Cluster of differentiation 36 (CD36) is a membrane-bound protein expressed on several cell types, and regulates uptake of long-chain fatty acids across the membrane as a transporter(Reference Newman, Haryono and Keast1,Reference Bonen, Campbell and Benton2) . Recent studies in humans and animals have proposed that CD36 is expressed in taste bud cells from circumvallate foliate and fungiform papillae(Reference Simons, Kummer and Luiken3,Reference Liu, Costanzo and Evans4) , suggesting that CD36 may play an important role in oral fat perception (OFP) and individual preferences for fatty foods. Previous studies in different ethnic subpopulations have revealed that CD36 SNP were significantly associated with CD36 mRNA expression level(Reference Love-Gregory, Sherva and Schappe5) and threshold values to detect fat content(Reference Pepino, Love-Gregory and Klein6–Reference Plesník, Šerý and Khan13). Following studies on CD36 genetic variants and OFP, the associations between CD36 polymorphisms and dietary intake of fatty acids have gained a great deal of attention. Although prior studies have focused on populations of Europeans(Reference Liu, Costanzo and Evans4) and Latin Americans(Reference Pioltine, de Melo and Santos14–Reference Ramos-Lopez, Panduro and Martinez-Lopez16), such associations in Asian populations remain unclear.
Along with the nutritional perspective, many CD36 SNP have been reported to be associated with various diseases and metabolic abnormalities in European and African American populations(Reference Love-Gregory, Sherva and Schappe5,Reference Keller, Liang and Sakimura8,Reference Mrizak, Šerý and Plesnik10,Reference Sayed, Šerý and Plesnik11,Reference Plesník, Šerý and Khan13–Reference Ramos-Lopez, Roman and Martinez-Lopez15,Reference Bokor, legry and Meirhaeghe17–Reference Ma, Bacci and Mlynarski24) . Based on these previous studies, recent case–control studies have investigated the associations of CD36 polymorphisms with ischaemic stroke(Reference Zang, Zang and Wang25,Reference Gong, Liao and Liu26) and CHD in Asian populations(Reference Zang, Ling and Deng27–Reference Yun, Song and Song29). Given the previous studies of CD36 polymorphisms, we hypothesised that fat perception based on CD36 polymorphisms could be an important factor for metabolic profile and disease onset in human. Therefore, a comprehensive analysis is needed to elucidate individual differences in nutritional intake and metabolic indices based on CD36 polymorphisms.
In the present study, two candidate SNP in the CD36 gene (rs1761667 and rs1527483) were selected based on previous studies that had identified significant associations with OFP(Reference Dauoudi, Plensnik and Sayed7). We aimed to investigate the associations between the genotypes of these two CD36 SNP and dietary fat intake and metabolic phenotypes in a Japanese population.
Materials and methods
Study subjects
A total of 525 community-dwelling individuals participated in a health check-up carried out in Hokkaido, Japan, at the end of August 2015. This cross-sectional study was conducted based on clinical information and lifestyle data collected during the health check-up. Twenty-nine participants were excluded, due to incomplete lifestyle information in the questionnaire (n 3) or lack of informed consent to participate in the present study (n 26). Genotyping was unable to be successfully performed in another individual, who was therefore excluded. As a result, a total of 495 residents (228 men and 267 women) were analysed. The study protocol was approved by the Ethics Committee at Nagoya University School of Medicine.
