Dyslipidaemia is an important risk factor for CVD(1). Overweight and obesity, in particular intra-abdominal fat deposition, is associated with decreased HDL-cholesterol (HDL-C) and hypertriacylglycerolaemia(1). Plasma concentrations of total cholesterol (TC), LDL-cholesterol (LDL-C), HDL-C and TAG are modified by diet, physical activity and smoking status(1). However, there are considerable inter-individual differences in metabolic susceptibility to these lifestyle factors(Reference Ye and Kwiterovich2, Reference Bluher3). These differences may partly be determined by genetic factors, and several genome-wide association studies (GWAS) have identified loci that are associated with blood lipid concentrations(Reference Aulchenko, Ripatti and Lindqvist4–Reference Teslovich, Musunuru and Smith15). Still, these loci only explain a part of the variance in lipid profiles, and this could be partly due to gene–gene and gene–environment interaction effects(Reference Zuk, Hechter and Sunyaev16), as previous candidate gene studies have suggested interaction effects between genes and lifestyle for dyslipidaemia(Reference Ordovas, Robertson and Cleirigh17, Reference Corella and Ordovas18).
The objective of the present study was to investigate whether SNPs in nutrient-sensitive genes involved in lipid metabolism modified lipid profile after weight loss and in response to a given diet. We analysed 240 SNPs in twenty-four presumed nutrient-sensitive candidate genes among overweight European adults participating in the Diet Obesity and Genes (DiOGenes) study. SNP main effects on TC, LDL-C, HDL-C and TAG were examined after an 8-week low-energy diet (LED), and both SNP main and SNP–diet interaction effects were examined after a 6-month ad libitum weight maintenance diet, with different contents of dietary protein or glycaemic index (GI).
Materials and methods
The DiOGenes study (www.DiOGenes-eu.org) is a Pan-European randomised dietary intervention study exploring the effect of diets with different contents of protein and GI on weight regain and metabolic health after weight loss. The study was conducted according to the guidelines laid down in the Declaration of Helsinki, and all procedures involving human subjects were approved by the local ethical committee in the respective country. Written informed consent was obtained from all subjects. The participants were overweight or obese (BMI 27–45 kg/m2) but otherwise healthy, with no heart disease, diabetes, severe dyslipidaemia or hypertension. The present trial has been registered at ClinicalTrials.gov (identification no. NCT00390637).
Design and methods have been described in detail previously(Reference Larsen, Dalskov and van Baak19). In brief, participants who had lost ≥ 8 % of their initial body weight after an 8-week LED (3400–4200 kJ/d) were randomised to one of five different 6-month ad libitum weight maintenance diets based on either combinations of low/high dietary protein (LP/HP) and low/high GI (LGI/HGI), or a control diet according to national dietary guidelines: (1) LP/LGI; (2) LP/HGI; (3) HP/LGI; (4) HP/HGI; (5) control diet. The target for the dietary intervention was a difference of 12 % of total energy consumed from protein between the HP and LP diets and a difference of 15 GI units between the HGI and LGI diets. The actual differences between the respective diets calculated from diet registrations were 5·4 % of energy consumed from protein and 5 GI units(Reference Larsen, Dalskov and van Baak20).
TC, HDL-C and TAG were analysed at the Department of Clinical Biochemistry, Gentofte University Hospital, Denmark, and LDL-C was calculated using Friedewald's equation(Reference Friedewald, Levy and Fredrickson21).
