Normal odd-chain SFA (OCSFA), particularly tridecanoic acid (n-13 : 0), pentadecanoic acid (n-15 : 0) and heptadecanoic acid (n-17 : 0), are normal components of ruminant products, specifically dairy products and beef( Reference Jensen 1 , Reference Pfeuffer and Jaudszus 2 ). They are also found in non-ruminant sources such as seafood( Reference Wang, Jackson and Twining 3 ). In recent years, n-15 : 0 and n-17 : 0 are considered biomarkers of dairy fat intake, mainly because their concentrations in serum and adipose tissue correspond with dairy product intake( Reference Albani, Celis-Morales and Marsaux 4 – Reference Wolk, Vessby and Ljung 9 ). Not only serum and adipose tissue, they are also found to incorporate in other human tissues such as plasma, erythrocytes and liver( Reference Pfeuffer and Jaudszus 2 ). Recent studies showed that n-15 : 0 and n-17 : 0 are biomarkers of not only dairy product but also seafood( Reference Wang, Jackson and Twining 3 ) and dietary fibre intake( Reference Weitkunat, Schumann and Nickel 10 ), which again indicates OCSFA is applicable in estimation of food intake. In addition, n-15 : 0 and n-17 : 0 are positively associated with insulin sensitivity and inversely associated with type 2 diabetes in both cohort( Reference Santaren, Watkins and Liese 6 , Reference Patel, Sharp and Jansen 11 ) and case–control studies( Reference Forouhi, Koulman and Sharp 12 – Reference Kroger, Zietemann and Enzenbach 14 ), which contrasts with links between incident diabetes and prevalent even-chain SFA (ECSFA) intake such as stearic acid (n-18 : 0). Circulating OCSFA and/or their relative concentrations (e.g. ratios) may be useful as a physiological index of health with careful attention to their origin and endogenous metabolism( Reference Pfeuffer and Jaudszus 2 , Reference Forouhi, Koulman and Sharp 12 , Reference Kroger, Zietemann and Enzenbach 14 , Reference Dawczynski, Kleber and Marz 15 ).
Concentrations of n-13 : 0, n-15 : 0 and n-17 : 0 in general milk products in the USA are about 0·1 %, 1·2 % and 0·6 % of total fatty acids (FA), respectively( Reference Jensen 1 ). They are likely to be originated primarily via ruminal bacteria. The ratio of n-15 : 0:n-17 : 0 in US dairy fat is 2:1( Reference Dewhurst, Moorby and Vlaeminck 16 – Reference Jenkins, West and Koulman 18 ). In contrast, it is approximately 1:2 in both freshwater and marine fish( Reference Wang, Jackson and Twining 3 , Reference Roubal 19 – Reference Njinkoue, Barnathan and Miralles 21 ) as well as in human plasma( Reference Weitkunat, Schumann and Nickel 10 , Reference Forouhi, Koulman and Sharp 12 , Reference Dawczynski, Kleber and Marz 15 , Reference Jenkins, West and Koulman 18 ). Comparable amounts of OCSFA are found in vegan erythrocytes( Reference Kornsteiner, Singer and Elmadfa 22 ). Because dairy fat is the predominant source of OCSFA in the USA, it is likely that endogenous FA interconversion alters their ratio via elongation, most importantly as n-15 : 0→n-17 : 0( Reference Pfeuffer and Jaudszus 2 , Reference Jenkins, West and Koulman 18 , Reference Riserus and Marklund 23 ); they may also arise by de novo synthesis. Current endogenous synthesis pathways of OCSFA are well summarised in previous reviews( Reference Pfeuffer and Jaudszus 2 , Reference Jenkins, West and Koulman 18 , Reference Riserus and Marklund 23 ). Among them, one theory proposes α-oxidation of ECSFA by intermediate hydroxylation/removal of one carbon from carboxylic end( Reference Jenkins, West and Koulman 18 ). However, few details are available on the gene products responsible for OCSFA biosynthesis, unlike well-known biochemical routes to ECSFA.
