As a macronutrient, lipids are a major source of energy and essential fatty acids and they are important structural components of biomembranes, act as carriers of lipid-soluble vitamins and function as precursors of eicosanoids, hormones and enzyme cofactors for aquatic animals including crustaceans(1). The optimal dietary lipid can have an effect of sparing protein, which can benefit growth and maximise N retention(Reference Helland and Grisdale-Helland2). Therefore, high-lipid diets have been increasingly used in aquaculture for cost-effective farming in recent years. However, excessive dietary lipid can retard growth, decrease digestive function and reduce feed utilisation(1). Furthermore, high-lipid diets may lead to excessive lipid deposition and metabolic disturbances in organisms(Reference Lu, Xu and Wang3). Lipid metabolism includes lipid uptake, transport, anabolism and catabolism and involves several biochemical pathways with key enzymes and transcriptional factors(Reference Chen, Luo and Huang4). Moreover, lipid is transported between tissues mainly in the form of lipoproteins(Reference Weil, Lefèvre and Bugeon5). Fatty acid transporter proteins in the cell membrane include fatty acid transport protein (FATP), fatty acid binding protein (FABP) and fatty acid translocase. Lipid catabolism is the enzymatic hydrolysis of TAG into fatty acids and glycerol and provides energy to all life activities in aquatic animals. The main pathway of fatty acid catabolism is β-oxidation in the mitochondrial matrix and peroxisome(Reference Lu, Xu and Wang3). Carnitine palmitoyltransferase (CPT) is a mitochondrial transferase catalysing the conversion of fatty acyl-CoA to fatty acyl-carnitine and plays an indispensable role in the process of β-oxidation, which releases energy, fatty acyl CoA and recyclable carnitine(Reference Price, van der Leij and Jackson6).
Fish oil is an important lipid source for aquatic feed, due to the high content of long-chain (≥C20) PUFA (LC-PUFA), especially EPA and DHA, which can satisfy the essential fatty acid requirements of aquatic animals, especially marine species(Reference Monroig and Kabeya7). However, high levels of dietary EPA and DHA might increase lipid oxidation(Reference McNamara and Strawn8), due to these fatty acids being susceptible to oxidation, producing lipid peroxides(Reference Visioli and Galli9). To protect cells and tissues from oxidative damage, animals have endogenous antioxidant defense systems to counteract the activity of free radicals produced during oxidation. Protective enzymes include catalase, superoxide dismutase (SOD) and glutathione peroxidase (GSH-Px)(Reference Locatelli, Canaud and Eckardt10,Reference Bhor, Raghuram and Sivakami11) . Lipid oxidation products and metabolites such as malondialdehyde (MDA) can serve as markers of endogenous lipid peroxidation in tissues(Reference Guéraud, Taché and Steghens12,Reference Zuo, Ai and Mai13) .
Swimming crab (Portunus trituberculatus) is an important crustacean species in mariculture and is widely distributed along the coastal waters of China, Korea and Japan(Reference Jin, Wang and Huo14). Due to its economic value, the culture of P. trituberculatus is now expanding rapidly in China and aquaculture production of this crab reached 119 777 tons in 2017(15). Until now, studies have focused mainly on dietary macronutrient requirements, such as protein and lipid, and optimal dietary lipid levels for P. trituberculatus have been reported to range from 4·2 to 13·8 %(Reference Han, Wang and Hu16–Reference Sun, Jin and Ding19). However, the relationship between dietary lipid level and the regulation of lipid uptake, anabolism and catabolism has not been studied in P. trituberculatus. Therefore, the objectives of the present study were to evaluate the effects of dietary lipid level on growth performance, tissue fatty acid profiles, antioxidant capacity and haematological characteristics and to determine the mechanism whereby dietary lipid regulates lipid metabolism in the hepatopancreas of juvenile swimming crab.
Methods
Ethics statement
The present study was performed in strict accordance with the Standard Operation Procedures of the Guide for Use of Experimental Animals of Ningbo University. The experimental protocol and procedures were approved by the Institutional Animal Care and Use Committee of Ningbo University.
Diet preparation
Three isonitrogenous (47 % crude protein) diets were formulated to contain 5·8, 9·9 and 15·1 % crude lipid by the addition of fish oil, with cellulose used to balance the lipid levels. Formulations and proximate compositions of the experimental diets are presented in online Supplementary Table S1, and fatty acid compositions of the diets and the fish oil utilised are shown in online Supplementary Table S2. White fishmeal, wheat gluten meal, soyabean protein concentrate and krill meal were used as protein sources; fish oil and soya lecithin were used as lipid sources, and dextrin was used as the main carbohydrate source. All dry ingredients were ground into fine powder with a particle size below 177 microns, and then micro-components such as vitamin and mineral premix were added by the progressive enlargement method. Finally, lipids and distilled water (40 %, w/w) were added to the premixed dry ingredients and mixed until homogenous in a Hobart-type mixer. Cold-extruded pellets of two uniform pellet sizes (3·0 mm in diameter and 5·0 mm in length, and 4·0 mm in diameter and 7·0 mm in length) were prepared using a granulating machine (G-250, machine factory of South China University of Technology), steamed for 30 min at 90°C and then air-dried to approximately 10 % moisture. The diets were stored at –20°C in sealed plastic bags until used in the feeding trial.
Crab rearing and experimental conditions
Juvenile P. trituberculatus were obtained from the Lai-Fa nursery farm. Prior to the start of the feeding trial, the juvenile crabs were reared in cement pools and fed commercial diet (45 % dietary protein, 8 % crude lipid, Ningbo Tech-Bank Corp.) for 1 week for acclimation to the experimental conditions. Before the feeding trial, crabs were fasted for 24 h and weighed individually. A total of 240 juvenile crabs (initial weight 21·09 (sem 1·11) g) were distributed randomly into 240 rectangular plastic baskets (36 cm × 21 cm × 35 cm; length × width × height) in two cement pools (7·0 m × 5·0 m × 1·8 m; length × width × depth) and fed once daily at 18.00 hours. Each experimental diet was randomly assigned to four replicates with twenty juvenile swimming crabs in each replicate and one crab in each basket. Each basket was divided into two equal parts by a nylon screen, with one area filled with sand as the crab habitat and the other area filled with seawater as the feeding area. All groups of crabs were fed at the same fixed rate of 6–8 % of wet body weight, with the daily ration adjusted every 2 weeks according to the weight of the crab in each basket, to maintain a level approaching apparent satiation, as described previously(Reference Jin, Wang and Huo14). Faeces and uneaten feed were removed, separated and the amount of uneaten feed recorded. To maintain water quality, 60 % of water in the cement pool was exchanged daily and any dead crabs were immediately removed, weighed and recorded.
The seawater in the cement pools was provided with continuous aeration through air stones to retain high dissolved oxygen levels. The pH, salinity, ammonia nitrogen and dissolved O2 were measured using the YSI Proplus instrument (YSI). During the experimental period, water temperature in the pool was 26–30°C, pH was 7·7–8·3, salinity was 26–28 mg/l, ammonia nitrogen was lower than 0·05 mg/l and dissolved oxygen was 6·7–7·1 mg/l. The feeding trial lasted for 8 weeks, starting on 7 July 2016 and finishing on 1 September 2016.
Sample collection
At the end of the 8-week feeding trial, crabs were counted and weighed to determine survival, weight gain, specific growth rate and feed conversion ratio. Haemolymph samples from three crabs of each replicate were collected immediately using the method described by Jin et al.(Reference Jin, Wang and Huo14). Haemolymph samples were placed into 1·5 ml Eppendorf tubes at 4°C for 24 h, followed by centrifugation (956 g , 10 min, 4°C) (Eppendorf, Centrifugal 5810 R). The supernatant was collected and stored at –80°C for haemolymph biochemistry and enzyme activity analysis. The hepatopancreas from three crabs in each replicate was dissected and weighed to calculate the hepatosomatic index, and hepatopancreas and muscle were quickly taken from these three crabs, frozen in liquid N2 and stored at −80°C prior to analysis of proximate composition and fatty acid profile. A further three crabs from each replicate were sampled to determine the antioxidant enzyme activities of the hepatopancreas. Finally, the hepatopancreas from four crabs in each replicate was collected into RNALater for the determination of expression of genes related to lipid metabolism.