Initially, sixty-nine candidate nutrient-sensitive genes were selected based on prior knowledge of whether the pathway, gene, gene transcript or SNP was implicated in obesity, weight loss, weight regain or associated diseases with emphasis on the interaction with dietary protein or GI, from literature search and the TUB database at IntegraGen (Evry, France) for the purpose of investigating their role in body weight regulation during dietary treatment. For the presumed nutrient-sensitive candidate genes, a comprehensive approach was used to ensure genetic coverage of the locus ( ± 5 kb) by selecting tagSNPs for each of the selected genes. TagSNPs were identified from the International HapMap data for European ancestry (release 20, NCBI build 35), and linkage disequilibrium (LD) structure was evaluated using Haploview software, version 3.32 (Broad Institute of MIT and Harvard, MA, USA)(Reference Barrett, Fry and Maller22). TagSNPs were selected using Tagger(23) with a single marker option with an LD threshold of r 2 0·7–0·8. SNPs located in exonic regions, frequently studied, or included in the Illumina HumanHap 300 were preferentially included as tagSNPs, while SNPs with an expected low genotyping success rate (in close proximity to another SNP (60 bp) or in a repeat region) were deselected. In total, 768 tagSNPs were selected for genotyping (Table S1, available online). Among the selected nutrient-sensitive genes, the genes that were known from the literature to be involved in lipid metabolism were included in the analyses; these twenty-four genes included: adiponectin (ADIPOQ)(Reference Shimabukuro, Higa and Asahi24); β2/β3-adrenergic receptor (ADRB2,3)(Reference Large, Hellstrom and Reynisdottir25); activating transcription factor 6 (ATF6)(Reference Zeng, Lu and Mori26); basic helix-loop-helix family, member e40 (BHLHE40)(Reference Iwata, Kawamoto and Sasabe27); caveolin 1 (CAV1)(Reference Park, Kim and Kim28); CCAAT/enhancer binding protein (CEBPB)(Reference Zhang, Tang and Vinson29); cathepsin S (CTSS)(Reference Taleb, Lacasa and Bastard30); fatty acid-binding protein 1 (FABP1)(Reference Newberry, Xie and Kennedy31, Reference Fisher, Weikert and Klapper32); fatty acid-binding protein 4 (FABP4)(Reference Cabre, Lazaro and Girona33); farnesyltransferase (FNTA)(Reference Capel, Viguerie and Vega34); leptin (LEP)(Reference Shimabukuro, Koyama and Chen35); lipin 1, 2, 3 (LPIN1, 2, 3)(Reference Reue and Dwyer36, Reference Reue and Zhang37); lipoprotein lipase (LPL)(Reference Wang and Eckel38); Max-like protein (MLX) interacting protein-like (MLXIPL)(Reference Hurtado del Pozo, Vesperinas-Garcia and Rubio39); matrix metallopeptidase 9 (MMP9)(Reference Chavey, Mari and Monthouel40); microsomal TAG transfer protein (MTTP)(Reference Xie, Newberry and Young41); nuclear receptor subfamily 1, group I, member 2 (NR1I2)(Reference Moreau, Vilarem and Maurel42); phosphoenolpyruvate carboxykinase 2 (PCK2)(Reference Shin, Park and Kim43); PPARγ co-activator 1-α (PPARGC1A)(Reference Bournat and Brown44); PPARδ (PPARD)(Reference Skogsberg, Kannisto and Cassel45); PPARγ (PPARG)(Reference Norris, Hirshman and Yao46).
Genotyping of all samples was performed using the Illumina® Bead Station System (Illumina, Inc.) by IntegraGen with CEPH (Human Polymorphism Study Center) controls (reproducibility: 100 %; concordance rate: 99·6 %). A total of 240 tagSNPs with a call rate ≥ 95 %, a minor allele frequency (MAF) >1 % and without significant (P>0·001) deviations from Hardy–Weinberg equilibrium were included in the main analyses. Genotype analyses were performed and reported with respect to the minor allele, here defined as the risk allele.
Statistical analyses
By multiple linear regression analyses, we examined the following: (1) SNP main effects on changes in blood lipids after an 8-week LED; (2) SNP main effects on changes in blood lipids after the 6-month ad libitum weight maintenance diet; (3) SNP–diet interaction effects on changes in blood lipids for HP v. LP and HGI v. LGI after the 6-month ad libitum weight maintenance diet. SNP main effects on TC, HDL-C, LDL-C and TAG at baseline were also examined but merely at an explorative level, as the present study was not initially designed to investigate baseline associations.
Before the multiple linear regression analyses, the five-level diet variables were recoded into indicator variables for levels of protein intake and GI (and for the control diet); additive genetic models were assumed and corresponding SNP main-effect, diet main-effect and SNP–diet (product-based) interaction variables were created and used for the analyses. Models were adjusted for baseline age, BMI and waist circumference, sex, smoking status, partner, weight loss (after the LED) and weight regain (after the ad libitum diet). Furthermore, we adjusted for period length of the LED and the ad libitum diet, where some variation from the intended duration occurred. The LGI or LP served as reference groups of main interest and control diet status was included in models but not of main focus, due to the variation in diets between countries.
