Most cognitive functioning declines with advancing age, and identifying the risk factors for age-related cognitive decline has become a topic of increasing interest. Previous research has indicated that a low folate status might increase the risk of cognitive impairment(Reference Kado, Karlamangla and Huang1). However, the potential biological mechanisms underlying this relationship remain to be elucidated.
One possible mechanism that might explain the involvement of folate status in cognitive performance is DNA methylation, which refers to the epigenetic modification of gene expression by the addition of methyl groups to cytosine residues in DNA(Reference Costello and Plass2). Recent studies on animals have suggested that DNA methylation may be involved in regulating synaptic plasticity in hippocampal neurons, thereby influencing learning and memory processes(Reference Miller and Sweatt3, Reference Levenson, Roth and Lubin4). In humans, both hypomethylation and hypermethylation of DNA have been implicated in psychiatric disorders, including schizophrenia(Reference Costa, Chen and Davis5); neurodegenerative disorders, such as Alzheimer's disease(Reference Gräff and Mansuy6); and syndromes associated with mental retardation, e.g. Fragile X syndrome(Reference Tsankova, Renthal and Kumar7).
Methyl groups for DNA methylation are provided by the universal methyl donor S-adenosylmethionine, which is synthesised from methionine(Reference Ulrey, Liu and Andrews8). Folic acid may increase the availability of S-adenosylmethionine by promoting the conversion of homocysteine into methionine, thereby influencing DNA methylation status(Reference Niculescu and Zeisel9). Indeed, an intervention study in older women has shown that low dietary folate intake was associated with global DNA hypomethylation, which could be reversed by folate repletion(Reference Jacob, Gretz and Taylor10). In addition, the common 5,10-methylenetetrahydrofolate reductase (MTHFR) 677 C → T polymorphism, which mimics folate deficiency by impairing the conversion of homocysteine into methionine, has also been related to DNA hypomethylation(Reference Friso, Choi and Girelli11).
Given the role of folate metabolism in generating methyl donors for methylation processes, and the involvement of DNA methylation in brain functioning, it seems reasonable to hypothesise that folate status might influence cognitive functioning by exerting effects on DNA methylation. However, the association between DNA methylation status and cognitive performance in the general population has not yet been investigated. Therefore, the present study examined whether leucocyte global DNA methylation was associated with cognitive performance in healthy older adults.
Methods
Study population
The present study was performed using data from the Folic Acid and Carotid Intima-Media Thickness (FACIT) study, a randomised, double-blinded, placebo-controlled trial, originally designed to investigate the effects of 3-year folic acid supplementation on the risk of CVD(Reference Durga, Van Boxtel and Schouten12). The study population consisted of 818 healthy men and women, aged 50–70 years at baseline. A detailed description of the study design and the selection of participants can be found elsewhere(Reference Durga, Van Boxtel and Schouten12).
Venous blood samples were collected at baseline. Leucocyte global DNA methylation was determined in a sub-sample of 216 participants. First, the study population was stratified by MTHFR 677 C → T genotype, to ensure equal distribution of MTHFR 677 C → T genotypes in the final sample. Thereafter, participants in the folate treatment group were randomly selected from the three strata and were individually matched with participants in the placebo group on the variables age, sex, smoking status and MTHFR 677 C → T genotype, as these variables may influence DNA methylation(Reference Friso, Choi and Girelli11, Reference Drinkwater, Blake and Morley13, Reference Brait, Ford and Papaiahgari14). Some samples were not measured due to human error in sample retrieval. Valid DNA methylation data were available for 111 participants in the treatment group and 105 participants in the placebo group. As valid data on cognitive functioning were lacking for one participant in the folate treatment group, the final study sample consisted of 215 individuals.
This study was conducted according to the guidelines laid down in the Declaration of Helsinki and all procedures involving human participants were approved by the Medical Ethics Committee of Wageningen University. Written informed consent was obtained from all the participants.
Cognitive functioning
Cognitive functioning on the domains of memory, sensorimotor speed, complex speed, information processing speed and word fluency was assessed by a comprehensive neuropsychological test battery, consisting of the Visual Verbal Word Learning Task, the Stroop Colour-Word Interference Test, the Concept Shifting Test, the Letter–Digit Substitution Test and the Verbal Fluency Test, as described earlier(Reference Durga, Van Boxtel and Schouten12).
DNA methylation status and genotyping
Genomic DNA was isolated from peripheral blood leucocytes at baseline. Global DNA methylation was determined by liquid chromatography-MS/MS, as described previously(Reference Kok, Smith and Barto15). Genomic DNA methylation status was calculated as the percentage of methylated cytosine to total cytosine (mCyt/tCyt) using the following formula(Reference Kok, Smith and Barto15): (nmol mCyt/(nmol mCyt+nmol Cyt)) × 100 %.
MTHFR 677 C → T genotype was determined by PCR with restriction fragment length polymorphism analysis with HinfI(Reference Frosst, Blom and Milos16), and was defined as a common variant (CC or CT genotype) or a rare variant (TT genotype).