Proximate composition analysis
Crude protein, crude lipid, ash and moisture contents of the diets and crab tissues (hepatopancreas and muscle) were analysed by standard methods of the AOAC(20). Crude protein (N × 6·25) was determined using the Dumas combustion method with an auto-protein analyser (FP-528, Leco). Crude lipid was determined by the diethyl ether extraction method using a Soxtec System HT (Soxtec System HT6, Tecator). Moisture content was determined by drying the samples to a constant weight at 105°C, and ash content was determined in a muffle furnace at 550°C for 8 h.
Fatty acid composition
The fatty acid profiles of fish oil, diets and crab tissues (hepatopancreas and muscle) were determined as described by Zuo et al.(Reference Zuo, Ai and Mai13) with minor modifications. The freeze-dried samples (approximately 80 mg hepatopancreas and 120 mg muscle) were added to 12 ml volumetric glass screw cap tubes (teflon gasket); 3 ml potassium hydroxide in methanol (1 N) was added and heated at 72°C in a water bath for 20 min. After cooling, 3 ml of 2 m-HCl in methanol was added and the mixture heated at 72°C in a water bath for 20 min. Finally, 1 ml hexane was added to the mixture, shaken vigorously for 1 min and then allowed to separate into two layers. Fatty acid methyl esters were separated and identified by GC-MS (Agilent technologies 7890B-5977A) as detailed fully in Jin et al.(Reference Jin, Lu and Yuan21). Results were expressed as relative percentages of each fatty acid, calculated as the proportion of the area under the considered peak to the total area of all peaks.
Haemolymph biochemical analysis
The total protein, cholesterol, TAG, glucose, LDL-cholesterol and HDL-cholesterol contents in haemolymph were determined with an automatic chemistry analyser (Hitachi 7600-110).
Oxidation and antioxidant parameter assays
All haemolymph samples were analysed undiluted, except for the GSH-Px analysis (double dilution). Hepatopancreas samples were homogenised in nine volumes (w/v) of cold saline (0·89 %, w/v) and centrifuged at 956 g for 10 min at 4°C. The supernatant was stored at −80°C until used. Activities of catalase, SOD and GSH-Px, as well as the contents of MDA and protein, were assayed by commercial kits purchased from Nanjing Jiancheng Bioengineering Institute, according to the manufacturer’s instructions by Multiskan spectrum (Thermo).
SOD activity was measured with the diagnostic reagent kits. Briefly, superoxide anion radical, produced in the reaction of xanthine with O2, catalysed by xanthine oxydase, reacts with hydroxylamine producing nitric ion. After nitric ion reacting with naphthalene diamine, sulphanilic acid produces a coloured product. The concentration of this mixture is proportional to the amount of produced superoxide anion radical. One unit of SOD activity was defined as the amount of enzyme required for 1 mg tissue proteins or 1 ml serum in 1 ml of a reaction mixture SOD inhibition rates to 50 % as monitored at 550 nm.
GSH-Px levels were measured by the dithiobisnitrobenzoic acid method. A yellow product which shows absorbance at 420 nm is formed as GSH-Px reacted with dithiobisnitrobenzoic acid.
Catalase activity in hepatopancreas was assayed by diagnostic reagent kits. One unit of catalase activity was defined as 1 mg tissue proteins consumed by 1 μmol H2O2 at 405 nm for a second.
MDA concentration was determined by the thiobarbituric acid method. A red adduct of MDA is formed with thiobarbituric acid, which shows absorbance at 532 nm.
Enzymatic activity assays
The activities of key enzymes of lipogenesis and lipolysis were determined in the hepatopancreas with three crabs per replicate. For this purpose, the hepatopancreas was homogenised (dilution 1:4) in ice-cold buffer (100 mm TRIS-HCl, 0·1 mm EDTA and 0·1 % Triton X-100 (v/v), pH 7·8). Homogenates were centrifuged at 30 000 g for 30 min at 4°C. After centrifugation, the resultant supernatant was collected and aliquots were stored at −80°C until analysis. Enzyme activities related to lipid metabolism, such as fatty acid synthase (FAS), glucose 6-phosphate dehydrogenase (G6PD), CPT-1 and lipoprotein lipase (LPL), were assayed using ELISA assay kits from Yuan Ye Biological Company. The enzyme activities were analysed by a Multiskan spectrum (Thermo) according to the manufacturer’s instructions.
Gene expression
Analyses of mRNA levels were performed on hepatopancreas samples with four crabs per replicate. Total RNA was extracted from the hepatopancreas using TRIzol reagent according to the manufacturer’s instructions. Quantity and quality of the isolated RNA were determined by spectrophotometry (Nanodrop 2000, Thermo Fisher Scientific) and on a 1·2 % (w/v) denaturing agarose gel, respectively. The cDNA was prepared from 1000 ng of DNAase-treated RNA and synthesised using a Prime Script™ RT Reagent Kit with gDNA Eraser (Perfect Real Time) (Takara). Four potential candidate genes, β-actin, 18s rRNA, RpL8, and RpL18, were selected for reference gene analysis. The primers for amplifying the four candidate reference genes are shown in online Supplementary Table S3.
The results confirmed that β-actin was very stable and was used as a reference gene to normalise the expression levels of the candidate genes. Specific primers of fad, elovl4, elovl, apod, lrp2, lpr1, lpr2, srb, fabp1, fabp3, fabp9, fatp4, cpt1, cpt2, acox1, acox2, acox3, fas, acc, g6pd, 6pgd, dgat1, gpat1, gpat3, hsl, lpl, il, tgl, pl, srebp1, hnf4a and rxr used for qPCR were designed using Primer Premier 5.0 (online Supplementary Table S3). The primer specificities of the candidate genes were checked as described by Bustin et al.(Reference Bustin, Beaulieu and Hugget22) by systematically running melting curve assays after the qPCR programme, running the qPCR products on a 1·2 % agarose gel, and DNA sequencing technology (BGI). Amplification was performed using a quantitative thermal cycler (Lightcycler 96, Roche). The qPCR assays were performed in a total volume of 20 μl, containing 1·0 μl of each primer, 10 μl of 2× conc SYBR Green I Master (Roche), 2 μl of 1/4 diluted cDNA and 6 μl DEPC-water. The thermal cycling conditions used for qPCR were as follows: 95°C for 2 min, followed by 45 cycles of 95°C for 10 s, 58°C for 10 s and 72°C for 20 s. Standard curves were generated using six different dilutions of the cDNA samples, and the amplification efficiency was analysed using the equation: E = 10(−1/Slope) − 1(Reference Jothikumar, Cromeans and Robertson23). The amplification efficiencies of all genes were approximately equal and ranged from 90 to 105 %. The relative expression levels were calculated using the 2−ΔΔCtmethod as described by Livak and Schmittgen(Reference Livak and Schmittgen24). Briefly, the relative expression ratio (R) of a target gene was calculated on the basis of real-time PCR efficiency (E) and the threshold cycle (Ct) deviation (ΔCT) of the unknown sample compared with a control sample and expressed in comparison with the β-actin reference gene:
 $${\rm{R}} = [({\rm{Etarget \ gene}})\Delta {\rm{CT \ target \ gene }}({\rm{mean \ control}} - {\rm{mean \ sample}})]/[\left( {{\rm{E}}\beta - {\rm{actin}}} \right)\Delta {\rm{CT}}\beta - {\rm{actin }}({\rm{mean \ control}} - {\rm{mean \ sample}})]$$
$${\rm{R}} = [({\rm{Etarget \ gene}})\Delta {\rm{CT \ target \ gene }}({\rm{mean \ control}} - {\rm{mean \ sample}})]/[\left( {{\rm{E}}\beta - {\rm{actin}}} \right)\Delta {\rm{CT}}\beta - {\rm{actin }}({\rm{mean \ control}} - {\rm{mean \ sample}})]$$
Efficiency of q-PCR was measured by the slope of a standard curve using serial dilutions of cDNA. Crab fed the 5·8 % lipid diet was used as the control group.
Statistical analysis
Results are presented as mean values with their standard errors. Data were checked for normality and homogeneity of variances, and they were normalised when appropriate. Proportional data were arcsine square root transformed before statistical analyses. Means were compared through one-way ANOVA followed by Tukey’s multiple-range test. The level of significance was set at P < 0·05. All statistical analyses were conducted using the SPSS 20.0 software package (IBM Crop.) for Windows.