Based on the available sample sizes (Table 1), we performed power calculations in the form of least detectable effects based on the assumption of significance levels and powers of 5 and 80 %, respectively. The analysis of SNP main effects associated with gains during the LED phase led to least detectable effects of 0·34 (MAF = 0·05) and 0·15 (MAF = 0·45) in units of standard deviations of the outcome. Similarly, the case of SNP main-effects analysis related to gains during the ad libitum diet gave least detectable effects of 0·40 (MAF = 0·05) and 0·18 (MAF = 0·45). Finally, considering the SNP–diet interaction analyses, the least detectable effects were 0·83 (MAF = 0·05) and 0·33 (MAF = 0·45), respectively. All power calculations were performed using QUANTO, version 1.2.4 (May 2009 (http://hydra.usc.edu/gxe/)).
Table 1 Characteristics of the participants included in the analyses for baseline, 8 weeks on the low-energy diet and 6-month ad libitum weight maintenance diet (Mean values, standard deviations and number of observations)

LP, low dietary protein; LGI, low glycaemic index; HGI, high glycaemic index; HP, high dietary protein; WC, waist circumference; Δ, change during the low-energy diet and during the 6-month ad libitum weight maintenance diet; TC, total cholesterol; LDL-C, LDL-cholesterol; HDL-C, HDL-cholesterol.
Bonferroni correction was used to adjust for multiple testing, concerning the 240 SNPs in genes presumed to be involved in lipid metabolism, in practice corresponding to an uncorrected significance level of α = 2·1 × 10− 4 ( = 0·05/240) at baseline/after the LED, and α = 6·9 × 10− 5 ( = 0·05/(3 × 240)) for main and interaction effects during the ad libitum diet period, accounting for SNP main effects, SNP–protein and SNP–GI interaction effects, when testing against a corrected α-level of 0·05. Analyses were performed using Stata 9·2/11·2 (StataCorp LP, 2007/2011).
Results
The characteristics of all participants at inclusion, after the 8-week LED period and after the 6-month ad libitum weight maintenance diet are summarised in Table 1. SNPs in lipid metabolism-related genes, with the strongest associations with blood lipids (corresponding to P values < 0·001) after the interaction with dietary protein and GI, are presented in Table 2.
Table 2 Associations of SNPs in the lipid metabolism genes with blood lipids after the interaction with dietary protein and glycaemic index, during the 6-month ad libitum diet* (Regression coefficients, 95 % confidence intervals and number of observations for the minor allele)
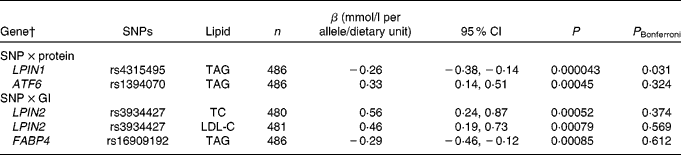
P, uncorrected P value; P Bonferroni, Bonferroni-corrected P value; LPIN1, lipin 1; ATF6, activating transcription factor 6; LPIN2, lipin 2; TC, total cholesterol; LDL-C, LDL-cholesterol; FABP4, fatty acid-binding protein 4.
* SNP–dietary protein and SNP–glycaemic index associations with blood lipids are estimated by the multiple linear regressions.
† The gene denotes the locus with the closest proximity to the SNP.
None of the SNPs in the lipid metabolism-related genes was found to modify TC, LDL-C, HDL-C or TAG after the 8-week LED, independent of weight loss (Table S3(A)–(D), available online).
After the 6-month ad libitum weight maintenance diet, a gene–dietary protein interaction effect on TAG was identified for LPIN1 rs4315495 with a decrease in TAG of − 0·26 mmol/l per A-allele/protein unit (95 % CI − 0·38, − 0·14, P= 0·000043; Fig. 1 and Table 2). We did not identify any other significant associations when correcting for multiple testing for SNP main effects (Table S4(A)–(D), available online) or diet interaction effects with dietary protein or GI after the 6-month ad libitum weight maintenance diet (Table 2; Tables S5(A)–(D) and S6(A)–(D), available online).

Fig. 1 Effect of the interaction between lipin 1 (LPIN1) rs4315495 and dietary protein on TAG during the 6-month ad libitum weight maintenance diet. Values are mean changes per genotype for low-protein (LP) v. high-protein diets (HP), with standard errors represented by vertical bars. The results for the two HP diets and for the two LP diets are pooled, combining the diets with a low/high glycaemic index. Regression analyses showed a significant interaction between SNP and dietary protein with a decrease in TAG concentration of − 0·26 mmol/l per A-allele/protein unit (95 % CI − 0·38, − 0·14, P= 0·000043). Outside each bar the genotype is listed, followed by the number of observations. Δ, Change during the 6-month weight maintenance diet.