Data collection
Anthropometric indices (height, weight and waist circumference) were measured during the health check-up to determine BMI (kg/m2) and obesity status. Obesity was defined as waist circumference ≥ 85 cm in men or ≥ 90 cm in women. Blood pressure was measured using an automatic sphygmomanometer. Mean blood pressure (MBP) was calculated using systolic blood pressure (SBP) and diastolic blood pressure (DBP) via the following formula: MBP = DBP + (SBP – DBP)/3. Hypertension was defined as SBP ≥ 130 mmHg and/or DBP ≥ 85 mmHg, or use of antihypertensive medications. Participants were instructed to fast overnight. Fasting blood samples were collected and centrifuged within an hour of sampling. Biochemical analyses of blood samples were performed in the laboratory of Yakumo General Hospital. Diabetes mellitus was defined as fasting blood glucose ≥ 100 mg/dl (5·55 mmol/l) or use of medications for diabetes mellitus. Dyslipidaemia was defined as TAG ≥ 150 mg/dl (1·69 mmol/l), HDL-cholesterol ≤ 40 mg/dl (1·03 mmol/l) or use of medications for dyslipidaemia. The self-reported questionnaire in this study included questions on medical history and lifestyle habits of participants, including smoking status (current, former or never), alcohol consumption (current, former or never) and exercise habit (almost none, 1–2 h/week, 3–4 h/week or ≥ 5 h/week). Dietary intake of energy, total fat, SFA, MUFA, PUFA, n-3 PUFA and n-6 PUFA was estimated using a valid and reliable short FFQ(Reference Tokudome, Goto and Imaeda30–Reference Imaeda, Goto and Tokudome33). Energy-adjusted nutrient intake using the residual method was utilised in our analyses to avoid effects of body size and weight on nutritional intake(Reference Willett, Howe and Kushi34). Because all items used for dietary fat intake showed positively skewed distributions, logarithmically transformed values were applied in our statistical analyses.
DNA collection and genotyping
Individual whole blood samples (350 μl) were collected at the health check-up and stored at 4°C until DNA extraction was performed. DNA was automatically extracted from the whole blood sample using a commercial kit and the BioRobot M48 Workstation (QIAGEN). In the genotyping step, we used PCR with confronting two-pair primers (PCR-CTPP) method, which is suitable for manually genotyping a number of samples in an epidemiological study in terms of running time and cost. To explain the PCR-CTPP method briefly, two pairs of allele specific primers (four primers: forward 1 (F1) and reverse 1 (R1) for X allele and F2 and R2 for Y allele) with different lengths are used. As shown in Supplementary Fig. S1, a PCR product with a length of a-bp, including X allele, is amplified between F1 and R1, although a product with a length of b-bp, including Y allele, is amplified between F2 and R2. A common band with a length of c-bp (a + b – 1) is amplified between F1 and R2. Therefore, a- and c-bp bands are observed in the XX genotype. Similarly, b- and c-bp bands are observed in the YY genotype. For the heterozygotes, all three bands are observed. The length of PCR products (a-bp and b-bp) should be different enough to distinguish by electrophoresis gel. Details and applications of this genotyping method have been reported elsewhere(Reference Hamajima, Saito and Matsuo35,Reference Hamajima36) . PCR primers for genotyping rs1761667 were: F1, 5’-TTG CCC TTT TTC CAT CTT CTG TGA-3’ and R1, 5’-CCA GGC TTT GAG CAT GGT-3’ for amplifying the A allele (155 base pairs (bp)); and F2, 5’-TTC ATC TTT GCA TGC CAG CG-3’ and R2, 5’-AGT CCA AGC TTC TAT CCT TCC TA-3’ for amplifying the G allele (341 bp). Primers for rs1527483 were: F1, 5’-TTC TGT ATG CAA GTC CTG ATG TT-3’ and R1, 5’-TCA CTT CAT AAA CCA TAG GAA GAA ATC-3’ for amplifying the C allele (259 bp); and F2: 5’-TCT TGA AAG TTA CTG AAA CTT AGG TCA-3’ and R2: 5’-TGC CCA ATC GAA TGT CAA-3’ for amplifying T allele (422 bp). These primers were obtained from Hokkaido System Science Co., Ltd. A total volume of 26 μl of PCR mixture comprising 17·4 μl of distilled water, 2·5 μl of 10× PCR Buffer (including 15 mm MgCl2, 2·0 μl dNTPs, 0·5 μl of each primer, 0·1 μl AmpliTaq Gold (Thermo Fisher Scientific Japan)) and 2·0 μl of template DNA was used to amplify specific DNA regions for rs1761667 and rs1527483. PCR amplification was performed using the Veriti Thermal Cycler (Applied Biosystems). Amplification for rs1761667 and rs1527483 was initiated with pre-denaturalisation at 95°C for 10 min, followed by 30 cycles of denaturalisation at 95°C for 1 min, annealing at 61°C (rs1761667) or 60°C (rs1527483) for 1 min and extension at 72°C for 1 min and finalised by post-extension at 72°C for 5 min. A 2 % agarose gel with 2 μl/100 ml of ethidium bromide was used to visualise all PCR products and determine the genotypes at each locus. Representative gels for the genotyping of rs1761667 and rs1527483 are shown in Fig. 1(a) and (b), respectively.