Results
Growth performance, feed utilisation and morphological index
Effects of dietary lipid levels on growth performance, feed utilisation and morphological indices are shown in Table 1. Crabs fed the 15·1 % lipid diet showed significantly lower FW, weight gain, specific growth rate and survival and also had higher feed conversion ratio, compared with crabs fed the 5·8 and 9·9 % lipid diets. There was no significant difference in hepatosomatic index among the treatments.
Table 1. Effects of dietary lipid level on growth performance and feed utilisation of swimming crab (Portunus trituberculatus) (n 4)*
(Mean values with their standard errors)
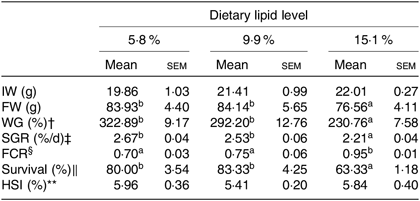
IW, initial weight; FW, final weight; WG, weight gain; SGR, specific growth rate; FE, feed efficiency; FCR, feed conversion ratio; HSI, hepatosomatic index.
* Mean values in the same row with unlike superscript letters are significantly different (P < 0·05) between treatments.
† WG (%) = 100 × (FW (g) − IW (g))/IW (g).
‡ SGR (%/d) = 100 × ((Ln FW (g) − Ln IW (g))/d).
§ FCR = feed consumed (g, dry weight)/weight gain (g, wet weight).
‖ Survival (%) = 100 × (final number of crabs/initial number of crabs).
** HSI (%) = 100 × (hepatopancreas weight/wet body weight).
Hepatopancreas and muscle composition
Moisture and ash contents in hepatopancreas and protein and ash contents in muscle were not significantly influenced by dietary lipid level (Table 2). Lipid content in muscle and hepatopancreas significantly increased as dietary lipid level increased from 5·8 to 9·9 %, but did not significantly increase when dietary lipid level increased from 9·9 to 15·1 %. Crabs fed the 15·1 % lipid diet had the lowest protein content in the hepatopancreas among the treatments.
Table 2. Proximate compositions (%, wet weight) of hepatopancreas and muscle of swimming crab (Portunus trituberculatus) fed the diets with different lipid contents (n 4)*
(Mean values with their standard errors)
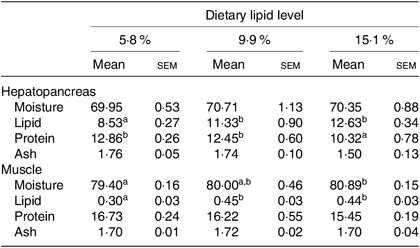
* Mean values in the same row with unlike superscript letters are significantly different (P < 0·05) between treatments.
Hepatopancreas and muscle fatty acid profiles
The fatty acid compositions of hepatopancreas and muscle are shown in Tables 3 and 4, respectively. Generally, the fatty acid composition of muscle and hepatopancreas resembled the fatty acid profiles of the diets. The hepatopancreas of crab fed the 15·1 % lipid diet presented a higher proportion of 14 : 0 and 16 : 1n-7 than those fed the other diets. The proportions of 18 : 2n-6 and total n-6 PUFA (n-6 PUFA) in hepatopancreas significantly decreased as dietary lipid level increased from 5·8 to 15·1 % (Table 3). Moreover, contents of 20 : 5n-3 (EPA), 22 : 6n-3 (DHA), total n-3 LC-PUFA, 20 : 4n-6 (arachidonic acid) and the n-3:n-6 ratio significantly increased as dietary lipid level increasing from 5·8 to 9·9 % (Table 3).
Table 3. Fatty acid compositions (% of total fatty acids) of hepatopancreas of juvenile swimming crab (Portunus trituberculatus) fed diets with different lipid contents (n 4)*
(Mean values with their standard errors)
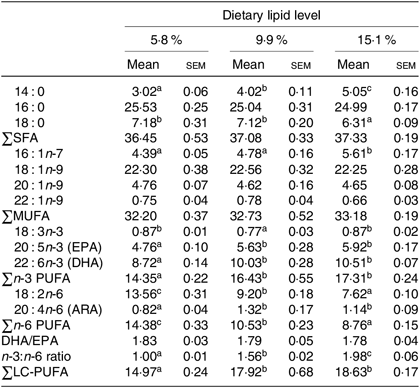
n-3 LC-PUFA, n-3 long-chain PUFA; ARA, arachidonic acid.
* Mean values in the same row with unlike superscript letters are significantly different (P < 0·05) between treatments. Some fatty acids in trace amount such as 20 : 0, 22 : 0, 24 : 0, 18 : 4n-3, 20 : 3n-3, 22 : 5n-3, 18 : 3n-6, 20 : 2n-6 and 22 : 5n-6 are not listed.
Table 4. Fatty acid compositions (% of total fatty acids) of muscle of juvenile swimming crab (Portunus trituberculatus) fed diets with different lipid contents (n 4)*
(Mean values with their standard errors)
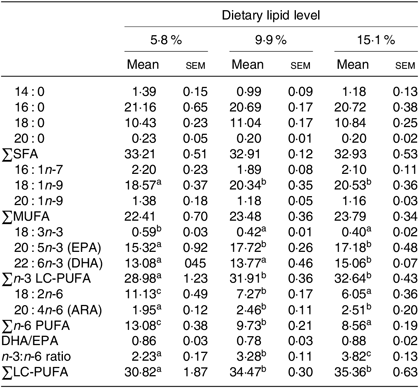
n-3 LC-PUFA, n-3 long-chain PUFA; ARA, arachidonic acid.
* Mean values in the same row with unlike superscript letters are significantly different (P < 0·05) between treatments. Some fatty acids in trace amount such as 22 : 1n-9, 18 : 4n-3, 20 : 3n-3, 22 : 5n-3, 18 : 3n-6, 20 : 2n-6 and 22 : 5n-6 are not listed.
Similar results were found in the muscle of juvenile swimming crab. Crabs fed the 5·8 % lipid diet had lower proportions of 18 : 1n-9, EPA, DHA, total n-3 LC-PUFA, arachidonic acid and lower n-3:n-6 PUFA ratio in muscle than those fed the diets containing 9·9 or 15·1 % lipid (Table 4). However, there was no significant difference in total SFA, MUFA or DHA:EPA ratio in muscle among the dietary treatments.
Haemolymph metabolites and antioxidant enzyme activities
The hematological parameters and enzyme activities in haemolymph and hepatopancreas of swimming crab fed different dietary lipid levels are presented in Table 5. Haemolymph total protein and LDL-cholesterol contents significantly decreased as the dietary lipid level increased from 5·3 to 9·9 %, whereas there were no significant differences in haemolymph total cholesterol, TAG, glucose and HDL-cholesterol contents among all treatments.
Table 5. Hematological characteristics, enzyme activities in serum and hepatopancreas of juvenile swimming crab (Portunus trituberculatus) fed diets with different lipid contents (n 4)*
(Mean values with their standard errors)
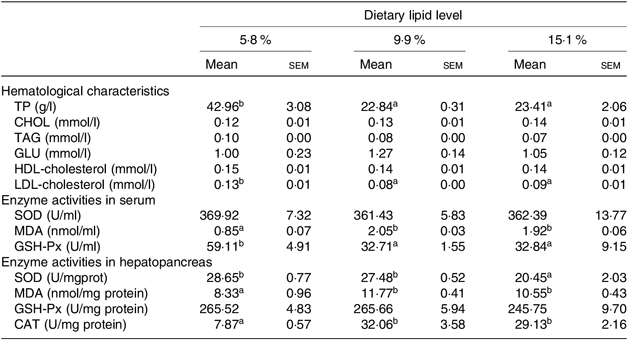
TP, total protein; CHOL, cholesterol; GLU, glucose; SOD, superoxide dismutase, MDA, malondialdehyde; GSH-Px, glutathione peroxidase; CAT, catalase.
* Mean values in the same row with unlike superscript letters are significantly different (P < 0·05) between treatments.