Blood measurements
Fasting venous blood samples were collected at baseline, processed directly and stored at − 80°C. Serum folate was measured using a chemiluminescent immunoassay (Diagnostic Products Corporation, Los Angeles, CA, USA). Erythrocyte folate was determined in duplicate and the average was taken to reduce measurement error. Erythrocyte folate concentrations were calculated using the following formula: (unadjusted erythrocyte folate/haematocrit) − ((1 − haematocrit)/haematocrit) × serum folate. Plasma total homocysteine was determined by HPLC and fluorimetric detection, as described previously(Reference Ubbink, Hayward-Vermaak and Bissbort17).
Demographic and lifestyle variables
Level of education (low/middle/high) was measured by classifying formal schooling according to the Dutch educational system(Reference De Bie18). Alcohol consumption (g/d) and current smoking (yes/no) were ascertained by means of self-report questionnaires. BMI (kg/m2) was calculated from height and weight, and physical activity was estimated using the Physical Activity Scale for the Elderly(Reference Washburn, Smith and Jette19).
Statistical analysis
Normality of data distributions was ascertained by normal P–P plots. Baseline data were used to assess the cross-sectional associations between global DNA methylation status and cognitive functioning. Independent samples t tests and univariate ANOVA were carried out to examine whether DNA methylation status varied according to sex, level of education, smoking status or MTHFR 677 C → T genotype.
Hierarchical linear regression analyses were performed for DNA methylation status in relation to each of the five cognitive performance indices. The analyses were corrected for sociodemographic and lifestyle variables that were considered potential confounders, i.e. age, sex, level of education, alcohol consumption, smoking status, physical activity, erythrocyte folate concentration and MTHFR 677 C → T genotype(Reference Friso, Choi and Girelli11, Reference Drinkwater, Blake and Morley13, Reference Brait, Ford and Papaiahgari14, Reference Yuasa, Nagasaki and Akiyama20).
To investigate the possibility of a non-linear relationship between global DNA methylation and cognitive performance, we repeated the analyses with the quadratic term for DNA methylation status as the independent variable, adjusted for covariates and the linear term for DNA methylation status. The quadratic term for DNA methylation status was expressed as the residuals of regressing (DNA methylation)2 on DNA methylation, i.e. the quadratic component that is orthogonal to the linear component of DNA methylation.
The statistical power for detecting associations between DNA methylation status and each of the dependent variables, assuming a small effect size of f 2 = 0·03, was 0·80. Statistical differences were considered significant at P < 0·05. All analyses were performed using SPSS 16.0 (SPSS, Inc., Chicago, IL, USA).
Results
Table 1 summarises the characteristics of the study population. The percentage of methylated cytosine to total cytosine residues in leucocyte DNA ranged from 4·0 to 5·6 %, which was comparable to the range reported by other population-based studies(Reference Friso, Choi and Girelli11, Reference Kok, Smith and Barto15). The extent of global DNA methylation did not vary according to sex (t = − 1·285, P = 0·200), level of education (F = 0·611, P = 0·544), smoking status (t = 1·611, P = 0·109) or MTHFR 677 C → T genotype (t = − 0·907, P = 0·365).
Table 1 Characteristics of the study population
(Mean values and 95 % confidence intervals)
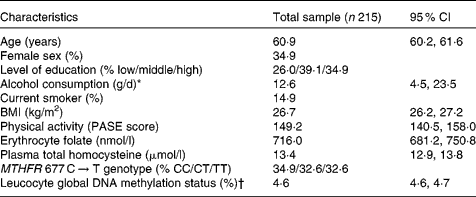
PASE, Physical Activity Scale for the Elderly; MTHFR, 5,10-methylenetetrahydrofolate reductase.
* Median (interquartile range) is given because of skewed data distribution.
† Percentage of methylated to total cytosine.
Hierarchical linear regression analyses corrected for age, sex, level of education, alcohol consumption, smoking status, physical activity, erythrocyte folate concentration and MTHFR 677 C → T genotype did not reveal any significant associations between leucocyte global DNA methylation and cognitive performance on any of the domains measured (Table 2). In addition, repeating the analyses with the quadratic term for DNA methylation status as the independent variable did not yield any significant results (data not shown), implying that global DNA methylation did not show a non-linear relationship with cognitive performance.
Table 2 Cross-sectional associations between leucocyte global DNA methylation and cognitive performance in older adults
(Regression coefficients and 95 % confidence intervals)
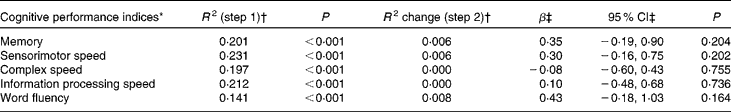
* Cognitive performance indices are expressed as Z-scores.
† R 2 represents the proportion of explained variance and R 2 change represents the change in the proportion of explained variance after each step in hierarchical linear regression analyses. The covariates age, sex, level of education, alcohol consumption, smoking status, physical activity, erythrocyte folate concentration and 5,10-methylenetetrahydrofolate reductase 677 C → T genotype were entered in step 1 and DNA methylation status in step 2.
‡ Unstandardised regression coefficient and 95 % CI for DNA methylation status in step 2.