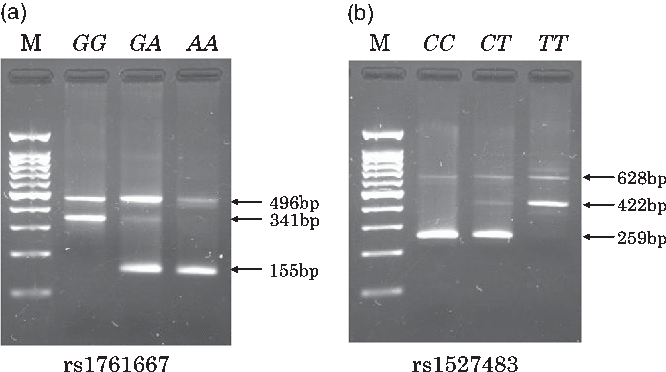
Fig. 1. Typical gels for genotyping the two cluster of differentiation 36 (CD36) polymorphisms (rs1761667 and rs1527483). Lane M contains a 100-bp ladder. For rs1761667 (a), lane GG, 341 bp band; lane GA, 341- and 155-bp bands; lane AA, 155-bp band. For 1527483 (b), lane CC, 259-bp band; lane CT, 259- and 422-bp bands; lane TT, 422-bp band.
Statistical analysis
The primary purpose of this study was to detect differences in dietary fat intake and metabolic profiles between three genotypes in rs1761667 and rs1527483. The required sample size for our analysis was calculated using the R package of pwr. For a type I error rate (α) of 0·05, the total sample size of 159 participants offered 80 % power (1 – β) to detect a moderate effect size (f = 0·25) of genetic variants on metabolic profiles and dietary fat intake(Reference Faul, Erdfelder and Lang37). Although normally distributed variables are expressed as mean values and standard deviations, TAG, HDL-cholesterol and all variables for dietary fat intake are presented as medians and 1st–3rd quartiles because of the skewed distributions. Fisher’s exact test was used to confirm the consistency of Hardy–Weinberg equilibrium in this population(Reference Hardy38,Reference Weinberg39) . Associations between the genotypes of two CD36 SNP, dietary fat intake and metabolic profiles were assessed using multiple linear regression analysis. Multiple logistic regression analysis was performed to estimate the OR for metabolic disorders, according to the genotypes. These regression analyses were performed after adjusting for sex and age and all statistical analyses were performed using R version 3.5.0 (R Foundation). Values of P < 0·05 were considered statistically significant.
Results
Basic characteristics of participants
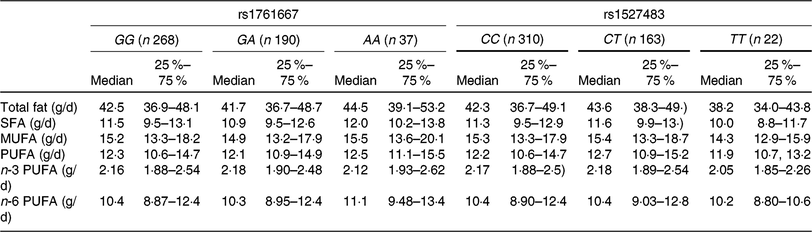
The basic clinical profiles of study subjects are summarised by the three genotypes of rs1761667 and rs1527483 (Table 1). No significant differences were observed between the genotypes of each polymorphism. The minor allele frequency (MAF) of rs1761667 and rs1527483 was 0·267 and 0·209, respectively, which was consistent with the result from a Japanese public database (Japan Multi Omics Reference Panel: JMorp) (MAF = 0·275 and 0·220, respectively)(Reference Tadaka, Saigusa and Motoike40). Genotype distributions of each SNP in this population were in accordance with Hardy–Weinberg equilibrium (P = 0·73 and 0·89, respectively). Table 2 shows dietary fat and fatty acid intake, according to the genotypes in the present study.
