Introduction
Using bio-inspiration during the conceptual design phase increases the number of novel solutions generated (Vandevenne et al., Reference Vandevenne, Pieters and Duflou2016; Keshwani and Chakrabarti, Reference Keshwani, Chakrabarti, Chakrabarti and Chakrabarti2017). In contrast with other design methods, which rely on the prior knowledge and experience of the designer, bio-inspired design offers a catalogue of proven designs, often using different working principles than the ones traditionally employed in a technical setting (Bogatyrev and Bogatyreva, Reference Bogatyrev, Bogatyreva, Azevedo, Brandenburg, Carvalho and Cruz-Machado2014). Despite such promising results, bio-inspired design has not yet been widely adopted as a design strategy and only a limited number of bio-inspired products are commercially available (Wanieck et al., Reference Wanieck, Fayemi, Maranzana, Zollfrank and Jacobs2017).
Most engineers do not possess biological background knowledge. They have a hard time identifying, filtering and understanding relevant biological strategies to apply them to their design problem, turning the search for relevant biological strategies into a frustrating and time-consuming task (Goel and Helms, Reference Goel, Helms, Chakrabarti and Blessing2014; Fayemi et al., Reference Fayemi, Wanieck, Zollfrank, Maranzana and Aoussat2017; Kruiper et al., Reference Kruiper, Vincent, Abraham, Soar, Konstas, Chen-Burger and Desmulliez2018; Graeff et al., Reference Graeff, Maranzana and Aoussat2019). Many methods and tools supporting engineers in retrieving relevant bio-inspiration for their design problem have been proposed (Lenau et al., Reference Lenau, Metze, Hesselberg and Lakhtakia2018; Hashemi Farzaneh and Lindemann, Reference Hashemi Farzaneh and Lindemann2019). However, most of these tools use a database of which the construction is not scalable (Vandevenne et al., Reference Vandevenne, Pieters and Duflou2016; Willocx et al., Reference Willocx, Ayali and Duflou2020).
In contrast with the multitude of bio-inspired design tools, only a few methods are used in a design context (Pentelovitch and Nagel, Reference Pentelovitch and Nagel2022). To more easily integrate with an existing design process used in a company, a tool integrated into the systematic engineering design process proposed by Pahl et al. (Reference Pahl, Beitz, Feldhusen and Grote2007) is useful (Nagel et al., Reference Nagel, Stone, McAdams, Goel, McAdams and Stone2014). Furthermore, a free-text search system ensures that the user does not have to learn a specific vocabulary, considerably lowering the difficulty of uptake. The literature review in section two highlights that no tool currently offers free-text search, automatic translation between engineering and biology, and access to the whole biological literature.
Furthermore, Broeckhoven and du Plessis (Reference Broeckhoven and du Plessis2022) highlight that most bio-inspired designs are based on a limited number of highly publicized organisms. To counteract this, they proposed to digitize existing Natural History collections and look at more biodiverse organisms in the search for inspiration. Another opportunity is to use the already digitally available biological literature, used for example in Shu (Reference Shu2010) or Vandevenne et al. (Reference Vandevenne, Verhaegen, Dewulf and Duflou2015).
The rise in open-access publishing (Laakso et al., Reference Laakso, Welling, Bukvova, Nyman, Björk and Hedlund2011) and more permissive text-mining policies on subscription content of large publishers (Van Noorden, Reference Van Noorden2014) allow the automated extraction of interesting insights from more content. This creates an opportunity to build a bio-inspired design tool on top of a large and ever-growing collection of scientific biological literature. By basing the tool on machine-processed publications and limiting the manual interaction required to update the underlying data, it would stay up to date with new developments in engineering or biology.
Research goals and scope
This study describes a natural language search method based on the essential function that the engineering design must fulfill. Figure 1 schematically presents the Functional Inspiration Search method (FISh). The essential function of a design is defined by Pahl et al. (Reference Pahl, Beitz, Feldhusen and Grote2007) as “the generalized crux of the problem”, and the definition thereof is part of the systematic design process. The proposed method automatically translates the functional engineering query into its biological equivalent and uses this to retrieve relevant biological publications. This allows FISh to be directly integrated into the systematic engineering design process. Furthermore, by automatically linking engineering and biology, the search tool can scale to encompass the complete biological literature and be easily updated with new literature.

Figure 1. Schematic representation of the approach proposed for the functional inspiration search method (FISh). Starting in the top left, a free-text functional query is converted to a vector representation (embedding) using the engineering language model and transformed to the biology domain using the linking method proposed in this work. Next, the most relevant organism aspects are selected and the documents retrieved are clustered based on their focus organisms and presented to the end user.
Figure 1 highlights the different areas for which a research effort is required. These research efforts are driven by the following research questions:
1) A literature review on the currently existing bio-inspired design support tools for the search phase and the size of their underlying datasets.
2) Apply machine translation methods to link the engineering and biology domain.
a. Create a dataset that is representative of the currently available biological literature.