In ECSFA metabolism, FA elongation follows a four-step cycle comprising condensation, reduction, dehydration and second time reduction which occurs in endoplasmic reticulum( Reference Kihara 24 ). Mammalian FA elongases elongation of very long-chain fatty acid (ELOVL)1–7 work in the first and rate-limiting condensation step and all have substrate specificities and tissue-specific expression distribution. Among them, ELOVL1, 3, 6 and 7 preferentially act on SFA and MUFA; ELOVL2 and 5 are known to work on PUFA; while ELOVL4 elongate very long-chain FA with more than twenty-four carbons regardless of unsaturation( Reference Kihara 24 – Reference Zhang, Kothapalli and Brenna 26 ). Currently established substrate specificities of ELOVL towards ECSFA are as follows: ELOVL6, n-12 : 0→n-14 : 0→n-16 : 0→n-18 : 0( Reference Guillou, Zadravec and Martin 25 , Reference Matsuzaka and Shimano 27 ); ELOVL1, 3 and 7, n-18 : 0→n-20 : 0→→→n-26 : 0; ELOVL4, n-26 : 0→→30 : 0( Reference Kihara 24 , Reference Guillou, Zadravec and Martin 25 ). ELOVL6 catalyses n-16 : 0→n-18 : 0 in ruminant mammary cells( Reference Shi, Wu and Zhu 28 ) and is expressed ubiquitously in bovine mammary epithelial cells( Reference Chen, He and Liu 29 ). ELOVL6 genetic variants are reportedly associated with insulin sensitivity in a Spanish population( Reference Morcillo, Martin-Nunez and Rojo-Martinez 30 ).
Unlike the well-studied ECSFA, endogenous metabolism of OCSFA once ingested is not well characterised with respect to the relevant genes encoding enzymes that catalyse their interconversion. As circulating n-15 : 0 and n-17 : 0 are regarded as biomarkers of dairy product, dietary fibre and seafood intake, we aimed to establish specificity of the ELOVL responsible for elongation of OCSFA of quantitative significance in the human diet. We tested the hypothesis that ELOVL6 is specific to n-13 : 0→n-15 : 0→n-17 : 0 compared with the other ELOVL known to operate on straight chain FA. We adopted an approach analogous to previous successful studies that established numerous novel functions for PUFA biosynthetic genes by transient or stable transfection of the open reading frame (ORF) into MCF7 cells and other models( Reference Park, Park and Kothapalli 31 – Reference Geay, Tinti and Mellery 38 ). To test ELOVLx (ELOVL1, 3, 6 and 7) function, we constructed expression vectors and transiently transfected them individually into MCF7 cells as a human cell host.
Methods
Chemicals and reagents
FA (n-13 : 0, n-15 : 0 and n-17 : 0) were purchased from Sigma-Aldrich. Solvents are HPLC grade for FA extraction and were purchased from Sigma-Aldrich and Burdick & Jackson. Cell culture media, fetal bovine serum (FBS) and other cell culture reagents were obtained from Life Technologies, Corning and Thermo Fisher Scientific.
Elongation of very long-chain fatty acid 6 sequence and phylogenetic analysis
The amino acid (AA) sequences of ELOVL6 from various vertebrate species are obtained from GenBank accession numbers (online Supplementary Table S1). The AA sequence of human ELOVL6 was aligned with several other vertebrate ELOVL6 sequences using ClustalX 2.1 software( Reference Larkin, Blackshields and Brown 39 ). The phylogenetic tree was constructed using the neighbour-joining method( Reference Saitou and Nei 40 ) with MEGA7( Reference Kumar, Stecher and Tamura 41 ). Confidence in the resulting phylogenetic tree branch topology was measured by bootstrapping test method with 1000 replicates( Reference Felsenstein 42 ).
Elongation of very long-chain fatty acid expression vector constructs
The ORF of ELOVL transcripts (ELOVL1, ELOVL3, ELOVL6 and ELOVL7) were cloned into a pcDNA3.1(+) expression vector (Thermo Fisher Scientific) containing cytomegalovirus promoter. The specific ELOVL gene synthesis and cloning was carried out by GenScript Service. The GenBank accession numbers of ELOVL mRNA (NM) and protein (NP) are provided in online Supplementary Table S1. Plasmid DNA used for transfection assays was extracted and purified using Plasmid Midi Kit (Qiagen). The extracted DNA was verified by DNA sequencing and stored at –20°C. DNA sequencing was performed at Cornell University Life Sciences Core Laboratories Center using the Applied Biosystems automated 3730 DNA analyser.