Discussion
The present study did not support the hypothesis that individual variation in cognitive functioning in older adults might be related to the extent of leucocyte global DNA methylation.
Although there are no previous studies investigating the relationship between global DNA methylation and cognitive functioning in healthy human subjects, aberrant DNA methylation has been implicated in neurodevelopmental disorders(Reference Tsankova, Renthal and Kumar7), psychiatric diseases(Reference Costa, Chen and Davis5) and neurodegenerative disorders(Reference Gräff and Mansuy6). In addition, animal research has suggested that DNA methylation status may be involved in learning and memory processes, e.g. by regulating synaptic plasticity in hippocampal neurons(Reference Miller and Sweatt3, Reference Levenson, Roth and Lubin4).
The observed lack of a relationship between global DNA methylation and cognitive performance in healthy adults might imply that there is no functional relationship between the extent of cytosine methylation within DNA and individual differences in cognitive performance in the general population. In line with earlier reports(Reference Kok, Smith and Barto15), we observed that global DNA methylation has a relatively narrow distribution in healthy individuals. These findings suggest that under non-pathological conditions, there appears to be little inter-individual variation in DNA methylation-based regulation of gene expression, which decreases the likelihood that individual differences in cognitive performances may be mediated by this epigenetic mechanism.
Although global DNA methylation might not be involved in cognitive functioning, the present results do not rule out the possibility that DNA methylation at specific loci may be related to cognitive performance. In human subjects, gene-specific alterations in DNA methylation patterns have been associated with a number of pathological conditions characterised by cognitive deficits. Animal studies have suggested that diet-induced folate deficiency may result in overexpression of the Presenilin 1 gene by causing hypomethylation of its promoter region(Reference Fuso, Nicolia and Pasqualato21). Increased expression of this gene, which leads to elevated production of β-amyloid peptide, has been implicated in the aetiology of Alzheimer's disease(Reference Scarpa, Fuso and D'Anselmi22). In addition, schizophrenia has been associated with reduced expression of the gene encoding the protein Reelin, which is involved in neurodevelopment and synaptic plasticity, due to hypermethylation of the gene's promoter region(Reference Costa, Chen and Davis5). However, although it may be speculated that gene-specific changes in DNA methylation might underlie part of the individual differences in non-pathological cognitive functioning, little is known about the genetic correlates of cognitive performance in healthy human subjects.
An alternative explanation for the present null findings is that cognitive performance might be related to short-term changes, i.e. within the range of hours, in DNA methylation patterns rather than individual variation on the level of global DNA methylation. Indeed, animal studies have reported that dynamic and reversible changes in DNA methylation, such as the transient methylation and demethylation of DNA, are crucial for synaptic plasticity, learning and memory processes(Reference Miller and Sweatt3, Reference Levenson, Roth and Lubin4). It might be complicated, however, to measure such short-term changes in DNA methylation in volunteers, which makes it rather difficult to test this possibility.
From a methodological perspective, the present study was limited by its cross-sectional nature. In addition, the fact that we determined global DNA methylation in leucocytes rather than brain tissue should also be considered a limitation, as the extent of DNA methylation might differ between cells derived from the periphery and the brain(Reference Schilling and Rehli23). However, no direct measures of DNA methylation status in the central nervous system were available, given the inability to measure cerebrospinal fluid or brain DNA methylation status in volunteers.
It might also be argued that because of the relatively small sample size, the present study might have been underpowered to detect very modest associations. However, it should be noted that the present study had 80 % power to detect a 3 % change in the proportion of explained variance, which may be considered a small effect size(Reference Cohen24).
The present study did not support the notion that folate metabolism might influence cognitive performance through the mechanism of global DNA methylation, as measured in leucocytes. In line with the present findings, we found that long-term supplementation with folic acid, which significantly improved cognitive performance in the FACIT population(Reference Durga, Van Boxtel and Schouten12), did not have any effect on leucocyte global DNA methylation status (A. Jung, Y. Smulders, P. Verhoef, F. J. Kok, H. Blom, R. Kok, E. Schouten, E. Kampman, J. Durga, 2010, unpublished results). This might be explained by the fact that methylation capacity is not exclusively dependent on folate status, as methyl groups may also be provided by dietary intake of methionine, or by betaine-mediated remethylation of homocysteine(Reference Niculescu and Zeisel9).
To the best of our knowledge, this is the first study to investigate the relationship between leucocyte global DNA methylation and non-pathological cognitive functioning in healthy older adults. Future studies focusing on gene-specific DNA methylation patterns or short-term changes in DNA methylation status might contribute further to identifying the epigenetic mechanisms involved in cognitive functioning.
Acknowledgements
The FACIT study was supported by the Netherlands Organization for Health Research and Development (grant number 200110002), Sanquin Blood Bank (grant number 02-001), Wageningen University, and Top Institute Food and Nutrition. The author contributions were as follows: O. J. G. S. and J. D. designed the study. F. J. K., P. V. and J. D. were responsible for data acquisition and management of the FACIT study. O. J. G. S. analysed and interpreted the data and wrote the manuscript. All the authors reviewed and approved the final manuscript. None of the authors had a personal or financial conflict of interest.