3) Automate the generation of organism aspects as described by Vandevenne et al. (Reference Vandevenne, Verhaegen, Dewulf and Duflou2015) to allow updating the system with new biological developments.
a. Automatically determine the number of necessary organism aspects.
b. Automatically filter the included lemmas to focus on functionality.
The scope of the tool here is limited to supporting mechanical engineers as a prototype. Furthermore, the validation of the search method is here limited to an assessment of the relevance of the retrieved biological publications and does not cover the potential to transfer this knowledge to the searching engineers.
The remainder of this research paper is structured as follows: first, the relevant literature on bio-inspired design support tools and natural language models is reviewed. Second, the proposed method of using statistical machine translation to support bio-inspired design is presented along with the used datasets to represent engineering and biology and align the spaces. Third, using existing bio-inspired designs and their inspiring organisms, the search method is validated. Finally, after providing conclusions, future research directions and possible alternative applications of the presented method are proposed.
Background
First, the relevant bio-inspired literature is reviewed to document the steps required to integrate bio-inspiration in the design process and highlight the shortcomings and pitfalls of the current methods. To introduce the automatic linking method between biology and engineering, a short literature review focused on the natural language models used is presented. The natural language model selection is explained and the literature on employing the characteristics of the model to enable machine translation is reviewed.
Bio-inspired design tools
Systematically applying biological inspiration requires a consistent process that aids the engineer in finding and applying relevant biological inspiration to a certain, given engineering problem. This process is known as the engineering pull (Helms et al., Reference Helms, Vattam and Goel2009), where the engineering problem is the start of the search for bio-inspiration. While different authors have defined several phases in the systematic process (e.g., Lenau et al., Reference Lenau, Metze, Hesselberg and Lakhtakia2018; Hashemi Farzaneh and Lindemann, Reference Hashemi Farzaneh and Lindemann2019), they can be reduced to the four phases schematically presented in Figure 2 (Vandevenne et al., Reference Vandevenne, Verhaegen, Dewulf and Duflou2015). After formulating the problem and searching for bio-inspiration, the different retrieved strategies are analyzed and selected. Finally, the working principles need to be identified and transferred analogically to the engineering domain.

Figure 2. The phases of the systematic bio-inspired design process as identified by Vandevenne et al. (Reference Vandevenne, Verhaegen, Dewulf and Duflou2015).
A major difficulty in bio-inspired design is retrieving and identifying relevant biological inspiration. The language used in engineering and biology is different (Fayemi et al., Reference Fayemi, Wanieck, Zollfrank, Maranzana and Aoussat2017) and engineers often lack the required biological terminology and knowledge to search for relevant biological systems effectively. This turns the search phase into a time-consuming task (Vattam and Goel, Reference Vattam and Goel2011). Furthermore, due to the large time investment required, engineers fixate on a single biological strategy, even when a better alternative is presented (Ahmed-Kristensen et al., Reference Ahmed-Kristensen, Christensen and Lenau2014).
A first method to provide engineers with biological inspiration during the design process are design books with a list of interesting biological systems, for example (Nachtigall and Wisser, Reference Nachtigall and Wisser2015). Another option would be to consult an expert biologist (Graeff et al., Reference Graeff, Maranzana and Aoussat2019). However, these time-consuming methods interrupt the engineering design process and are thus not likely to be employed.
In the following paragraphs, the existing bio-inspired design support tools will be reviewed concerning their search input or query, the type and size of the dataset they can search and their current availability. To limit the scope of the review, only recent supportive methods or tools that have an explicit contribution to supporting the search for relevant bio-inspiration are considered. Table 1 presents the results of this review.
Table 1. Overview of the different bio-inspired design support methods that describe a method for finding bio-inspiration. For each method, the input query and the search method are summarized. The type and size of the dataset that is accessible via the search method are based on the most recent publicly available data or the data that have effectively been prepared and described by the authors of the method.
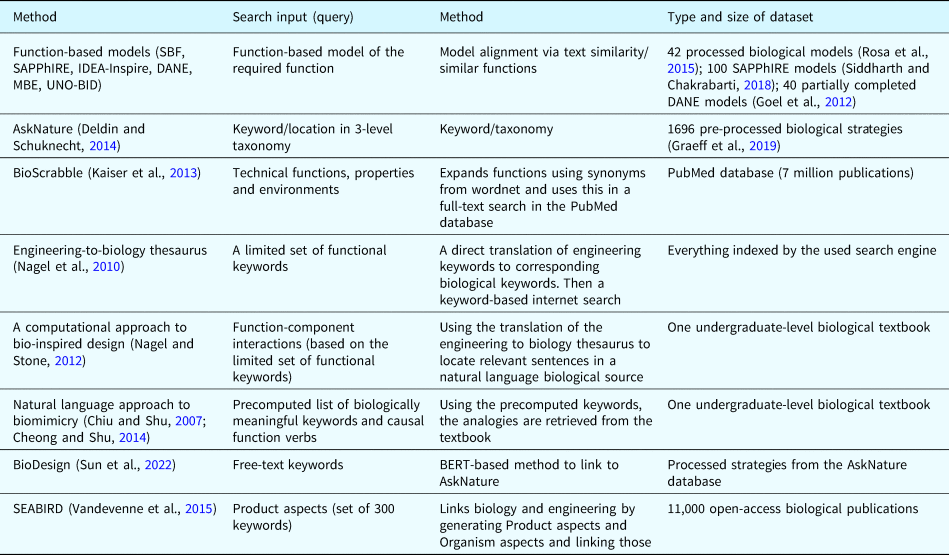
In the function-based model tools, the engineering problem and the biological strategies are expressed in a functional model, and then, a model alignment is used to match the biological strategies with the model of the problem. The time-consuming nature of the modeling process (e.g., creating a single model takes between 40 and 100 h (Vattam et al., Reference Vattam, Wiltgen, Helms, Goel, Yen, Taura and Nagai2011)) causes the databases of, for example, UNO-BID to be limited to 42 biological models. Furthermore, before a biological strategy search can be started, the engineering problem will also have to be modeled.