Mammalian cell culture, transfection and fatty acid supplementation
MCF7 human breast cancer cells were grown at 37°C in a humidified environment with 5 % CO2, using minimum essential medium α with 10 % FBS and 10 mm buffer (HEPES), as described previously( Reference Park, Kothapalli and Park 32 , Reference Park, Kothapalli and Lawrence 34 ).
MCF7 cells were seeded at 1×106 cell density into 60 mm cell culture dishes. After 48 h, when they reach 60–80 % confluency, cells were washed with 1× PBS and ELOVL (ELOVL1, 3, 6 and 7) transcripts were transfected individually using Polyplus jetPRIME transfection reagent. Empty vector was used as control. According to the jetPRIME reagent kit protocol, 4 µg of vector (control) or ELOVL DNA was transfected into cells along with 200 µl jetPRIME buffer, 8 µl jetPRIME reagent and 5 ml growth media. After 24 h, the transfected MCF7 cells were supplemented with 80 µm of bovine serum albumin (BSA) bound OCSFA substrates (n-13 : 0, n-15 : 0 and n-17 : 0). Briefly, to make BSA-bound substrates, n-13 : 0, n-15 : 0 and n-17 : 0 were dissolved in absolute ethanol to make 100 mm FA stock. FA stock (200 µl) was then mixed with FA-free BSA in 1× PBS (4·4 % w/w) and incubated overnight at 37°C. BSA-bound OCSFA were filtered using 0·22 µm syringe, and diluted to 80 µm with non-FBS MCF7 media and then added to cells. After additional 24 h incubation, cells were washed twice with 1× PBS, harvested by trypsinisation and supernatant removed after centrifuging.
RNA isolation and complementary DNA synthesis
RNA was isolated from the harvested MCF7 cell pellets using E.Z.N.A. Total RNA Kit I (Omega Bio-tek Inc.). The RNA quantity and quality were verified by micro spectrophotometer Nanodrop 2000 (Thermo Scientific). Complementary DNA (cDNA) was synthesised from 1 µg of RNA using high-capacity cDNA reverse transcription kit (Life Technologies). Synthesised cDNA was then used as template for real-time PCR (RT-PCR) reactions( Reference Yan, Wang and Greenwald 43 ).
RT-PCR
Gene-specific primers ELOVL1–7 were designed using PrimerQuest software (Integrated DNA Technologies), and primer sequences and annealing temperatures are shown in online Supplementary Table S2. RT-PCR amplification reactions were performed using EmeraldAmp GT PCR Master Mix (Clontech) using gradient thermal cycler (Eppendorf). PCR products were separated by 2 % agarose gel electrophoresis stained with ethidium bromide, and bands were visualised under UV light. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control gene.
Fatty acid extraction and analysis
Fatty acid methyl esters (FAME) from the harvested MCF7 cell pellets were prepared according to the modified one-step method of Garces & Mancha( Reference Garces and Mancha 44 ). FAME were structurally identified by GC – chemical ionisation, electron ionisation (EI) MS and EIMS/MS using a Saturn 2000 mass spectrometer attached to a Varian Star 3400 gas chromatograph( Reference Ran-Ressler, Lawrence and Brenna 45 ). FAME were quantified by GC-flame ionisation detector (GC-FID; Hewlett-Packard). An equal weight FAME mixture, GLC462 (Nu-Check Prep Inc.), was used to calculate response factors of all FA. Percentage conversion of substrates (S) to products (P) was calculated as: ((P)/(S+P))×100, and normalised to the control group.
Statistical analysis
All treatments were performed using two biological replicates; the mean of three technical replicate GC-FID analyses were used for each biological replicate and no data were excluded. In numerous earlier studies, we used an analogous approach with MCF7 transfection and FA treatment with 2–3 biological replicates for functional characterisation of PUFA biosynthetic genes( Reference Park, Park and Kothapalli 31 – Reference Park, Kothapalli and Lawrence 34 ); this approach is comparable to functional characterisation studies conducted by others( Reference Morais, Castanheira and Martinez-Rubio 35 – Reference Geay, Tinti and Mellery 38 ). Values generated using averages across the two biological replicates are expressed as means and standard deviations. Statistical analysis of comparisons between multiple groups was performed using OriginPro 8 Software (OriginLab Corporation). One-way ANOVA with post hoc Tukey honest significant difference test was used to analyse significant differences between groups. When P<0·05 the differences are considered significant.