The contents of MDA both in the haemolymph and hepatopancreas significantly increased as dietary lipid level increased from 5·8 to 9·9 %. However, haemolymph GSH-Px and hepatopancreas SOD activities decreased as dietary lipid level increased, with highest GSH-Px and SOD activities observed in crabs fed the 5·8 % lipid diet. There were no significant differences in the activities of haemolymph SOD and hepatopancreas GSH-Px.
Hepatopancreas enzyme activity involved in lipid metabolism
The lipid metabolic enzyme activities in hepatopancreas of juvenile swimming crabs fed different lipid levels are shown in Fig. 1. LPL enzyme activity in hepatopancreas was not significantly influenced by dietary lipid level. Crabs fed the 15·1 % lipid diet had significantly lower FAS and G6PD activities in hepatopancreas than those fed the 5·8 and 9·9 % lipid diets. In contrast, the activity of CPT-1 was significantly higher in crabs fed the 15·1 % lipid diet than those fed the 5·8 % lipid diet.
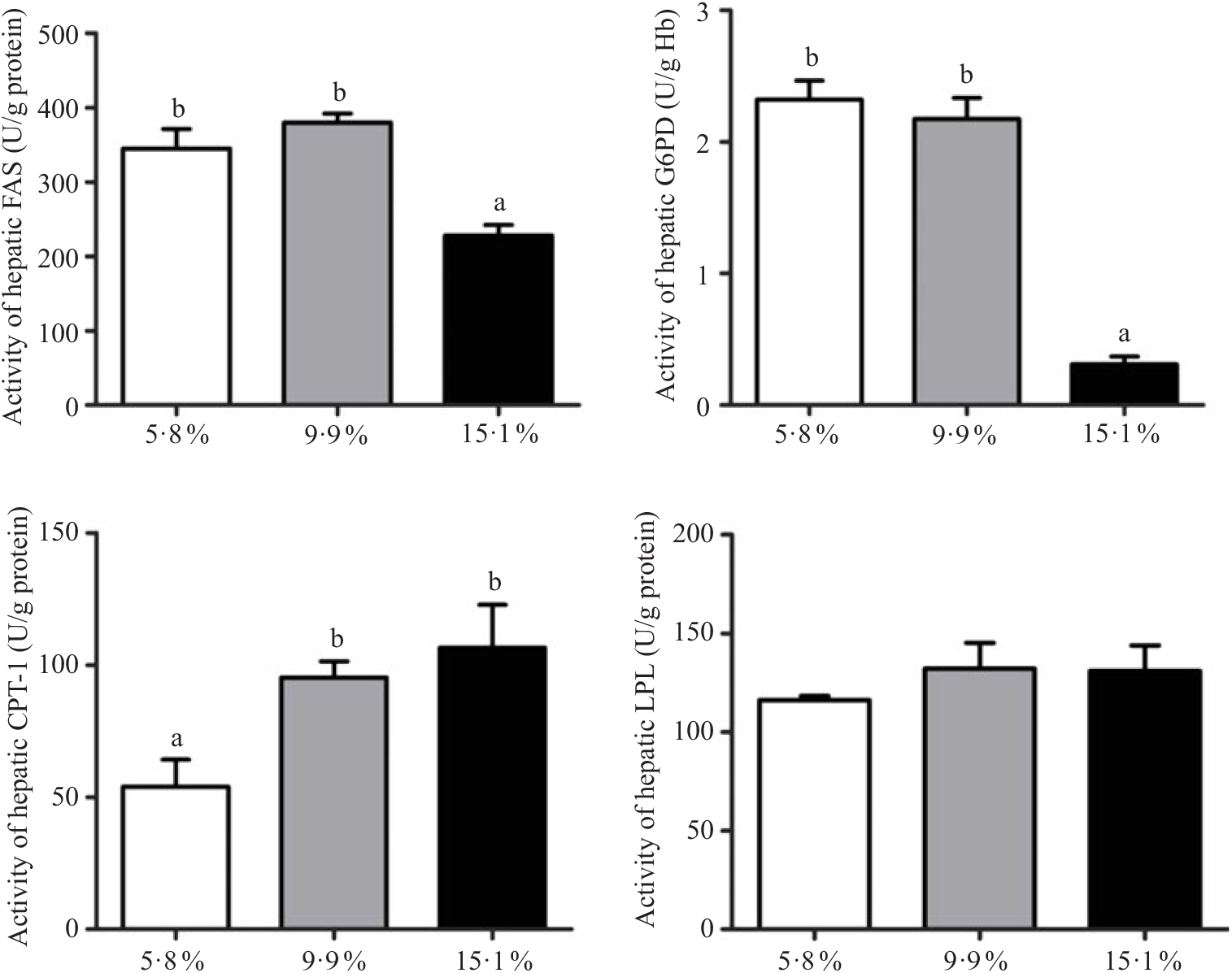
Fig. 1. Activities of lipid metabolic enzymes (anabolism and catabolism) in hepatopancreas of juvenile swimming crab (Portunus trituberculatus) fed diets containing different lipid levels. Bars represent the means and standard errors of four replicates. a,b Within the same enzyme, unlike letters denote significant differences as determined by ANOVA and Tukey’s test (P < 0·05). FAS, fatty acid synthase; G6PD, glucose 6-phosphate dehydrogenase; CPT-1, carnitine palmitoyltransferase-1; LPL, lipoprotein lipase.
Gene expression
The relative expression of genes involved in lipid metabolic pathways, such as LC-PUFA biosynthesis, lipoprotein assembly and receptors, fatty acid uptake, fatty acid oxidation, lipid anabolism and lipid catabolism in hepatopancreas, is presented in Figs. 2–7, respectively. The expression of genes involved in LC-PUFA biosynthesis, fad, elovl4, elovl, srebp1, hnf4a and rxr, was significantly up-regulated in crabs fed the 5·8 % lipid diet compared with those fed the other diets. The lowest expression of fad, elovl4, elovl, srebp1, hnf4a and rxr genes was found in hepatopancreas of crabs fed the 15·1 % lipid diet (Fig. 2).
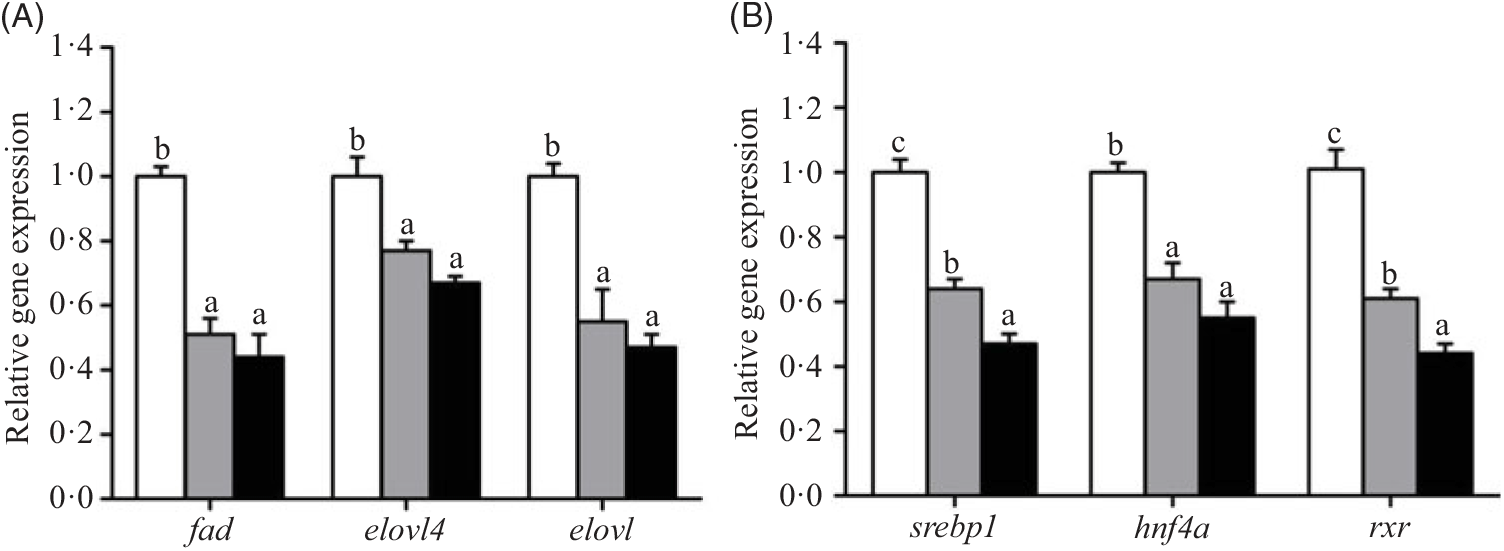
Fig. 2. Effects of dietary lipid level on relative expression of genes involved in long-chain PUFA (LC-PUFA) biosynthesis (A) and transcription factors (B) in hepatopancreas of Portunus trituberculatus. ![]() , 5·8 % lipid diet;
, 5·8 % lipid diet; ![]() , 9·9 % lipid diet;
, 9·9 % lipid diet; ![]() , 15·1 % lipid diet. Values are means (n 4), with their standard errors represented by vertical bars. a,b,c Mean values for the same gene with unlike letters were significantly different as determined by ANOVA and by Tukey’s test (P < 0·05). fad, Fatty acyl desaturase; elovl4, elongation of very long-chain fatty acid protein 4; elovl, elongation of very long-chain fatty acid protein; srebp-1, sterol regulatory element-binding protein-1; hnf4a, hepatocyte nuclear factor 4-α; rxr, retinoid X receptor.