The AskNature database uses a functional taxonomy to organize the biological strategies, which allows engineers to systematically find these by locating the corresponding function in the taxonomy. As noted by Deldin and Schuknecht (Reference Deldin, Schuknecht, Goel, McAdams and Stone2014), the functional categories have been arbitrarily chosen. A more recent version of the website also allows searching based on keywords (Hooker and Smith, Reference Hooker and Smith2016). However, this runs into the difficulty that keyword-based search engines are not designed for cross-domain retrieval (Rugaber et al., Reference Rugaber, Bhati, Goswami, Spiliopoulou, Azad, Koushik, Kulkarni, Kumble, Sarathy, Goel, Goel, Belén Díaz-Agudo and Roth-Berghofer2016). Each biological strategy that is entered into the database is pre-processed to be useful in an engineering design exercise. Again, this time-consuming process limits the number of biological strategies that can be added to the database.
BioScrabble overcomes the limited size of a prepared database by searching directly in PubMed Central, an open archive containing full-text articles from biomedical and life-sciences journals. The support tool expands the functional query using the synonyms present in WordNet and uses these extra keywords to ameliorate the query (Kaiser et al., Reference Kaiser, Farzaneh and Lindemann2013). Each query retrieves a large number of publications, requiring the design engineer to manually scan them (Kaiser et al., Reference Kaiser, Hashemi Farzaneh and Lindemann2014).
The engineering to biology thesaurus provides a translation guide between the functional (engineering) basis and biological language use (Nagel et al., Reference Nagel, Stone and McAdams2010). The functional basis consists of a limited vocabulary of 42 terms that are claimed to be a standard necessary to describe all engineering problems (Hirtz et al., Reference Hirtz, Stone, McAdams, Szykman and Wood2002). This method was later expanded by using the biological keywords to search directly in natural language biological texts, extracting the relevant sentences directly (Nagel and Stone, Reference Nagel and Stone2012).
SEABIRD (Vandevenne et al., Reference Vandevenne, Verhaegen, Dewulf and Duflou2015) links biology and engineering by generating so-called product and organism aspects from patents and biological publications for representing respectively the engineering and biological domains. These aspects were then manually linked together, allowing for the input of one or more product aspects to yield the most relevant organism aspects and their associated publications. This search tool allows to access 11,000 biological publications. To formulate a query, the engineer must be familiar with the 300 product aspects and map the engineering design problem on those. Major drawbacks of the method are that, when updating the dataset with new engineering information, the product aspects will change to follow new developments in engineering (requiring retraining of the engineer) and the manual linking must be performed again.
Despite these drawbacks, the grouping property of organism aspects based on the functionality described in the biological documents is useful for the work at hand. This work will utilize the concept of organism aspects, but improve the generation by automating the lemma selection and build a theoretical foundation to automatically pick a relevant number of organism aspects. Furthermore, the automatic translation method proposed eliminates the need for product aspects and manual linking between engineering and biology.
A systematic approach that can deal with free text queries has the advantage of dealing with out-of-vocabulary words, not requiring the engineer to be familiar with a predefined vocabulary. Furthermore, when the engineering domain moves forward, the predefined vocabularies will age and not be able to represent new developments. For example, the domain of additive manufacturing is a recent development and cannot be reasonably modeled with the engineering-to-biology thesaurus. This illustrates the difficulty of keeping up to date with new engineering developments.
While Sun et al. (Reference Sun, Xu, Meng and Lu2022) provided a free-text search with keywords, there is no provision taken for the difference in vocabulary between engineering and biology. This is not necessary with the use of the pre-processed strategies for engineering application in AskNature, however, given the slow pace of growth of AskNature, it is currently unclear if this method can scale beyond the AskNature database.
In conclusion, while several search support tools have been built, no tool offers free-text search, automatic translation, and access to the whole biological literature.