Table 1. Clinical characteristics of participants (n 495) according to the three genotypes of cluster of differentiation 36 (CD36) SNP
(Mean values and standard deviations; numbers of participants and percentages; medians and interquartile ranges)
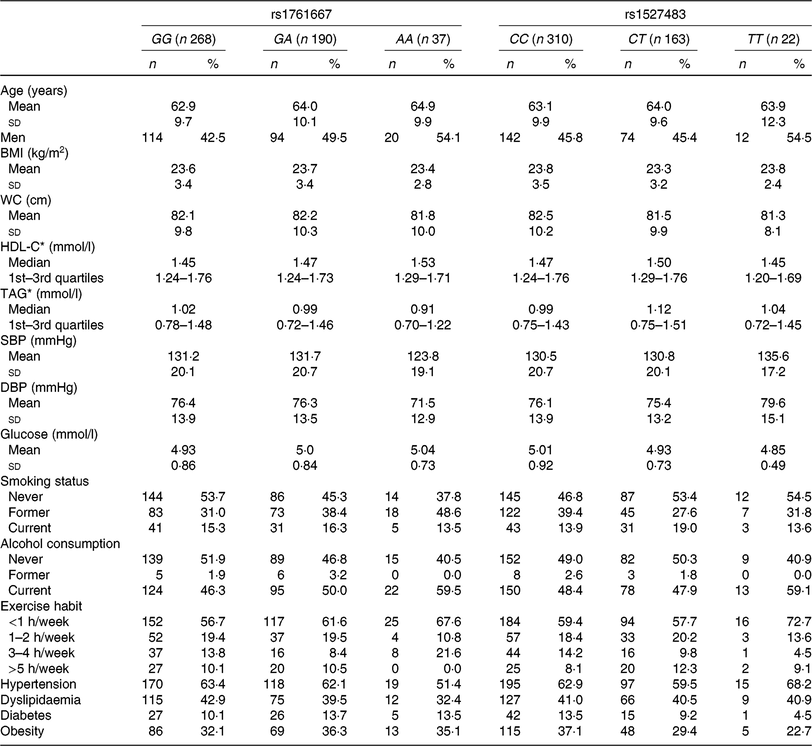
WC, waist circumference; HDL-C, HDL-cholesterol; SBP, systolic blood pressure; DBP, diastolic blood pressure.
* HDL-C and TAG are presented as medians and 1st–3rd quartiles because of skewed distributions.
Table 2. Dietary fat and fatty acid intakes of participants according to the three genotypes of cluster of differentiation 36 (CD36) SNP
(Medians and interquartile ranges)
Genotypes of two CD36 polymorphisms and dietary fat intake
Table 3 shows the results of adjusted linear regression analyses for associations between three genotypes of two CD36 polymorphisms (rs1761667 and rs1527483) and dietary fat intake. Individuals with the AA genotype showed higher fat and MUFA intake (standardised β = 0·110 and 0·087, P = 0·01 and 0·05, respectively) compared with the GG and GA genotypes, whereas no significant associations between the genotypes of rs1761667 and intake of other fatty acids were observed in the present study. For rs1527483, SFA intake in the TT genotype was significantly lower compared with the CC and CT genotypes (standardised β = −0·087, P = 0·05, respectively). However, no significant association was observed between the genotypes of rs1527483 and intake of other dietary fat.
Table 3. Multiple linear regression analysis* for the association between two cluster of differentiation 36 (CD36) SNP and dietary fat intake
(β-Coefficients and 95 % confidence intervals)
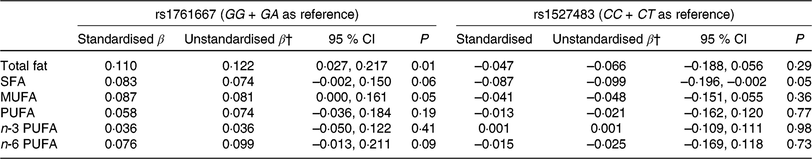
* Adjusted for sex and age.
† The β-coefficient indicates an increase/a decrease of dietary fat intake (g/d) for the AA (rs1761667) or TT genotype (rs1527483).