Results
Amino acid sequence and phylogenetic analysis of elongation of very long-chain fatty acid 6
The human ELOVL6 cDNA (NM_024090.2) consist of a 798 bp ORF, encoding a protein of 265 AA (NP_076995.1). As shown in Fig. 1, human ELOVL6 shares >90 % AA sequence identity with other vertebrate ELOVL6 sequences. The human ELOVL6 shared 98·11, 96·98, 96·60 and 93·16 % identity, with monkey, mouse, rat and cattle sequences, respectively. All vertebrate ELOVL6 possessed five transmembrane domains (I–V) found among elongases( Reference Matsuzaka, Shimano and Yahagi 46 ) as well as the conserved histidine box HXXHH motif characteristic of elongase families( Reference Jin, Monroig and Navarro 47 – Reference Zheng, Li and Han 49 ).

Fig. 1 Alignment of amino acid (AA) sequences of elongation of very long-chain fatty acid 6 (ELOVL6) from human and other vertebrates. The AA sequences of various species obtained from GenBank accession numbers were aligned using ClustalX 2.1. The well conserved histidine motif HXXHH is depicted in the box. The dotted lines with roman numerals indicate the putative transmembrane regions( Reference Matsuzaka, Shimano and Yahagi 46 ).
A phylogenetic tree was constructed by comparing the AA sequences of ELOVL6 from various vertebrates (Fig. 2). As expected, the human ELOVL6 grouped with primates, while rodents (rat and mouse) and fish (catfish and zebrafish) grouped together.
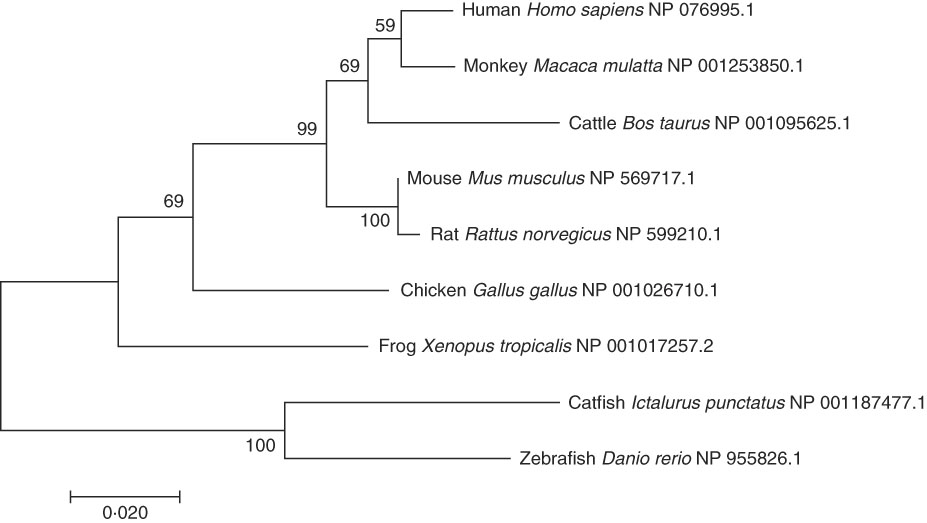
Fig. 2 Phylogenetic tree of ELOVL6 from human and other organisms. The tree was constructed using the neighbour-joining method with 1000 bootstrap replicates in MEGA7. The numbers represent the frequencies (%); and horizontal branch length is proportional to amino acid substitution rate per site. Each species was followed by its National Center for Biotechnology Information (NCBI) reference sequence (NP).