Natural language models
To automate the free-text translation from engineering vocabulary to biology, a method to mathematically represent both domains is required. This section first explains the main idea of representing language using embeddings, then reviews the recent language models created to perform this task. Next, the choice for the specific language model is underpinned. Finally, the literature on leveraging the characteristics of the chosen language model to translate between different languages is reviewed. This will later be applied between domains within the same language.
Word embeddings are a method of representing words in machine learning tasks. To create a word embedding, the given word is associated with a mathematical object which allows us to perform a natural language processing task. Often this is a vector in which each of the dimensions includes information about the word, its meaning or context (Turian et al., Reference Turian, Ratinov and Bengio2010). For example, using the naïve method of one-hot coding the words cat, dog, and house would result in the respective vectors <1,0,0>, <0,1,0>, and <0,0,1>.
The distributional hypothesis assumes that the meaning of words is determined by the context in which they occur (Landauer et al., Reference Landauer, McNamara, Dennis and Kintsch2007). Recent developments in language representation for AI models allow converting a word to a vector representation which captures the meaning using the context in which the word occurs (Karani, Reference Karani2018). Different methods for creating this vector exist and a more comprehensive overview is given by Devlin et al. (Reference Devlin, Chang, Lee and Toutanova2018). Here, the focus is placed on the methods that will be employed in the remainder of the study.
Mikolov et al. calculated the vector for each word by performing a word prediction task with a shallow neural network as represented in Figure 3. The network must predict the word based on the given context. The hidden projection layer for each word is assigned to be the word embedding, enabling the resulting vector to capture the information about the context in which the word is used in the text on which the training is performed. This causes synonymous words and words with a similar conceptual meaning to be grouped in the space spanned by the vectors of the language model (Mikolov et al., Reference Mikolov, Chen, Corrado and Dean2013a).

Figure 3. Word prediction neural network as employed by Mikolov et al. The tokens surrounding a masked token in a sentence are presented to the neural network. The training objective is to predict the masked lemma, leading the neural network to form an internal representation of the masked token. This internal representation becomes the word embedding for a given token.
Bojanowski et al. (Reference Bojanowski, Grave, Joulin and Mikolov2016) expand the word prediction task from Mikolov et al. (Reference Mikolov, Chen, Corrado and Dean2013a) to consider subword information by using character n-grams instead of the complete word. This has the consequence of extending the method to be able to deal with unseen (out-of-vocabulary) words. Due to this property and the public availability of the code to create language models, this method is selected to be used. A known disadvantage of this method is that lemmas with multiple meanings only are assigned one vector.
Here, a conscious choice to use context-free word embeddings is made. For example, the more recent BERT word representation system takes the context of the word into account when assigning a context vector to a word (Devlin et al., Reference Devlin, Chang, Lee and Toutanova2018). However, as the expected search queries will consist of functional keywords and possibly a limited set of related words, a query does not set up a context. Furthermore, by using a larger context vector, aligning the spaces (as described in the next section) would require a larger parallel corpus. Finally, comparing two vectors with the cosine similarity rapidly becomes less useful when the dimensionality increases (Houle et al., Reference Houle, Kriegel, Kröger, Schubert, Zimek, Gertz and Ludäscher2010).
By utilizing a small bilingual corpus shared between two different languages, the transformation necessary to link the vectors of the known words from one space to another can be calculated. Mikolov et al. (Reference Mikolov, Le Quoc and Sutskever2013b) proposed to calculate the transformation matrix by solving the least squares problem with the embeddings of the word pairs in both language models. This transformation allows the translation of any word vector from one model to another. By selecting the closest known word in the target language, machine translation between both languages is achieved. Expanding on this work, Smith et al. (Reference Smith, Turban, Hamblin and Hammerla2017) proved that the required transformation is orthogonal and provide a robust calculation of the transformation matrix using the singular value decomposition of the product of the matrices of the embeddings of the known word pairs.
From the success of using this method to reliably translate between two similar languages (Smith et al., Reference Smith, Turban, Hamblin and Hammerla2017), here it is theorized that this method can also be used to translate between two domains using different jargon in the same language. Assembling a parallel corpus to align the spaces is a challenge since there is no clear “translation” possible between the domain of engineering and biology. This challenge is tackled in the next paragraphs.
Linking engineering and biology
As the language used in the engineering and biology domains is different (Nagel, Reference Nagel, Goel, McAdams and Stone2014; Helfman Cohen and Reich, Reference Helfman Cohen and Reich2016) and literature about both areas is prevalent, the hypothesis that machine translation between both domains can translate a query between engineering and biology is posited. It is proposed here to train a language model representing the biological domain using biological publications. Similarly, the engineering domain's representative language model is trained on publicly available patents, representing the latest engineering innovations.
As both language models represent different domains but are rooted in English, the assumption that the most used words retain their meaning between both models is made. Now, the most used words can be used to calculate the transformation between both models, which allows the translation of the functional query to the biology domain.
Figure 4 schematically presents the proposed alignment of both spaces by using the top words and an illustrative example of the translation of an engineering query. Via the alignment, the functional query for a filtration system is transferred from the engineering language model to the conceptually similar location in the biological model.