Genotypes of two CD36 polymorphisms and metabolic phenotypes
In adjusted linear regression analyses, the AA genotype of rs1761667 was significantly associated with 7·0 mmHg lower MBP compared with the GG genotype (standardised β = −0·123, P = 0·02), while the GA genotype was not significantly associated with lower MBP (standardised β = −0·053, P = 0·32) (Table 4). No significant association was observed between the genotypes of CD36 polymorphisms and other metabolic profiles (Supplementary Tables S1 and S2). Multivariable logistic regression also showed that the adjusted OR for hypertension was marginally lower in the AA genotype in rs1761667 than in the GG genotype (OR = 0·49, 95 % CI 0·24, 1·02, P = 0·06), but not in the GA genotype (OR = 0·85, 95 % CI 0·57, 1·28, P = 0·44) (Table 5). No significant associations between hypertension and the genotypes of rs1527483 were observed in our logistic regression analysis. Associations between the genotypes of CD36 polymorphisms and other metabolic disorders can be seen in Supplementary Tables S3 and S4.
Table 4. Multiple linear regression analysis* for associations between the genotypes of cluster of differentiation 36 (CD36) SNP and mean blood pressure (n 349)
(β-Coefficients and 95 % confidence intervals)

* This analysis was performed after excluding those who took antihypertension medications (n 146).
† The β-coefficient indicates an increase/a decrease of blood pressure (mmHg) in each genotype compared with the GG (rs1761667) or CC (rs1527483) genotype.
Table 5. Multiple logistic regression analysis for the associations between the genotypes of two cluster of differentiation 36 (CD36) SNP and hypertension (HT)
(Odds ratios and 95 % confidence intervals)

Discussion
The primary goal of this study was to elucidate the associations of the genotypes of CD36 polymorphisms with dietary fat intake and metabolic phenotype. We found that the AA genotype of rs1761667 in the CD36 was significantly associated with higher fat and MUFA intake and lower blood pressure in the Japanese population.
The two polymorphisms (rs1761667 and rs1527483) examined in the present study are non-coding intronic SNP located at chromosome 7, and do not cause structural changes in the CD36 protein. The AA genotype of rs1761667 is known to be involved in reduced CD36 mRNA expression(Reference Love-Gregory, Sherva and Schappe5). Furthermore, previous studies in humans and experimental animals have reported CD36 expression level as significantly associated with individual differences in OFP(Reference Pepino, Love-Gregory and Klein6). According to these results, rs1761667 is assumed to play an important role in human OFP. Keller et al.(Reference Keller, Liang and Sakimura8) originally reported that African American participants with the AA genotype of rs1761667 had a preference for adding fats and oils. In a similar finding described by Pepino et al.(Reference Pepino, Love-Gregory and Klein6), the threshold for detecting oleic acid, one of the major MUFA, was eightfold higher in a Caucasian population with the GG genotype than in those with the AA genotype(Reference Love-Gregory, Sherva and Schappe5). Obese women with the AA genotype exhibited a lower sensitivity for oleic acid compared with those with the G allele(Reference Mrizak, Šerý and Plesnik10). Such results have suggested that individuals with the AA genotype of rs1761667 have reduced sensitivity to fat content, which can lead to increased fat intake. In fact, Hispanic subjects with the AA genotype of rs1761667 showed higher fat intake compared with subjects with the GG genotype(Reference Ramos-Lopez, Roman and Martinez-Lopez15), although few studies have reported no significant association between the genotypes of rs1761667 and dietary fat intake in Asian populations. Given these previous studies, greater dietary intake of fat and MUFA in the AA genotype of rs1761667 appears reasonable. With regard to rs1527483, we found no significant associations with dietary fat and fatty acid intake. In a Malaysian population that consisted mostly of ethnic Chinese, individuals with the TT genotype perceived significantly greater fat content independent of fat concentrations than C allele carriers(Reference Ong, Tan and Say12). One study in African Americans also found that participants with the minor allele of rs1527483 (T) rated fat content higher than those with the CC genotype(Reference Keller, Liang and Sakimura8). A recent study among Czech young adults reported that the CC genotype of rs1527483 had higher sensitivity for linoleic acid detection than the CT and TT genotypes(Reference Plesník, Šerý and Khan13). On the other hand, the genotypes of rs1527483 was not significantly associated with the capability in a Caucasian population to detect oleic acid(Reference Mellis, Sollai and Muroni9). Compared with rs1761667, the association of rs1527483 with OFP seems invalid, and comprehensive studies are needed to better characterise the relationship between the genotypes of rs1527483, OFP and dietary fat intake in either ethnic group.