Distribution of SFA in MCF7 cells
MCF7 cells are high in ECSFA n-16 : 0 (16·63 (sd 0·43) %) and n-18 : 0 (12·98 (sd 0·52) %), but contain only trace amount of OCSFA n-17 : 0 (0·22 (sd 0·02) %), and n-13 : 0 and n-15 : 0 are at undetectable amounts (Z Wang, KSD Kothapalli and JT Brenna, unpublished results). Table 1 summarises when MCF7 cells are dosed with 80 µm of specific OCSFA. Cells readily uptake OCSFA in the order of n-17 : 0>n-15 : 0>n-13 : 0.
Table 1 Uptake efficiency of odd-chain SFA (OCSFA) by MCF7 cellsFootnote * Footnote † (Mean values and standard deviations)
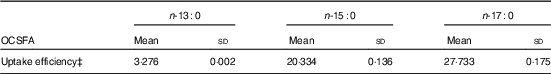
* MCF7 cells have trace amount of basal OCSFA; control cells readily uptake OCSFA when treated with 80 µm of albumin-bound individual OCSFA, and was in the order of n-17 : 0>n-15 : 0>n-13 : 0.
† Data from two biological replicates.
‡ Uptake efficiency (%)=wt (OCSFA uptake)/wt (total fatty acids excluding OCSFA)×100 %.
Transient transfection, elongation of very long-chain fatty acid gene expression
Online Supplementary Fig. S1 shows ELOVL (ELOVL1 to ELOVL7) gene expression in wild type, control (empty pcDNA3.1(+) vector) and ELOVLx transiently transfected cells. The cells were incubated with (a) 80 µm n-13 : 0; (b) 80 µm n-15 : 0 and (c) 80 µm n-17 : 0. ELOVL1 expression was higher in ELOVL1 transfected cells; similarly ELOVL3, ELOVL6 and ELOVL7 expression are higher in ELOVL3, 6 and 7 transfected cells. These results show transfections were successful.
Elongation, n-13 : 0→n-15 : 0
MCF7 cells transiently expressing ELOVL1, ELOVL3, ELOVL6 and ELOVL7 and control (empty vector) were incubated with 80 µm of albumin-bound n-13 : 0 OCSFA. MCF7 cells have native elongase activity, so the gain of function is compared with control (Fig. 3). The cells expressing ELOVL6 showed significantly increased activity towards n-13 : 0 (131·19 (sd 5·52) %). No gain of activity was seen with other elongases.
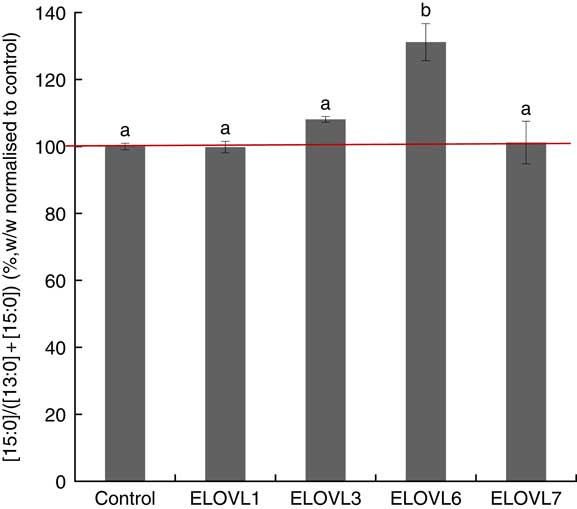
Fig. 3 Elongation of very long-chain fatty acid (ELOVL)x activity, n-13 : 0→n-15 : 0. Percentage conversion of fatty acid substrate n-13 : 0 into elongation product n-15 : 0 (ratios shown, calculated as (n-15 : 0)/(n-13 : 0)+(n-15 : 0)) was measured in MCF7 cells and normalised to the control group. ELOVL1, 3 and 7 showed no activity towards n-13 : 0 (n-13 : 0→n-15 : 0). ELOVL6 had significantly higher catalytic activity towards n-13 : 0 (n-13 : 0→n-15 : 0). Data from two biological replicates. a,b Mean values with unlike letters were significantly different (P<0·05).