, 15·1 % lipid diet. Values are means (n 4), with their standard errors represented by vertical bars. a,b,c Mean values for the same gene with unlike letters were significantly different as determined by ANOVA and by Tukey’s test (P < 0·05). fad, Fatty acyl desaturase; elovl4, elongation of very long-chain fatty acid protein 4; elovl, elongation of very long-chain fatty acid protein; srebp-1, sterol regulatory element-binding protein-1; hnf4a, hepatocyte nuclear factor 4-α; rxr, retinoid X receptor.
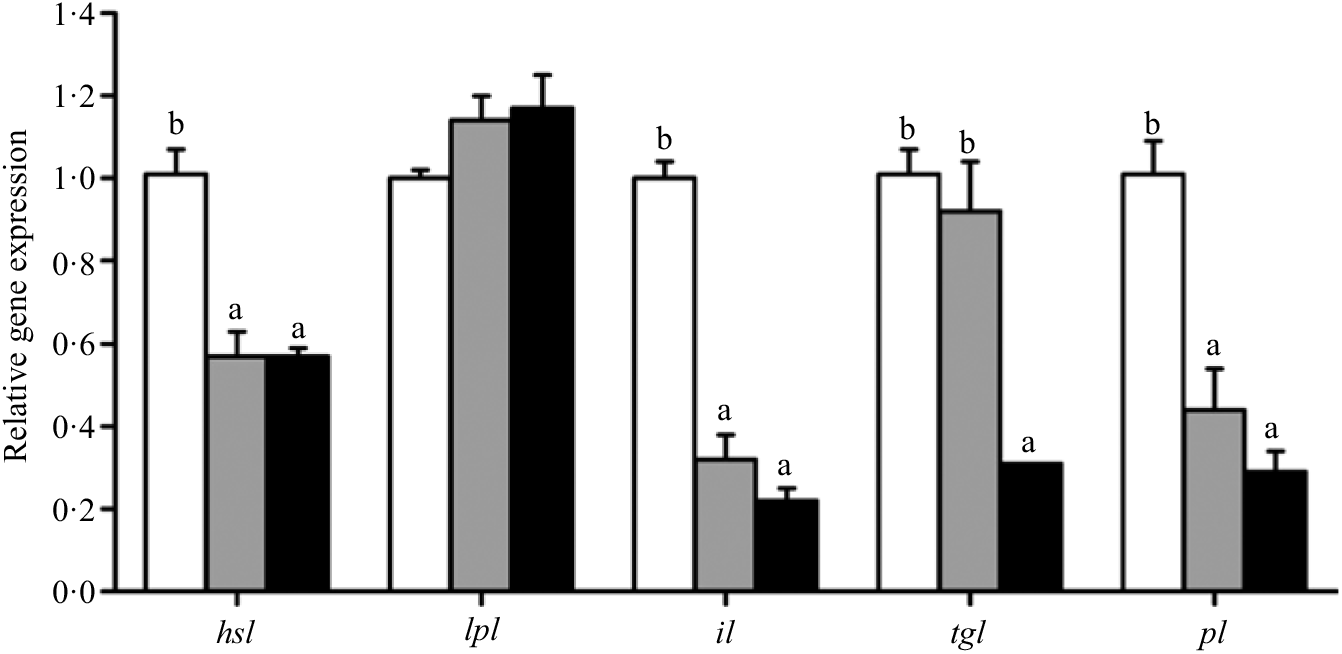
Fig. 7. Effects of different dietary lipid levels on relative gene expression involved in lipid catabolism in the hepatopancreas of Portunus trituberculatus. ![]() , 5·8 %;
, 5·8 %; ![]() , 9·9 %;
, 9·9 %; ![]() , 15·1 %. Values are means (n 4), with their standard errors represented by vertical bars. a,b Mean values for the same gene with unlike letters were significantly different among all treatments by Tukey’s test (P < 0·05). hsl, Hormone-sensitive lipase; lpl, lipoprotein lipase; il, intracellular lipase; tgl, TAG lipase; pl, pancreatic lipase.
, 15·1 %. Values are means (n 4), with their standard errors represented by vertical bars. a,b Mean values for the same gene with unlike letters were significantly different among all treatments by Tukey’s test (P < 0·05). hsl, Hormone-sensitive lipase; lpl, lipoprotein lipase; il, intracellular lipase; tgl, TAG lipase; pl, pancreatic lipase.
The expression of the apod gene was up-regulated in crabs fed the 15·1 % lipid diet. In contrast, the lowest expression of lrp2, lpr1 and srb genes was observed in crabs fed the 15·1 % lipid diet. Expression of the lpr2 gene showed no significant differences among the treatments (Fig. 3).
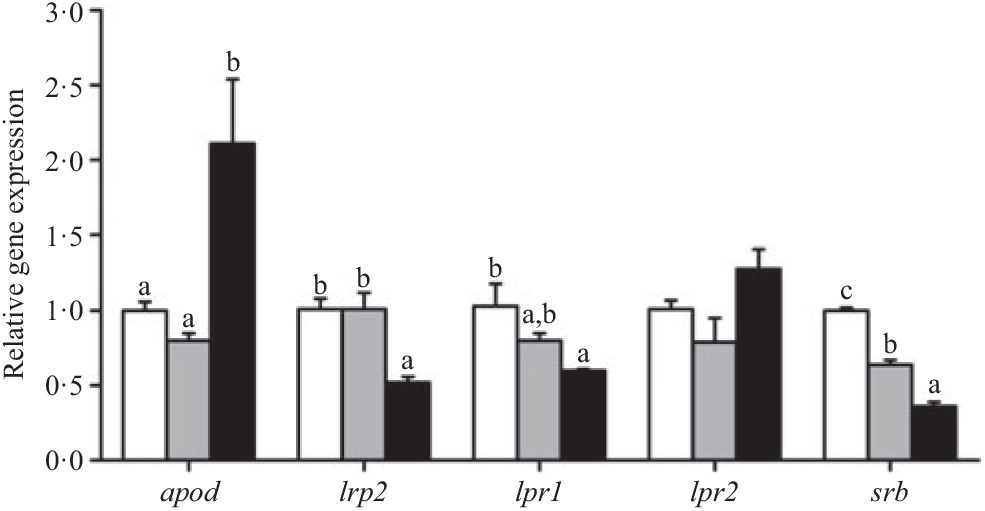
Fig. 3. Effects of dietary lipid level on relative expression of genes involved in lipoprotein assembly and lipoprotein receptors in the hepatopancreas of Portunus trituberculatus. ![]() , 5·8 % lipid diet;
, 5·8 % lipid diet; ![]() , 9·9 % lipid diet;
, 9·9 % lipid diet; ![]() , 15·1 % lipid diet. Values are means (n 4), with their standard errors represented by vertical bars. a,b,c Mean values for the same gene with unlike letters were significantly different as determined by ANOVA and Tukey’s test (P < 0·05). apod, apo D; lrp2, LDL receptor-related protein 2; lpr, lipoprotein receptor; srb, scavenger receptor class.