We also examined the associations between SNPs in the lipid metabolism-related genes and baseline blood lipid profile. We identified the associations between LPL rs328 and HDL-C with an increase of 0·09 mmol/l per G-allele (95 % CI 0·05, 0·13, P= 0·000044; Table S2(C), available online), PPARGC1A rs10002521 and TC with an increase of 0·19 mmol/l per G-allele (95 % CI 0·09, 0·29, P= 0·00013; Table S2(A), available online), and FABP1 rs894194 and TAG with an increase of 0·13 mmol/l per G-allele (95 % CI 0·06, 0·20, P= 0·00018; Table S2(D), available online). No other SNPs in the lipid metabolism pathways were associated with baseline lipid profile after adjusting for multiple testing (Table S2(A)–(D), available online).
Discussion
In the present randomised dietary intervention study with statistically significant separation of the LP and HP intake groups and the LGI and HGI groups(20), and repeated sampling providing detailed phenotypic characterisation of all participants, we identified an association with plasma TAG for an interaction between LPIN1 rs4315495 and dietary protein after the ad libitum weight maintenance diet. LPIN1 acts as a phosphatidate phosphatase in TAG synthesis(Reference Reue and Dwyer36), and LPIN1 SNPs have previously been associated with obesity-related phenotypes, insulin sensitivity and the metabolic syndrome(Reference Csaki and Reue47, Reference Loos, Rankinen and Perusse48). Enhanced LPIN1 expression in transgenic mice promotes increased lipid storage in the adipose tissue and decreased TAG secretion from the liver, whereas LPIN1-deficient mice exhibit lipodystrophy and increased hepatic TAG secretion(Reference Csaki and Reue47). The present finding indicates that minor allele carriers are more likely to decrease their concentrations of circulating TAG on a high-protein diet than on a low-protein diet. A reduction in the dietary intake of carbohydrates have previously been shown to reduce circulating levels of TAG(Reference Samaha, Iqbal and Seshadri49), and participants on the HP diets, in the present study, consumed a lower amount of carbohydrate than participants on the LP diets, which supports this finding.
LPIN1 is located on chromosome 2p25.1, and rs4315495 is located in intron 1. In these analyses, the SNP was not in high LD with any of the other LPIN1 SNPs (r 2< 0·5), nor were the other fifteen LPIN1 SNPs associated with changes in TAG by the interaction with dietary protein. The function of rs4315495 is not known, but it is in high LD (r 2 0·87) with rs13412852. The rs13412852 is also an intronic SNP, which has previously been associated with BMI(Reference Fawcett, Grimsey and Loos50) and the severity of non-alcoholic fatty liver disease in children(Reference Valenti, Motta and Alisi51). The association with obesity- and lipid-related phenotypes observed for these two linked SNPs could indicate that they could be linked to another SNP with functional effect or work by affecting as yet unidentified regulatory regions in the loci. Expression of LPIN1 is regulated by adipocyte differentiation factors, PPARGC1A, sterol regulatory element-binding protein 1, TNF and, possibly, LPIN2(Reference Csaki and Reue47). SNPs in PPARGC1A, TNF and LPIN2 were also included in the present analyses, but no interactions with dietary protein for the modification of TAG were identified for any of these (Table S4(D), available online).
We did not identify any SNP main effects on blood lipids after the 8-week LED. Previous human intervention studies of gene–diet interaction effects on blood lipids have shown that among overweight individuals, SNPs in CD36 modified HDL-C and LDL-C by an interaction with a LED(Reference Goyenechea, Collins and Parra52), and TAG and HDL-C by an interaction with fish oil supplements(Reference Madden, Carrero and Brunner53), and SNPs in MTTP modified TC and TAG by an interaction with the Mediterranean diet(Reference Lairon, Defoort and Martin54). In addition, several SNPs in genes in lipid metabolism pathways have been shown to modulate blood lipids in response to interventions with varying amounts and types of dietary fat(Reference Ye and Kwiterovich2, Reference Robitaille, Brouillette and Lemieux55) and to modulate postprandial blood lipid levels in response to high-fat test meals(Reference Lopez-Miranda, Williams and Lairon56, Reference Delgado-Lista, Perez-Martinez and Perez-Jimenez57). No studies have addressed interactions between SNPs and dietary protein or the GI in relation to blood lipids.