Elongation, n-15 : 0→n-17 : 0
MCF7 cells transiently expressing ELOVL1, ELOVL3, ELOVL6 and ELOVL7 and control (empty vector) were incubated with 80 µm of albumin-bound n-15 : 0 OCSFA. MCF7 cells have native elongase activity, so the gain of function is compared with control (Fig. 4). The cells expressing ELOVL6 showed significantly increased activity towards n-15 : 0 (129·51 (sd 1·74) %), followed by moderate activity in ELOVL7 cells (113·99 (sd 1·30) %). No gain of activity was seen with ELOVL1. Elongation activity was lower than control in the ELOVL3 cells though not different than ELOVL1 cells.
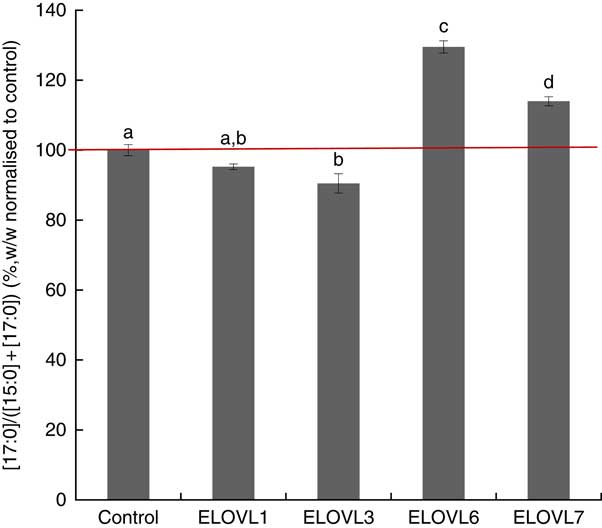
Fig. 4 Elongation of very long-chain fatty acid (ELOVL)x activity, n-15 : 0→n-17 : 0. Percentage conversion of fatty acid substrate n-15 : 0 into elongation product n-17 : 0 (ratios shown, calculated as (n-17 : 0)/((n-15 : 0)+(n-17 : 0))) was measured in MCF7 cells and normalised to the control group. ELOVL1 showed no activity towards n-15 : 0 (n-15 : 0→n-17 : 0). ELOVL3 reduced activity towards n-15 : 0 (n-15 : 0→ n-17 : 0) compared with control though it was not different than ELOVL1. ELOVL6 had significantly higher catalytic activity towards n-15 : 0 (n-15 : 0→n-17 : 0). ELOVL7 showed moderate activity towards n-15 : 0 (n-15 : 0→n-17 : 0). Data from two biological replicates. a,b,c,d Mean values with unlike letters were significantly different (P<0·05).
Interconversion, n-17 : 0→n-15 : 0 but not n-19 : 0
MCF7 cells transiently expressing ELOVL1, ELOVL3, ELOVL6 and ELOVL7 and control (empty vector) were incubated with 80 µm of albumin-bound n-17 : 0 OCSFA. All MCF7 cells (ELOVL1, ELOVL3, ELOVL6 and ELOVL7 and control) readily uptake n-17 : 0 but none of the cells showed elongation activity towards n-17 : 0; n-19 : 0 is not detected. While n-15 : 0 is not detectable in untreated cells, incubation with n-17 : 0 results in n-15 : 0 appearance at approximately equal amounts ((15:0)/((15:0)+(17:0)) approximately 3 %) regardless of ELOVLx transfection, suggesting that β-oxidation is a route of n-17 : 0 conversion.
Discussion
Our results confirm the hypothesis that ELOVL6 is the primary ELOVL with activity towards OCSFA, specifically catalysing n-13 : 0→n-15 : 0 and n-15 : 0→n-17 : 0. Furthermore, ELOVL7 has moderate activity towards n-15 : 0→n-17 : 0. ELOVL1 had no activity towards any OCSFA, and no ELOVL catalysed n-17 : 0→n-19 : 0, consistent with the trace amount of n-19 : 0 in human tissue. ELOVL3 significantly reduced activity compared with control but not compared with ELOVL1, which was not different than control. MCF7 cells have native elongation activity, thus it is plausible that ELOVL3 may have inhibited that native conversion, possibly by non-active substrate binding. The magnitude of the effect is small, however.
Our AA sequence alignments of ELOVL6 showed a high degree of conservation among evolutionarily distant-related genomes (human to zebrafish). The histidine-rich motif and all five transmembrane regions are conserved from human to zebrafish. The pattern of ELOVL6 homology among several vertebrate species even with distant phylogeny points to similar metabolic conservation.