, 15·1 % lipid diet. Values are means (n 4), with their standard errors represented by vertical bars. a,b,c Mean values for the same gene with unlike letters were significantly different as determined by ANOVA and Tukey’s test (P < 0·05). apod, apo D; lrp2, LDL receptor-related protein 2; lpr, lipoprotein receptor; srb, scavenger receptor class.
Crabs fed the 5·8 % lipid diet presented significantly higher mRNA gene expression of fabp3, fabp9 and fatp4, whereas the highest gene expression of fabp1 was observed at crabs fed the 9·9 % lipid diet (Fig. 4). Down-regulation of cpt2, acox1, acox2 and acox3 and up-regulation of cpt1 were found as the dietary lipid levels increased (Fig. 5), with significant values at 5·8 %. Gene expression of fas, acc, g6pd, 6pgd, dgat1, gpat1 and gpat3 involved in lipid anabolism was down-regulated with increasing dietary lipid levels (Fig. 6). Meanwhile, expression of genes involved in lipid catabolism decreased with increasing dietary lipids, except for lpl that was not affected by dietary lipids. Down-regulation of tgl in the crabs fed the diet containing 15·1 % lipid was significantly lower than that of the other dietary groups (Fig. 7).
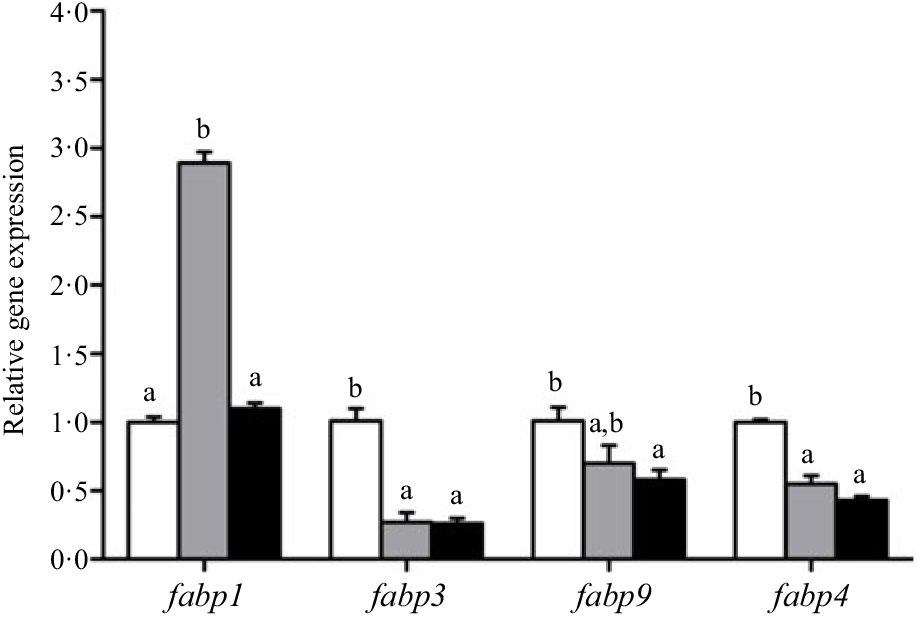
Fig. 4. Effects of dietary lipid levels on relative gene expression involved in fatty acid uptake in the hepatopancreas of Portunus trituberculatus. ![]() , 5·8 %;
, 5·8 %; ![]() , 9·9 %;
, 9·9 %; ![]() , 15·1 %. Values are means (n 4), with their standard errors represented by vertical bars. a,b Mean values for the same gene with unlike letters were significantly different in all the graphs by Tukey’s test (P < 0·05). fabp, Fatty acid binding protein; fatp, fatty acid transport protein.
, 15·1 %. Values are means (n 4), with their standard errors represented by vertical bars. a,b Mean values for the same gene with unlike letters were significantly different in all the graphs by Tukey’s test (P < 0·05). fabp, Fatty acid binding protein; fatp, fatty acid transport protein.
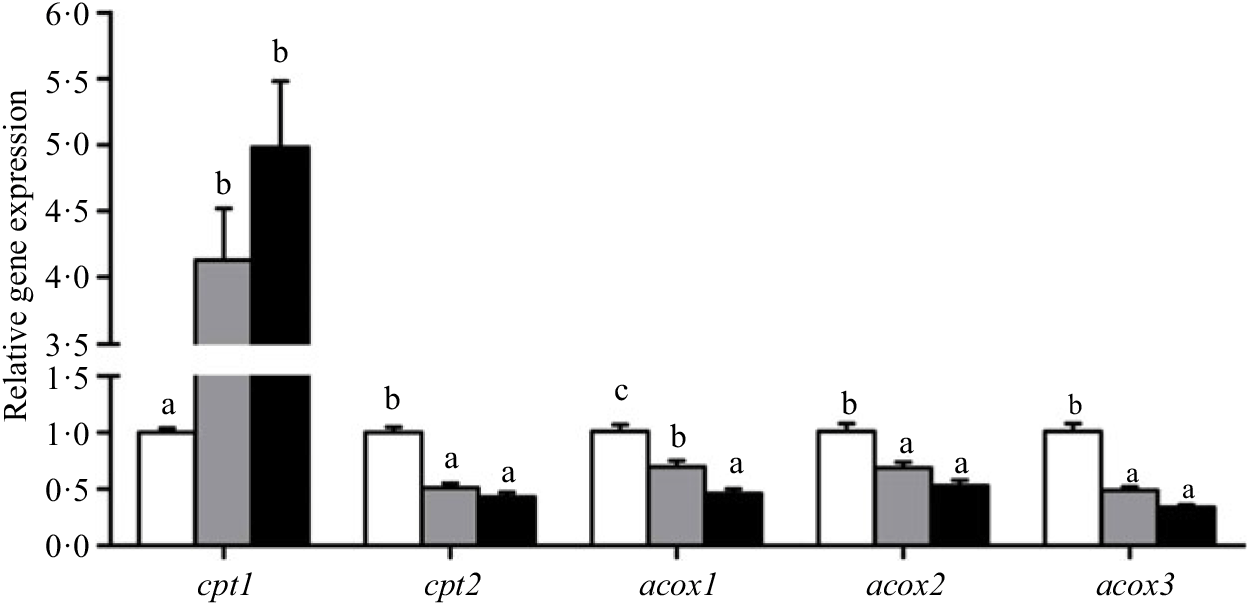
Fig. 5. Effects of dietary lipid levels on relative gene expression involved in fatty acid oxidation in the hepatopancreas of Portunus trituberculatus. ![]() , 5·8 %;
, 5·8 %; ![]() , 9·9 %;
, 9·9 %; ![]() , 15·1 %. Values are means (n 4), with their standard errors represented by vertical bars. a,b,c Mean values for the same gene with unlike letters were significantly different among treatments by Tukey’s test (P < 0·05). cpt, Carnitine palmitoyltransferase; acox, acyl-CoA oxidase.
, 15·1 %. Values are means (n 4), with their standard errors represented by vertical bars. a,b,c Mean values for the same gene with unlike letters were significantly different among treatments by Tukey’s test (P < 0·05). cpt, Carnitine palmitoyltransferase; acox, acyl-CoA oxidase.