Figure 4. Schematic representation of the alignment of the biological and engineering language models using the most frequent words in both languages. (a) Illustrates using the vectors of the most used words in English to determine the transformation required to go from model to model. Here, the alignment of both spaces is illustrated as a 90° clockwise rotation from engineering to biology. In reality, both domains are represented by a 300-dimensional space and this translation is performed by executing a matrix multiplication using the transformation matrix. (b) To illustrate the transformation of a query from engineering to biology, an example query related to additive manufacturing is presented. The colored polygons illustrate the contexts of terms related to additive manufacturing and growing for engineering and biology, respectively. By using the transformation determined in the previous step, the query vector is transferred from engineering to biology and ends up near the biological context related to growth.
The functions described in the biological publications are grouped by performing the organism aspect generation procedure described in Vandevenne et al. (Reference Vandevenne, Verhaegen, Dewulf and Duflou2015). These organism aspects are then located in the biological space, completing the link. After mapping the query from engineering to biology, the closest organism aspects are selected and used in a weighted calculation to determine the most relevant biological documents. By using the organism aspects instead of the content of the publication, the focus is placed on the relevant functions.
The next sections detail the data and methods used to represent biology and engineering and the pre-processing of the language and alignment data.
Corpora
To compile the corpus of biological documents, the reference list of biological journals from Scopus is downloaded and processed to exclude journals not published in English. Furthermore, as the scope of useful inspiration in the biology corpus is limited to mechanical engineering in this research, any journals covering molecular biology or human health are also excluded. By leveraging the metadata service Crossref, 2.5 million potential biological documents were identified. Of these documents, not all are available in electronic form, nor have a license that allows text mining. To limit the number of individual text-mining agreements that had to be reviewed, the largest publishers, Elsevier, Springer, Wiley, and Nature, were selected. Additionally, the Journal of Experimental Biology is also included in the dataset. This results in a mix of open-access and subscription publications.
The next selection is based on the content of the documents. First, using a language classifier, texts not in English are excluded. Second, texts shorter than 500 words are also excluded. Third, the focus organism detection algorithm described by Vandevenne et al. (Reference Vandevenne, Verhaegen and Joost Duflou2014) is used to detect the focus organisms in the title and abstract of the publication. The organism detected in the title or most mentioned in the abstract is taken as the focus organism of the publication. Documents not containing a focus organism are finally also excluded. In the end, this procedure yields a corpus of 161,342 publications.
To compile the engineering corpus, patents were downloaded from the United States Patent Office (USPTO). To create a representative corpus which does include the latest innovations but also takes older patents into account, the dataset was built biased toward newer patents. Starting from 2001, for each year, a random month was selected to download all the patents published in that month. Next, all the patents of 2020 were also included in the dataset. To ensure that both models contain approximately the same amount of training data, patents are randomly drawn from the set of available patents until the same amount of text as contained in the biological corpus is attained. Another option would be to use engineering publications to represent the engineering domain, however, this lowered the performance during screening experiments. It is theorized that the research publications do not allow the model to represent functions as well as patents. For future research, it might be interesting to compare the effectiveness of the models for the representation of other types of queries.
Pre-processing and model generation
Before training the language models, the corpora are pre-processed to remove organism mentions, group common bigrams and reduce the vocabulary. Organism mentions are removed by using the organism detection described in Vandevenne et al. (Reference Vandevenne, Verhaegen and Joost Duflou2014) so the language model does not use the species to define the context.
Common bigrams are joined together to better capture meaning in the resulting language model. This is done based on a statistical model where the presence of one word strongly implies the other. For example, when “additive” and “manufacturing” are together, their meaning is compounded, and they are joined together. This is done for both the patents and the biological documents based on mutual information with a cut-off value determined on a knee point analysis of the mutual information of lemmas in the biological documents (Mei et al., Reference Mei, Shen and Zhai2007).
Finally, lemmatization is performed by the Spacy package (Montani et al., Reference Montani, Honnibal, Honnibal, Van Landeghem, Boyd, Peters and McCann2022). The stemming combines different versions of the same root word, collating more word use information together, allowing the language model to better model the meaning of the word.
Using the processed texts, the language models are trained using the code provided by Bojanowski et al. (Reference Bojanowski, Grave, Joulin and Mikolov2016). Here, the dimension of 300 is chosen since this is the same as reported by Bojanowski et al. (Reference Bojanowski, Grave, Joulin and Mikolov2016) and this gave the best results in a screening experiment with the complete system.
The proposed procedures are automated and can be repeated, for example, every 6 months, to keep the corpora and models up to date with new biological research and engineering developments. By updating this model with the latest released patents, the search functionality will be able to understand queries along the state of the art.
