Article contents
Assessing priorities for conservation in Tuscan cattle breeds using microsatellites
Published online by Cambridge University Press: 22 August 2011
Abstract
Preservation of rare genetic stocks requires assessment of within-population genetic diversity and between-population differentiation to make inferences on their degree of uniqueness. A total of 194 Tuscan cattle (44 Calvana, 35 Chianina, 25 Garfagnina, 31 Maremmana, 31 Mucca Pisana and 28 Pontremolese) individuals were genotyped for 34 microsatellite markers. Moreover, 56 samples belonging to Argentinean Creole and Asturiana de la Montaña cattle breeds were used as an outgroup. Genetic diversity was quantified in terms of molecular coancestry and allelic richness. STRUCTURE analyses showed that the Tuscan breeds have well-differentiated genetic backgrounds, except for the Calvana and Chianina breeds, which share the same genetic ancestry. The between-breed Nei's minimum distance (Dm) matrices showed that the pair Calvana–Chianina was less differentiated (0.049 ± 0.006). The endangered Tuscan breeds (Calvana, Garfagnina, Mucca Pisana and Pontremolese) made null or negative contributions to diversity, except for the Mucca Pisana contribution to allelic richness (CT = 1.8%). The Calvana breed made null or negative within-breed contributions (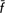 W = 0.0%; CW = −0.4%). The Garfagnina and Pontremolese breeds made positive contributions to between-breed diversity but negative and high within-breed contributions, thus suggesting population bottleneck with allelic losses and increase of homozygosity in the population. Exclusion of the four endangered Tuscan cattle breeds did not result in losses in genetic diversity (
W = 0.0%; CW = −0.4%). The Garfagnina and Pontremolese breeds made positive contributions to between-breed diversity but negative and high within-breed contributions, thus suggesting population bottleneck with allelic losses and increase of homozygosity in the population. Exclusion of the four endangered Tuscan cattle breeds did not result in losses in genetic diversity (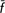 T = −0.7%; CT = −1.2%), whereas exclusion of the non-endangered breeds (Chianina and Maremmana) did (
T = −0.7%; CT = −1.2%), whereas exclusion of the non-endangered breeds (Chianina and Maremmana) did (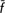 T = 2.1%; CT = 3.9%); the simple exclusion of the Calvana breed from the former group led to losses in genetic diversity (
T = 2.1%; CT = 3.9%); the simple exclusion of the Calvana breed from the former group led to losses in genetic diversity (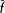 T = 0.47%; CT = 2.34%), indicating a diverse significance for this breed. We showed how quantifying both within-population diversity and between-population differentiation in terms of allelic frequencies and allelic richness provides different and complementary information on the genetic backgrounds assessed and may help to implement priorities and strategies for conservation in livestock.
T = 0.47%; CT = 2.34%), indicating a diverse significance for this breed. We showed how quantifying both within-population diversity and between-population differentiation in terms of allelic frequencies and allelic richness provides different and complementary information on the genetic backgrounds assessed and may help to implement priorities and strategies for conservation in livestock.
Keywords
Information
- Type
- Full Paper
- Information
- Copyright
- Copyright © The Animal Consortium 2011
Footnotes
These authors contributed equally to this work and shall share the first authorship.
References
- 13
- Cited by

