD (30 mg/kg) relieved depressive-like behaviors but did not restore dendritic spine density in vCA1 following stress-induced depression (Ma et al., Reference Ma, Li, Wang, Zhang, Li, Zhang, Huang and Zhang2021). However, repeated CBD treatment promotes dendritic spine remodelling in the hippocampus of chronically stressed mice (Fogaça et al., Reference Fogaça, Campos, Coelho, Duman and Guimarães2018). CB1/CB2 antagonists abolished the remodelling effect, suggesting that endocannabinoid receptors mediate the alteration in synaptic density (Fogaça et al., Reference Fogaça, Campos, Coelho, Duman and Guimarães2018). Similar to our findings, changes in plasticity and cytoskeleton gene expression were observed exclusively with repeated doses, not with a single dose. Thus, changes in plasticity via gene expression require repeated CBD administration, while single-dose behavioural effects may relate to brain circuit excitatory/inhibitory balance.
Chromatin regulatory genes
Previous studies emphasize the importance of DNA methylation via DNMTs for neuronal plasticity, including learning and memory (Levenson et al., Reference Levenson, Roth, Lubin, Miller, Huang, Desai, Malone and Sweatt2006; Feng et al., Reference Feng, Zhou, Campbell, Le, Li, Sweatt, Silva and Fan2010). CBD (10 mg/kg) induced antidepressant-like effects and reduced global methylation in prefrontal cortex and hippocampus of mice submitted to forced swimming tests (Sales, et al., Reference Sales, Guimarães and Joca2020). Moreover, CBD effects in DNMT activity are linked to modified DNA methylation levels and antidepressant-like effects (Sales, et al., Reference Sales, Guimarães and Joca2020). Our data indicate increased Dnmt3a (DNA methyltransferase 3A) expression in both CA1 sub-regions after CBD administration. Dnmt3a plays a role in modulating dendritic spine growth and behavioural traits (LaPlant et al., Reference LaPlant, Vialou, Covington, Dumitriu, Feng, Warren, Maze, Dietz, Watts, Iñiguez, Koo, Mouzon, Renthal, Hollis, Wang, Noonan, Ren, Eisch, Bolaños, Kabbaj, Xiao, Neve, Hurd, Oosting, Fan, Morrison and Nestler2010). These findings may indicate that CBD influences synapse organization by upregulating DNMT enzymes but further research is required.
We are the first to provide evidence that CBD regulates the expression of numerous enzymes involved in histone modification. We found several upregulated methyltransferases and demethylases (Kmt2a, Kmt2b, Kmt2c, Nsd3, Kdm2a, Kdm4b, and Kdm7a) which control lysine methylation in histone tails (Hyun et al., Reference Hyun, Jeon, Park and Kim2017). Our results show that CBD increases gene expression of chromatin and histone modification transcripts in CA1, shedding light on new players for CBD epigenetic effects.
The role of these enzymes is increasingly associated with cancer (Roy et al., Reference Roy, Walsh and Chan2014), as well as neurodegenerative diseases (Karpova et al., Reference Karpova, Sales and Joca2017) and neurodevelopmental disorders (Kim et al., Reference Kim, Lee, Lee, Lee and Cho2017). Moreover, emerging evidence regarding the potential of CBD or other cannabinoids in treating many diseases has also grown (Elsaid et al., Reference Elsaid, Kloiber and Le Foll2019; Britch et al., Reference Britch, Babalonis and Walsh2021; Iffland & Grotenhermen, Reference Iffland and Grotenhermen2017). Although the mechanisms by which CBD acts in these diseases are not fully understood, the epigenetic alterations generated by its administration may be a potential target for study to explore CBD cellular effects in disease models and therapeutic application.
In conclusion, the transcriptome data generated here reveal that repeated CBD administration leads to significant modifications in dorsal and ventral CA1 neuronal expression. These results provide evidence for CBD cellular effects and molecular mechanisms under healthy conditions, as well as to understand the potential therapeutic effects of its use. Additionally, new potential signalling pathways and genes that are altered after CBD administration in the CA1 sub-region of mice hippocampus were identified with high statistical confidence. These findings improve our understanding of CBD effects on gene expression and its potential impact on molecular mechanisms in the hippocampus.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/neu.2024.13
Acknowledgements
We acknowledge the use of artificial intelligence (AI) tools, specifically OpenAI ChatGPT, to refine the quality of the English language used in the present paper. The original English text from the authors was corrected by the AI tool and was still revised by the authors to ensure accuracy and clarity.
Authors’ contribution
João P. D. Machado: Conceptualization, methodology, investigation, visualization, formal analysis, and writing – original draft. Valéria de Almeida: Conceptualization, methodology, formal analysis, and writing – original draft. Antonio W. Zuardi: Conceptualization and methodology. Jaime E. C. Hallak: Conceptualization and methodology. José A. Crippa: Conceptualization and methodology André S. Vieira: Conceptualization, methodology, project administration, supervision, and writing – original draft.
Financial support
This work was supported by grants from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Fundação de Amparo à Pesquisa do Estado de São Paulo, SP, Brazil (FAPESP; grant number 2013/07559-3)
Competing interests
The authors declare they have no conflicts of interest.
Animal welfare ethical
All procedures complied with ethical standards for animal experimentation at the University of Campinas-UNICAMP (Brazilian federal law 11.794 (10/08/2008 - Animal Use Ethics Committee protocol 2903-1).
Ethical standard
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.