ELOVL6 belongs to a highly conserved microsomal enzyme family that is involved in FA biosynthesis. ELOVL6 activity characterised by cloning of mammalian ELOVL6 ORF into expression vector and expressing the vector in mammalian cells shows that ELOVL6 specifically catalyses the elongation of ECSFA and MUFA with chain lengths of 12, 14 and 16 carbons( Reference Matsuzaka, Shimano and Yahagi 46 , Reference Moon, Shah and Mohapatra 50 ). ELOVL6 is found to be ubiquitously expressed in mice and bovine tissues( Reference Chen, He and Liu 29 , Reference Matsuzaka, Shimano and Yahagi 46 ), and plays a role in energy metabolism and insulin sensitivity( Reference Matsuzaka and Shimano 27 ), nonalcoholic steatohepatitis( Reference Laggai, Kessler and Boettcher 51 ), breast cancer( Reference Feng, Chen and Kuo 52 ), pulmonary fibrosis( Reference Sunaga, Matsui and Ueno 53 ) and squamous cell carcinoma of the lung( Reference Marien, Meister and Muley 54 ).
The endogenous synthesis of OCSFA via α-oxidation of ECSFA has been previously reported( Reference Jenkins, West and Koulman 18 , Reference Roberts, Virtue and Vidal-Puig 55 , Reference Kondo, Ohno and Yamagata 56 ). In an adipocyte differentiation study, both kinetic and 13C palmitate labelled experiments showed significant increase in the synthesis of OCSFA via α-oxidation( Reference Roberts, Virtue and Vidal-Puig 55 ). The differentiating adipocytes converted 13C palmitate (n-16 : 0) to n-15 : 0, showing that OCSFA were endogenously synthesised from n-16 : 0 by α-oxidation in this cell type. This conversion happened only in the cells and not in cell culture media( Reference Roberts, Virtue and Vidal-Puig 55 ). Similarly, Su et al. ( Reference Su, Han and Yang 57 ) showed differentiating adipocytes when incubated with [9,-10-3H]n-16 : 0 that resulted in the production of radiolabelled [3H]n-15 : 0 via α-oxidation. Casteels et al. ( Reference Casteels, Sniekers and Fraccascia 58 ) proposed that α-oxidation mechanism might play a role in the formation of OCSFA in the brain. An in vivo experiment with rats infused with n-18 : 0 showed approximately 70 % (P<0·001) increase in the n-17 : 0 levels in the serum compared with a control rat group( Reference Jenkins, Seyssel and Chiu 59 ). Our results show that specific ELOVL6 or 7 may operate on these nascent products α-oxidation n-13 : 0 and n-15 : 0 but not on n-17 : 0 to further alter OCSFA profile (Fig. 5).
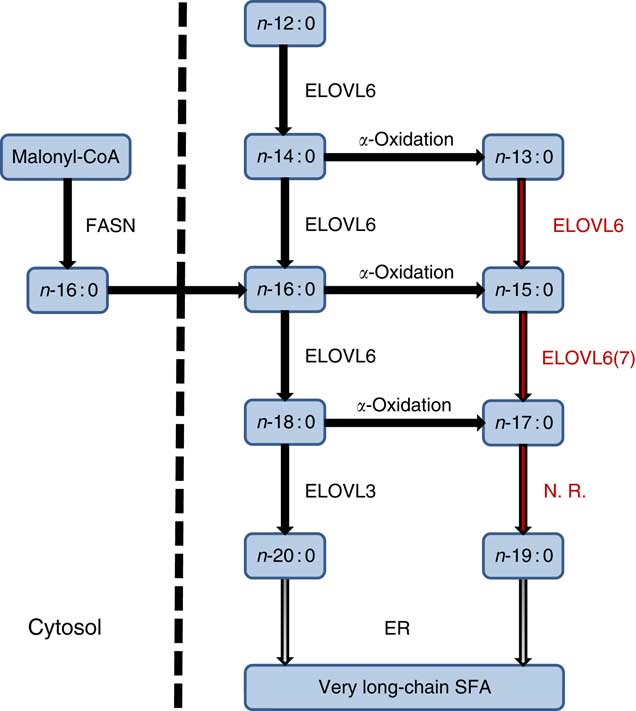
Fig. 5 Even-chain SFA (ECSFA) and odd-chain SFA (OCSFA) biosynthesis in vertebrates. Elongation of very long-chain fatty acid (ELOVL)6 catalyses elongation of n-12 : 0 to n-18 : 0, whereas ELOVL3 catalyses elongation of n-18 : 0 to n-20 : 0. Depicted in red font colour: ELOVL6 elongates OCSFA n-13 : 0→n-15 : 0 and n-15 : 0→n-17 : 0. ELOVL7 (shown as 7) has moderate activity (n-15 : 0→n-17 : 0). Palmitic acid (n-16 : 0), the major product of fatty acid synthase (FASN), and α-oxidation of ECSFA to OCSFA are also shown. ER, endoplasmic reticulum; N.R., no reaction.