Organism aspects
For the generation of the organism aspects, the vocabulary in the processed texts is further filtered based on the part-of-speech tagging provided by the Spacy package and the usefulness to explaining functions of the words: verbs (Shu, Reference Shu2010), nouns (Vandevenne et al., Reference Vandevenne, Verhaegen, Dewulf and Duflou2015), and adjectives (Ke et al., Reference Ke, Chiu, Wallace and Shu2010) are kept. Adverbs are explicitly removed as they tend to dominate an aspect without adding information. Lemmas that appear less than ten times in the corpus are also excluded.
While Vandevenne et al. (Reference Vandevenne, Verhaegen, Dewulf and Duflou2015) relied on a further manual categorization of the lemmas that were useful for the transfer from the biological domain to further filter the lemmas included in the organism aspect generation, here an automatic categorization method is proposed by leveraging the hierarchy of concepts found in WikiData (Wikimedia Foundation, 2020). A hierarchy of concepts groups words into their respective conceptual categories. For instance, the abdomen of an arthropod is, climbing the taxonomy, a tagma, an anatomical structure, a biological component …
The hierarchy of concepts allows defining a few rules to filter out unwanted categories of lemmas, as visualized in Figure 5. Using the manual annotation work of Vandevenne allowed to start finding the categories that must be excluded. This resulted in a list of 35 categories and their subcategories that are excluded from the vocabulary used to generate the organism aspects. This list was compiled in September 2020 and is included in Appendix A.

Figure 5. Lemma filtering based on a taxonomy of concepts. The taxonomy shown is based on the WikiData taxonomy, excluding chemical compounds and anatomical structures, based on the lemmas excluded by Vandevenne et al. (Reference Vandevenne, Pieters and Duflou2016).
Finally, it is important to determine the number of organism aspects that can be generated, balancing ignoring nonsense aspects and leaving behind aspects that describe real functions. Vandevenne et al. (Reference Vandevenne, Verhaegen, Dewulf and Duflou2015) arbitrarily chose 300 aspects, however, the number has to be adjusted to the information contained in the dataset. The optimum is thus expected to be dependent on the number of biological publications present in the dataset.
To determine an upper limit on the number of organism aspects to generate, a parallel analysis using the real data and synthetic data based on the real dataset is used (Franklin et al., Reference Franklin, Gibson, Robertson, Pohlmann and Fralish1995). These synthetic data are generated based on the frequencies of the lemmas in the real data, but they are randomly spread over the different documents. This causes the synthetic data to represent documents which are indiscernible from noise but are drawn from the same distribution as the real data.
The generation of the organism aspects is based on a principal component analysis, where cumulatively each additional aspect captures more variance than the last. By comparing the variance captured in the organism aspect generated by the real data and the synthetic data, the cut-off point where the aspects start to capture noise is determined. When the corrected variance captured from the generated data is more than that of the real data (Jolliffe, Reference Jolliffe2002), the cut-off number of organism aspects is reached (Horn, Reference Horn1965; Buja and Eyuboglu, Reference Buja and Eyuboglu1992). This analysis is repeated several times to determine the 5% quantile where the cut-off is placed. For the current corpus of biological documents, this cut-off is determined to be at 3403 organism aspects.
Each organism aspect is loaded on a limited number of functional lemmas, which are used to represent the organism aspect in the biological space. The weighting of the lemmas is considered when determining the embedding by weighting the contribution of each lemma's vector. The matching between an engineering query and the organism aspects is performed based on the cosine distances between the translated query and the vectorial representations of the organism aspects.
By presenting the automatic selection method of the vocabulary that is relevant to represent the functionality in the biological literature and selecting the number of organism aspects that must be generated, this work contributes an automatic method of recalculating the organism aspects. Furthermore, this method could also be applied to the product aspects presented in Verhaegen et al. (Reference Verhaegen, D'hondt, Vandevenne, Dewulf, Duflou and Bernard2011) to keep them up to date.
Space alignment and representation
The 1500 most used lemmas in the Corpus of Contemporary American English (COCA) (Davies, Reference Davies2019) are used to align the engineering and biology space. While approximately 40% of the most used lemmas in engineering and biology are also present in the most used words overall, the remainder of the most used lemmas differ between both domains. This motivates the use of the independent COCA corpus (Davies, Reference Davies2019) to determine the lemmas used to determine the transformation between both models. The transformation matrix is determined using the procedure presented by Smith et al. (Reference Smith, Turban, Hamblin and Hammerla2017).
The assumption that these lemmas are mapped to the same context in both domains was verified by using a 20/80% test–train split where the test set was mapped with a precision@5 (Manning et al., Reference Manning, Raghavan and Schuetze2009) score near the same lemmas of 89% using the transformation trained on the train set.
The contributions mentioned above allow the translation system to be updated without manual intervention other than supplying the new source documents for both domains. This is important to keep up with the ever-changing language use. Delobelle et al. (Reference Delobelle, Winters and Berendt2022) for example, find that a 3-year-old language model underperforms on more recent benchmarks. However, this could be solved by adding new tokens to the model (reflecting the changing word use) and training the model with newer data.
Interface
The search method presented in this work returns multiple biological publications. As this can cause information overload on the processing engineer (Willocx et al., Reference Willocx, Ayali and Duflou2020), an interface based on the work of (Vandevenne et al., Reference Vandevenne, Verhaegen, Dewulf and Duflou2015) was added to the tool.