Previous studies in rodents found that Cd36 genetic background was linked to the regulation of blood pressure(Reference Aitman, Glazier and Wallace41,Reference Pravenec, Zidek and Simakova42) . Abnormal expression of CD36 in the kidney was involved in the genetic basis of hypertension(Reference Pravenec, Churcill and Churcill43). In humans, CD36 polymorphisms have been reported to be associated with CVD such as ischaemic stroke(Reference Zang, Zang and Wang25,Reference Gong, Liao and Liu26) , CHD(Reference Zang, Ling and Deng27,Reference Yun, Song and Song29) and essential hypertension(Reference Liu, Meng and Yang28) in Asians. Such findings supported consistent evidence of a significant association between CD36 and CVD in both humans and animals. In the present study, the AA genotype of rs1761667 had a lower blood pressure than the GG genotype, but no significant relationship was observed in the genotypes of rs1527483. Taking our findings together, the AA genotype of rs1761667 was associated with higher dietary intake of MUFA, which may be associated with decreased blood pressure and lower risk of CVD(Reference Gillman, Cupples and Millen44,Reference Rasmussen, Thomsen and Hansen45) . Although significant associations between the genotypes of CD36 polymorphisms (rs1761667 and rs1527483) and anthropometric and lipid indices have been observed in many studies in Caucasian(Reference Bokor, legry and Meirhaeghe17,Reference Choquet, Labrune and De Graeve18) and African American populations(Reference Heni, Müssig and Machicao21,Reference Boghdady, Arafa and Sabet22) , we did not find any such associations among community-dwelling Japanese adults in Japan. Our result is in line with previous studies in Asians, showing no significant relationship between the two polymorphisms and those phenotypes(Reference Ong, Tan and Say12,Reference Zang, Zang and Wang25) . These discrepancies could be attributable to different ethnicities, which may indicate that rather than obesity or dyslipidaemia, hypertension based on the genotypes of rs1761667 plays an important role in CVD onset in Asian populations.
The strength of this study was the comprehensive analysis used to examine the associations of CD36 polymorphisms with individual dietary intake and clinical profiles in an Asian population. However, several limitations must also be considered. The key consideration in this study was that CD36 deficiency is relatively common in Asian populations (2–3 %) but not in Caucasians (less than 0·3 %)(Reference Kashiwagi, Tomiyama and Honda46 – Reference Curtis and Aster48). CD36 deficiency has been well investigated for several pathophysiological conditions(Reference Yanai, Chiba and Morimoto49 – Reference Furuhashi, Ura and Nakata52). One study in a Japanese population demonstrated that patients with CD36 deficiency showed an atherogenic effect(Reference Yuasa-Kawase, Masuda and Yamashita53). Future work in Asian populations is thus needed to clarify the effects of CD36 deficiency on blood pressure and hypertension. In our study, subjects were genetically homogeneous and community-dwelling people living within a single administrative area. The sample size was relatively small for exploring the influences of genetic variants on several indices, but the estimated sample size seemed sufficient. Our results must therefore be confirmed in other Asian populations with larger sample sizes. Furthermore, transethnic studies are warranted to clarify the molecular mechanism underlying the associations between CD36 polymorphisms and diseases across different ethnicities.
In conclusion, we found that the AA genotype of rs1761667 was significantly associated with higher dietary fat and MUFA intake and a lower blood pressure compared with the GG genotype. This result could shed light on the individual differences in dietary fat intake based on the genotypes of CD36 polymorphisms and its effect on blood pressure in an Asian population.
Acknowledgements
The authors thank Mrs. Yoko Mitsuda and Mrs. Keiko Shibata from the Department of Preventive Medicine at Nagoya University Graduate School of Medicine for their technical assistance. The authors are also grateful to Mr. Kensuke Suzuki at the Department of Economics, Pennsylvania State University, for his expert advice in statistical method. We are also grateful to the participants and staff of the health examination programme for residents of Yakumo Town, Hokkaido, Japan.
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
R. F. drafted the article and performed statistical analyses; R. F., A. H., K. S., K. W. and T. K. contributed for data acquisition; N. H. and K. W. were responsible for the conception and design; N. I. and C. G. contributed to the interpretation of results. All the authors reviewed the manuscript.
The authors declare no conflicts of interest with respect to this study.
Supplementary material
For supplementary material/s referred to in this article, please visit https://doi.org/10.1017/S0007114519000679