Here, we found overlapping activity for ELOVL6 and ELOVL7; both had substrate specificity for n-15 : 0. Elongases in several species are known to share common overlapping functions( Reference Zheng, Li and Han 49 , Reference Lee, Stephens and Paul 60 , Reference Wang, Botolin and Christian 61 ). Previously it has been shown that mammalian ELOVL1, ELOVL3 and ELOVL7 share common substrates( Reference Guillou, Zadravec and Martin 25 , Reference Zheng, Li and Han 49 , Reference Ohno, Suto and Yamanaka 62 , Reference Sassa and Kihara 63 ). ELOVL1 elongates ECSFA of chain lengths n-18 : 0 to n-26 : 0, with the highest activity towards n-22 : 0 FA, whereas, ELOVL3 and ELOVL7 were found to elongate n-16 : 0 to n-22 : 0 FA, with the highest activity towards n-18 : 0 FA( Reference Ohno, Suto and Yamanaka 62 – Reference Naganuma, Sato and Sassa 64 ).
The relatively small biological replicate size (n 2) in our study has consistently yielded reliable results in our previous work( Reference Park, Park and Kothapalli 31 – Reference Park, Kothapalli and Lawrence 34 ). For functional characterisation studies, others have used a single assay to support conclusions based on identification of a novel FA product by chromatography with subsequent calculation of a conversion ratio( Reference Morais, Castanheira and Martinez-Rubio 35 – Reference Geay, Tinti and Mellery 38 ), thus greater confidence in matching biological duplicates of the present results is warranted. Our uses of duplicates with consistently small standard deviations in FAME analysis, as well as validation of ELOVLx transfection efficiency via RT-PCR, support a similar level of confidence in the present results. Finally, we note that our data on normal PUFA( Reference Park, Park and Kothapalli 31 – Reference Park, Kothapalli and Lawrence 34 ) and ECSFA (Z Wang, KSD Kothapalli and JT Brenna, unpublished results) faithfully replicates that of others.
In conclusion, we provide the first molecular evidence demonstrating ELOVL6 is the major elongase acting on OCSFA, with specificity towards n-13 : 0 and n-15 : 0. Modest activity was found for ELOVL7 towards n-15 : 0. The present study expands ELOVL substrate specificity range to OCSFA. Nutrition studies considering these FA as markers of specific food intake should consider interconversion of OCSFA in light of these findings, particularly genome-wide association studies and targeted gene studies as they associate circulating OCSFA levels with SNP and other polymorphisms.
Acknowledgements
The authors thank Dr Hui Gyu Park, Dr Jiyao Zhang and Dr Lei Liu for assistance with experiment protocols.
This work was supported by National Institutes of Health (NIH) grant R01 AT007003 from the National Center for Complementary and Integrative Health and the Office of Dietary Supplements. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
J. T. B., K. S. D. K. and Z. W. formulated the research questions and designed the study; Z. W., D. H. W., Y. G. and Y. Y. executed the research; J. T. B., K. S. D. K., Z. W. and P. L. analysed and interpreted the data; and J. T. B., K. S. D. K. and Z. W. wrote the first draft and all authors approved the final draft.
The authors declare that there are no conflicts of interest.
Supplementary material
For supplementary material/s referred to in this article, please visit https://doi.org/10.1017/S0007114518003185