Vandevenne et al. (Reference Vandevenne, Verhaegen, Dewulf and Duflou2015) assumed that the publications on the same focus organism retrieved for a search query deal with the same biological strategy. To exploit this, the documents are clustered based on the detected focus organism and the location of the organism in the NCBI taxonomy (Schoch et al., Reference Schoch, Ciufo, Domrachev, Hotton, Kannan, Khovanskaya and Leipe2020). To avoid having too specific clusters with too few documents, the clustering per organism is performed on the family level. If a focus organism is specified more specifically, the taxonomy tree is climbed until the family level is reached. This groups the more specific levels of the tree together. For broad publications located higher in the taxonomy, for example, treating a whole class of organisms, the tree is not descended. This has the drawback of creating small clusters containing broad publications.
Furthermore, this clustering method does not group organisms that have arrived at the same working principle by parallel evolution, nor the working principles that are broadly adopted in the taxonomy (Willocxand Duflou, Reference Willocx and Duflou2021). Organisms that employ multiple working principles are also grouped together, as shown in the discussion and in Willocx et al. (Reference Willocx, Ayali and Duflou2020). However, for the current study, the authors feel that this is a better balance than not clustering the documents at all. An interesting future research direction can be to cluster the documents based on the working principle and not the focus organism.
In the interface, these clusters are represented by an image retrieved from Wikimedia Commons, in contrast to the previous work by Vandevenne et al. This image does not necessarily present the biological strategy employed by the organism, however, serves to give the designer a visual cue. Figure 6 presents the results of the search query for “reversible attachment surface”. By clicking on the image of the focus organisms, the designer is presented with the list of biological publications linked to the focus organism, along with links to read them.

Figure 6. Representation of the focus organisms each representing a cluster of documents retrieved for the search query for "reversible attachment surface". The organism images were retrieved from Wikimedia Commons under either a public domain or Creative Commons license.Footnote 1
Validation using existing biologically inspired designs
The proposed search tool is validated by performing a search for the functionality provided by known bio-inspired designs and comparing the output of the tool with the original inspiring organism. The resulting rank of the inspiring organism is reported in the search output for the different functional queries aiming for the functionality of the bio-inspired design. Note that the goal here is not to place those organisms directly at rank one: other strategies might also be (more) relevant for the targeted function. The original inspiring organism should be returned with a reasonable rank to assert that the proposed mapping method returns functionally relevant documents linked to known solutions.
As noted previously, the query is the essential function that is performed by the bio-inspired design. The essential function was determined after consulting (Nachtigall and Wisser, Reference Nachtigall and Wisser2015), which contains examples of bio-inspired design and explains the function that is taken from the biological system. From this explanation, one or two essential functions were distilled. To avoid overly specific queries biased toward the bio-inspired solutions, care was taken to keep the functional queries used in the validation simple. Table 2 presents the resulting essential functions that were extracted.
Table 2. A list of bio-inspired designs, their inspiring organisms, the extracted essential function, and the rank of the inspiring organism's cluster in the results for the given queries. Furthermore, the label of the AskNature functional category containing the strategy was also used as a query and the rank of the organism's cluster for this query is also reported.

Furthermore, the label of the functional category that contains the inspiring strategy in the AskNature database is also taken as a functional query. The functional labels used in the AskNature taxonomy are quite broad and were assigned by independent contributors (Deldin and Schuknecht, Reference Deldin, Schuknecht, Goel, McAdams and Stone2014). This makes the label of the functional categories an unbiased functional query. The resulting rank of using the label as a query is reported in Table 2.
The bio-inspired designs used in the validation were chosen for their prevalence as examples given when discussing bio-inspired design. As they have been used many times as an example of bio-inspired design, they stood the test of time. Table 2 lists the selected existing bio-inspired designs, their essential function, the inspiring organisms, and the rank of these organisms in the clustered search results.
The ranks of the inspiring organisms in most of the validation cases are within a reasonable number of organism clusters to be checked by the engineer. Again, the goal of the search tool is not to present the single best biological strategy but to offer a list of strategies that the designer can use to quickly select the strategy that resonates most with the problem at hand, causing them to quickly go through 10–15 groups of organisms.
The last column reports on the rank of the organism's cluster when using the relatively broad labels of the functional categories presented in the AskNature taxonomy. These ranks are well within the number of strategies already present in some categories in the AskNature database. Since a search using the taxonomy also requires the designer to evaluate an equal number of strategies, this is a reasonable result.
To illustrate the contents of the formed organism clusters and highlight the difficulties that still arise when processing this bio-inspiration, Tables 3 and 4 present the clusters formed for the butterfly family for the queries “modify color” and “display color”, respectively. These clusters were formed by grouping the documents based on the family of the focus organism, as described in the previous section. Here, it is noted that while some documents describe the coloration of butterflies and how to control it on a biological level, other documents also describe using colors to perform cooling and color vision in the focus organism. The grouping based on the focus organism mixes different working principles. Furthermore, the current presentation format of a list of documents requires a lot of processing for the designer to find out what documents apply to their design problem.