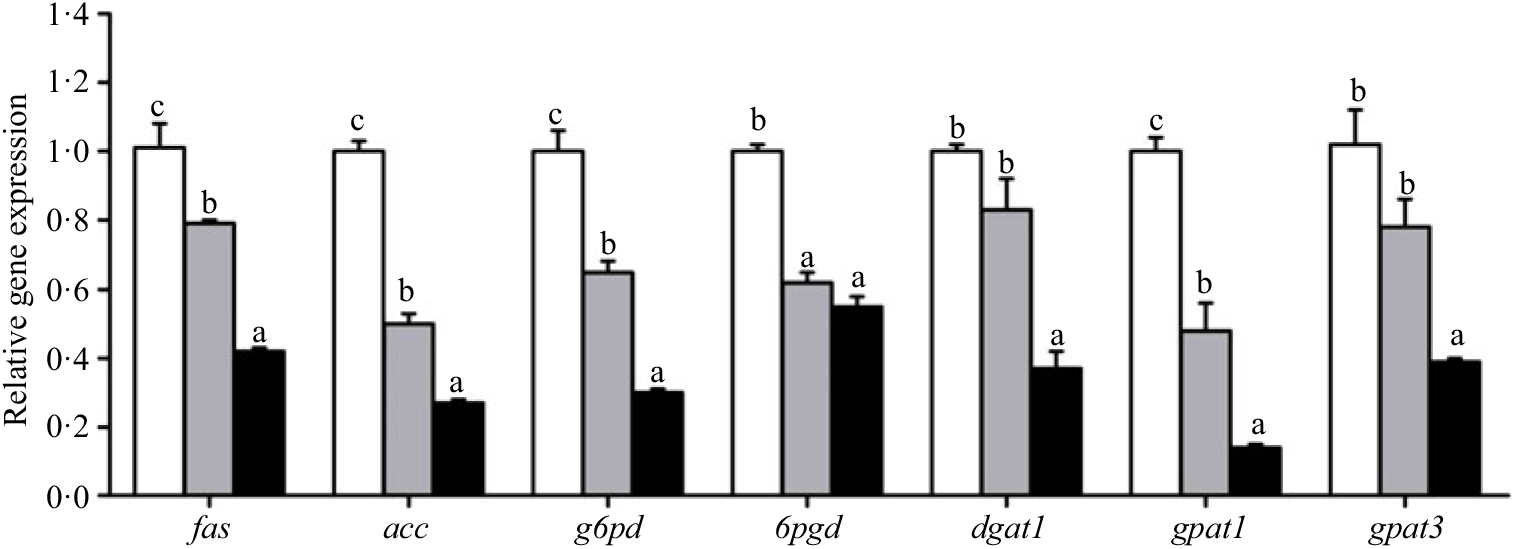
Fig. 6. Effects of different dietary lipid levels on relative gene expression involved in lipid anabolism in the hepatopancreas of Portunus trituberculatus. ![]() , 5·8 %;
, 5·8 %; ![]() , 9·9 %;
, 9·9 %; ![]() , 15·1 %. Values are means (n 4), with their standard errors represented by vertical bars. a,b,c Mean values for the same gene with unlike letters were significantly different among all treatments by Tukey’s test (P < 0·05). fas, Fatty acid synthase; acc, acetyl-CoA carboxylase; g6pd, glucose 6-phosphate dehydrogenase; 6pgd, 6-phosphogluconate dehydrogenase; dgat1, acyl CoA diacylglycerol acyltransferase 1; gpat, glycerol-3-phosphate acyltransferase.
, 15·1 %. Values are means (n 4), with their standard errors represented by vertical bars. a,b,c Mean values for the same gene with unlike letters were significantly different among all treatments by Tukey’s test (P < 0·05). fas, Fatty acid synthase; acc, acetyl-CoA carboxylase; g6pd, glucose 6-phosphate dehydrogenase; 6pgd, 6-phosphogluconate dehydrogenase; dgat1, acyl CoA diacylglycerol acyltransferase 1; gpat, glycerol-3-phosphate acyltransferase.
Discussion
In the present study, the results indicated that the lower dietary lipid levels promoted growth and feed utilisation, whereas the highest dietary lipid level retarded growth and decreased survival. These results were similar to previous studies for marine crustacea that demonstrated negative relationships between growth performance and dietary lipid level(Reference Huo, Jin and Zhou17,Reference Xu, Liu and Shen25 ,Reference Zhang, Li and Wu26). The dietary lipid requirement for juvenile swimming crab had been previously estimated to range from 4·2 to 13·8 %(Reference Han, Wang and Hu16). Moreover, higher dietary lipid levels (>10 %) are known to impair growth and survival of shrimp, probably due to a reduction in consumption caused by high energetic content or an inability to efficiently metabolise high levels of lipid (reduced digestibility)(1). Aquatic animals can effectively digest a certain amount of dietary lipid and, when this is exceeded, the excess lipid is wasted in faeces(Reference Izquierdo, Socorro and Arantzamendi27).
The results of the present study indicated that lipid content significantly increased in the muscle and hepatopancreas when dietary lipid increased from 5·8 to 9·9 %. This was in agreement with previous findings of Han et al.(Reference Han, Wang and Hu16) who reported that dietary lipid level influenced lipid retention in muscle and hepatopancreas for the same crab species. Therefore, excess dietary lipid may result in increased lipid deposition in liver or hepatopancreas, as observed previously in other crustaceans(Reference Catacutan28–Reference Zhao, Wen and Li31). In contrast, Sheen & Wu(Reference Sheen and Wu32) indicated that mud crab fed a diet containing 12 % lipid had significantly lower body lipid deposition compared with crabs fed diets with lower lipid. The reason for this discrepancy might be ascribed to different species, life stage, culture conditions and/or experimental diet formulations.
Dietary lipid level and source are known to regulate lipogenesis and fatty acid bioconversion pathways, which may affect tissue lipid deposition and fatty acid composition. The fatty acid composition of tissues in crustaceans is influenced by the fatty acid profile of the diet(1,Reference Copeman, Daly and Eckert33) and, in the present study, the fatty acid composition in hepatopancreas and muscle of swimming crab reflected the dietary fatty acid profile. Due to its content of LC-PUFA, especially EPA and DHA, fish oil is widely used in aquatic feed, especially in mariculture of marine fish and crustaceans(Reference Shiau34). In the present study, crabs fed the 9·9 % lipid diet had a higher proportion of DHA or EPA in hepatopancreas and muscle than those fed the 5·8 % lipid diet. However, crabs fed the 15·1 % lipid diet had similar levels of LC-PUFA compared with those fed the 9·9 % lipid diet, suggesting that dietary lipids at relatively higher levels were not efficiently digested. However, the fatty acid profiles of hepatopancreas and muscle were not entirely consistent with each other, with the proportions of n-3 LC-PUFA, especially EPA and DHA, being higher in hepatopancreas than in muscle, similar to results found in mud crab (Scylla paramamosain)(Reference Zhao, Wen and Li31).
SOD, as a first line of enzymatic-antioxidant defense, and GSH-Px, which may reduce organic lipid peroxides, were two important antioxidant enzymes that could protect cells from oxidative damage(Reference Locatelli, Canaud and Eckardt10,Reference Winston and Di Giulio35) . In the present study, the activity of GSH-Px in haemolymph was lower when dietary lipid level exceeded 5·8 % and crabs fed the 15·1 % diet had lower activity of SOD in the hepatopancreas than crabs fed the lower lipid diets. These results indicated that the antioxidant capacity of tissues decreased with increased dietary lipid level. Tissue MDA concentrations reflect the extent of lipid peroxidation in the animal and indirectly indicate the degree of potential damage to cells(Reference Jin, Wang and Huo14). In the present study, the contents of MDA both in haemolymph and hepatopancreas increased as dietary lipid increased from 5·8 to 15·1 %, indicating that lipid peroxidation products increased with increased dietary lipid level, possibly reflecting higher n-3 LC-PUFA levels. Overall, these results demonstrated that relatively high dietary lipid levels led to an increased risk of peroxidation and imposed a peroxidation burden(Reference Ji, Li and Liu36, Reference Tian, Lei and Ji37).
In the present study, key genes involved in LC-PUFA biosynthesis, such as fad, elovl4, elovl, srebp1, hnf4a and rxr, were significantly down-regulated when dietary lipid level increased from 5·8 to 15·1 %, which was in agreement with previous studies in teleost fish when subjected to similar experimental conditions(Reference Monroig, Tocher, Castro and Burdge38). Moreover, some studies in crustaceans showed that fad and elovl4 expression were inhibited by the level of dietary fish oil(Reference Lin, Hao and Zhu39–Reference Wu, Huang and Liu41). This result may suggest that high dietary fish oil could inhibit the synthesis of LC-PUFA in hepatopancreas of crab or, alternatively, that low dietary LC-PUFA activates endogenous production of these key fatty acids. In the present study, the expression levels of genes of transcription factors (srebp1, hnf4 and rxr) decreased with increased dietary lipid level, indicating that high dietary lipid (and especially high LC-PUFA) inhibited the transcriptional regulation of LC-PUFA biosynthesis pathways in the hepatopancreas of swimming crab. Functional characterisation of fad and elovl4 cloned from swimming crab indicated that the conversion ratios were much lower than for vertebrates, (unpublished) and so their LC-PUFA biosynthesis activity may be very low.