Table 3. Documents contained in cluster 10 retrieved for the query “modify color”

This cluster was formed based on the NCBI taxonomy.
Table 4. Documents contained in cluster 12 retrieved for the query “display color”

A further validation where the output of the FISh search tool is compared with the bio-inspiration found by an expert biologist and AskNature is presented in Willocx et al. (Reference Willocx, Ayali and Duflou2020). The results of that evaluation indicate that the FISh search tool does retrieve all of the biological strategies present in AskNature and proposed by the biologist, but that there is too much information for the designer to identify all of the relevant strategies on the first pass (Willocx et al., Reference Willocx, Ayali and Duflou2020). A second attempt at interpreting the documents did reveal the strategies mentioned in AskNature.
An interesting avenue to support the engineer to better identify the functional relevance of the biological strategy could consist of a summary generated from the biological documents in the organism cluster. By allowing the engineer to grasp the content quickly, the time investment for each strategy can be reduced.
Conclusion
The presented FISh system allows a free-text search with an engineering functional query in the biological literature. Each component can be updated automatically, allowing the system to be kept up to date with new engineering developments and biological publications.
Key contributions are the transformation between two domain-specific languages based on the most common English words, the automatic filtering of relevant lemmas and automatically determining the required number of organism aspects. Finally, the proposed system was validated using already existing biologically inspired designs.
Care should be taken to comply with all the different copyright restrictions, however, with the rise of open-access publishing with permissive licenses, the automated remixing of biological publications will open exciting avenues for bio-inspired design.
Outlook
As noted previously and in Willocx et al. (Reference Willocx, Ayali and Duflou2020), the search method returns a list of biological documents. While they are relevant to the query, this is often unclear to the designer due to the information overload of receiving too many documents. A future research goal is to process these documents further into a more manageable summary for the engineer. However, care should be taken to comply with the “No Derivatives” (ND) licence some papers are published under Willocx (Reference Willocx2021).
The developed method could also be applied to different domains. For example, palaeontology can be considered a source domain for other analogies, resulting in the recently coined Paleomimetics. Perricone et al. (Reference Perricone, Grun, Raia and Langella2022) describe a conceptual framework to transfer biomimetics to the palaeontology domain. This method still suffers from difficulties in identifying the organism from which the principles will be used. The authors propose to create a database consisting of extinct organisms, but with a corpus consisting of palaeontology publications, the proposed search system could be integrated and the time investment of creating a new database avoided.
Furthermore, with a focus on different innovations, we speculate that a transfer between the chemical engineering domain and the – here explicitly excluded – domain of biochemistry might yield some interesting results, certainly in the domain of catalysts.
Finally, the very recent developments in the field of chatbots powered by large language models open some interesting avenues for use in bio-inspired design. Despite the enormous potential, the currently available chatGPT is a black box model that might supply wrong answers (Kitamura, Reference Kitamura2023). While asking the model directly for an analogy risks running into a confident, but untrue answer, asking the model to extract the working principles from the retrieved documents is a more certain route. By carefully crafting the right prompt, the chatbot might be able to annotate the summary with a reference to the source of the idea. A similar approach is already in use in the perplexity.ai search engine (Srinivas, Reference Srinivas2022), which generates a short answer or summary annotated with references from the search results for a general search query.
Data availability statement
The code, DOI identifiers, and other links to the content used in this study are available upon reasonable request from the corresponding author. The full-text content used to generate the models presented in this study is available from their respective publishers. Restrictions apply to the availability of these data, which were used under licence for this study.
Acknowledgements
This work was supported by the Fraunhofer-Gesellschaft Think Tank project BioMANU II.
Competing interests
The authors declare no competing interests.
Mart Willocx is a researcher at the Centre for Industrial Management at KU Leuven. After finishing his studies as a mechanical engineer, he was promptly convinced by the inspiring genius of nature. His current research focuses on bridging the gap between engineering and biology, giving mechanical engineers the tools to find relevant biological strategies.
Joost R. Duflou is a Professor in the Department of Mechanical Engineering at KU Leuven. He has master degrees in architectural and electromechanical engineering and a PhD in engineering from KU Leuven. He is a member of CIRP and has been published in over 200 international publications. His principal research activities are in the field of design support methods and methodologies, with special attention for systematic innovation, ecodesign, and life cycle engineering.
Appendix A
To automate the generation of the organism aspects, a rule-based approach is presented based on the location of the lemma in the concept hierarchy in WikiData. Based on the target of identifying strategies and the manual annotation by Vandevenne, the entities presented in Table A1 and their descendants are excluded from the organism aspects. This list was compiled in September 2020.
Table A1. List of entities which along with their descendants will be excluded in the generation of the organism aspects. This list was built based on the contents available in WikiData in September 2020.