Scavenger receptor class (Srb) is an integral membrane protein that can act as a receptor for lipoproteins(Reference Yang, Zhou and Wang42). The relative expression of the srb gene was down-regulated in crabs fed the 15·1 % lipid diet, which may be the main reason that leads to a reduction of LDL in haemolymph. In mice, transport rates of LC-PUFA varied among members of the fatps family with relative rates of 8, 5, 2, 13, 2, 0 for fatp1 to fatp6 (Reference DiRusso, Li and Darwis43). Similarly, fabp1, fabp3 and fabp9 have different specificities in the transport of fatty acids. For instance, fabp1 has a close relationship with the transport and uptake of LC-PUFA, and it specifically binds LC-PUFA in the cytoplasm(Reference McArthur, Atshaves and Frolov44). It has been demonstrated that fabp3 has high affinity to LC-PUFA (especially EPA), but fabp9 affinity is reduced as the length of fatty acids increases in Eriocheir sinensis (Reference Zheng45). In addition to fabp1, the expression of lipoprotein receptor genes (lrp2, lpr1 and srb) and fatty acid uptake and transport genes (fabp3, fabp9 and fatp4) was down-regulated in crabs fed the 15·1 % lipid diet. The down-regulated expression of fabp family genes may be due to negative feedback regulation by high dietary lipid in order to prevent lipid deposition in the cells or tissues(Reference Bonen, Parolin and Steinberg46). The down-regulation of lrp2 and lpr1 expression inhibited the absorption of lipoprotein, and the expression of fabp3, fabp9 and fatp4 also decreased, which reduced fatty acid transport and absorption in hepatopancreas when crabs were fed the 15·1 % lipid diet. The expression of the fabp1 gene increased when dietary lipid level increased from 5·8 to 9·9 % and then decreased as dietary lipid further increased to 15·1 %. Apod is a member of the apo family, which function in lipid transport to maintain lipid balance of the organism(Reference Jiang, Cong and Zhang47). But, it is also part of the antioxidant defense system, which protects cells and tissues from oxidative damage(Reference Skerra48). Thus, apod is also a sensitive pathological marker and an early prediction index of disease(Reference Wang, Su and Ming49). In the present study, crabs fed the 15·1 % lipid diet presented higher expression of the apod gene than crabs fed the other diets, which indicated that to protect hepatopancreas from oxidative damage, the antioxidant defense systems must be activated by up-regulated expression of apod.
Both CPT and acyl-CoA oxidase (ACO) are key enzymes of fatty acid β-oxidation. In the present study, the relative expression of cpt1 was up-regulated as dietary lipid level increased from 5·8 to 9·9 %, which was consistent with results reported in large yellow croaker(Reference Yan, Liao and Wang50). In contrast, the relative expression of cpt2 decreased with increased dietary lipid level, suggesting that the accumulation of acyl-carnitine between the inner and outer membranes of mitochondria reduced the entry of long-chain fatty acids into the mitochondrial matrix and thus led to reduced β-oxidation. Meanwhile, the expression of the β-oxidation genes (cpt2, acox1, acox2 and acox3) in the hepatopancreas was also down-regulated, which may lead to a further accumulation of lipid in the hepatopancreas of crab. It is possible that β-oxidation of mitochondria (cpt2) and peroxisome (acox1, acox2 and acox3) was inhibited by high lipid (or n-3 LC-PUFA) in the diet. Lu et al.(Reference Lu, Xu and Wang3) also demonstrated that reduced CPT1 and ACO activities mainly contributed to lower hepatic β-oxidation capacity in fish fed a high lipid diet. Therefore, the increased proportion of LC-PUFA in hepatopancreas may be related to fabp1, and the up-regulation of cpt1 may also be related to the high level of fabp1. The accumulation of LC-PUFA in cells may therefore inhibit the transport of fabp1.
TAG anabolism and catabolism are crucial factors affecting lipid accumulation in hepatopancreas. FAS is a critical enzyme in de novo lipogenesis, and acyl-CoA diacylglycerol acyltransferase (DGAT) catalyses the final reaction step in the synthesis of TAG(Reference Yen, Stone and Koliwad51). The present study showed that lipogenic genes (fas, acc, g6pd, 6pgd, dgat1, gpat1 and gpat3) were down-regulated when dietary lipid level increased from 5·8 to 15·1 %. Down-regulation of lipogenic genes of TAG synthesis may be due to excessive lipid deposition in hepatopancreas, which leads to negative feedback regulation. However, in addition, high n-3 LC-PUFA can also inhibit lipogenesis. Van Herpen and Schrauwen-Hinderling(Reference Van Herpen and Schrauwen-Hinderling52) demonstrated that the down-regulation of fas and dgat inhibited lipogenesis and lipid deposition in cells to prevent lipo-toxicity.
Dietary lipid level can affect the expression of lipolytic genes such as hsl and lpl (Reference Lu, Xu and Wang3,Reference Yan, Liao and Wang50) . In the present study, the relative expression of hsl was down-regulated as dietary lipid level increased from 5·8 to 9·9 %, which was in agreement with studies in the teleost black seabream(Reference Jin, Lu and Yuan21). However, lpl was not significantly influenced by dietary lipid level, which was also in agreement with previous studies in red sea bream(Reference Liang, Oku and Ogata53) and large yellow croaker(Reference Yan, Liao and Wang50,Reference Wang, Li and Hou54) . Moreover, the lipolytic genes tgl and pl were down-regulated in hepatopancreas when the crabs were fed diets containing 15·1 and 9·9 % lipid, respectively. In addition, high dietary lipid appeared to inhibit the expression of intracellular lipase (il), which may suggest decreased ability for digestive hydrolysis of lipid droplets in hepatopancreas. Excessive dietary lipid also inhibited the catabolic response of TAG in hepatopancreas, reducing the capability for lipid digestion and utilisation, and thus promoting lipid deposition in hepatopancreas and muscle.
Conclusion
The results of the present study indicated that dietary lipid level affected growth performance, feed utilisation, fatty acid profiles of hepatopancreas and muscle and enzyme activities related to lipid metabolism of swimming crab. Specifically, high dietary lipid level retarded growth and reduced feed utilisation. Furthermore, dietary lipid level regulated lipid metabolic pathways in the hepatopancreas impacting lipid anabolism and catabolism of swimming crab. High dietary lipid level decreased lipoprotein clearance, reduced lipid digestion and lipolysis, inhibited mitochondrial and peroxisomal β-oxidation and resulted in increased hepatopancreatic lipid deposition. The present study provides new perspectives on the use of nutritional strategies for inducing LC-PUFA biosynthesis in marine crabs.
Acknowledgements
The authors expressed our thanks to the technical staff of FSN Laboratory involved in this experiment. We would also like to thank T. T. Zhu, C. C. Li and P. Ruan for their valuable assistance during the feeding trial, sampling and chemical analysis.
The present study was supported by National Key R & D Program of China (2018YFD0900400), China Agriculture Research System (CARS-48), National Natural Science Foundation of China (41476125), Nature Science Foundation of Zhejiang Province (LY17C190002), Key Research Program of Zhejiang Province of China (2018C02037), Zhejiang Aquaculture Nutrition & Feed Technology Service Team (ZJANFTST2017-2) and Major Agriculture Program of Ningbo (2017C110007). This research was also sponsored by the K. C. Wong Magna Fund in Ningbo University.
P. S. formulated the research question, designed the study, carried out the study, analysed the data and wrote the article. M. J. designed the study, assisted in the correction and developed the questions. L. J. assisted in the correction and developed the questions. O. M. designed the study, assisted in the correction and revised the manuscript. J. C. N. developed the questions and revised the manuscript. D. R. T. designed the study, assisted in the correction and developed the questions. M. B. B. developed the questions and revised the manuscript. X. W. was involved in data analysis. Y. Y. was involved in the laboratory assessments and data analysis. Q. Z. formulated the research question, designed the study and revised the manuscript. All the authors read and approved the final version of the manuscript.
The authors declared that there were no conflicts of interest.
Supplementary material
For supplementary material referred to in this article, please visit https://doi.org/10.1017/S0007114519002563