Among the selected lipid metabolism genes, only LPL and MLXIPL have previously been shown to be associated with lipid concentrations in GWAS(Reference Teslovich, Musunuru and Smith15). We identified a significant association of LPL rs328 with baseline HDL-C (Table S2(C), available online). LPL encodes the enzyme LPL with the primary function to hydrolyse TAG in circulating lipoproteins(Reference Wang and Eckel38). The present finding is consistent with previous reports in terms of effect size, risk allele and MAF(Reference Kathiresan, Melander and Guiducci6, Reference Sabatti, Service and Hartikainen11), and generally in line with GWAS that have found associations between SNPs in LPL and lipoprotein metabolism, including associations between several LPL SNPs and HDL-C(Reference Aulchenko, Ripatti and Lindqvist4, Reference Chasman, Pare and Mora5, Reference Middelberg, Ferreira and Henders10, Reference Wallace, Newhouse and Braund13), with the reported SNPs rs17482753(12), rs12678919(Reference Kathiresan, Willer and Peloso7, Reference Willer, Sanna and Jackson14, Reference Teslovich, Musunuru and Smith15) and rs10503669(Reference Ma, Yang and Runesha9, Reference Willer, Sanna and Jackson14) in complete LD with rs328.
In GWAS, MLXIPL have been associated with LDL-C(Reference Chasman, Pare and Mora5), HDL-C(Reference Teslovich, Musunuru and Smith15) and TAG(Reference Aulchenko, Ripatti and Lindqvist4, Reference Kathiresan, Melander and Guiducci6–Reference Kooner, Chambers and Aguilar-Salinas8, Reference Sabatti, Service and Hartikainen11, Reference Teslovich, Musunuru and Smith15, Reference Lanktree, Anand and Yusuf58). MLXIPL encodes the carbohydrate-responsive element-binding protein, which is an essential transcription factor for lipogenesis(Reference Hurtado del Pozo, Vesperinas-Garcia and Rubio39). The MLXIPL SNPs included in the present study are different from the ones that were found to be associated with lipid profile in these GWAS, and LD was not strong (pairwise r 2< 0·8 for all SNPs). However, the present analyses do suggest that MLXIPL rs1051921 could be associated with lower baseline levels of TAG (Table S2(D), available online).
Baseline associations were also identified between PPARGC1A rs10002521 and TC and between FABP1 rs894194 and TAG. FABP1 encodes the liver fatty acid-binding protein that plays a key role in fatty acid metabolism, and variation in this gene has previously been associated with circulating TAG and LDL-C in human subjects(Reference Fisher, Weikert and Klapper32). PPARGC1A encodes the protein PPARGC1A with a regulatory role in fatty acid metabolism and mitochondrial function(Reference Bournat and Brown44). The identified association for PPARGC1A has not previously been shown, but variants in PPARGC1A have previously been associated with type 2 diabetes(Reference Yang, Mo and Chen59).
The present study is limited by the relatively low number of participants in the DiOGenes study. While high for an intervention study, it is low in terms of conducting this type of genetic analyses. The candidate gene approach, although with a coverage of the genetic variation of the loci within 5 kb up/downstream and with a tagSNP disequilibrium limit of 0·7–0·8, limits the study particularly in detecting baseline associations, but also in identifying SNP–diet interactions, and the selected candidate genes in the study do not represent all known lipid-associated genes, nor do the SNPs include all of the recently identified lipid-associated SNPs(Reference Teslovich, Musunuru and Smith15).
In conclusion, in the present analyses of 240 SNPs in presumed nutrient-sensitive lipid metabolism genes among overweight and obese European adults, we identified an interaction between LPIN1 rs4315495 and dietary protein that resulted in a decrease in TAG concentration for minor allele carriers on the high-protein weight maintenance diet. Adjusting for multiple testing, no other effects of SNPs or SNP–diet (protein content or GI) interactions on blood lipid profile were detected after weight loss or after the 6-month ad libitum weight maintenance diet.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S0007114512006058
Acknowledgements
The DiOGenes project is funded by a grant from the European Union Food Quality and Safety Priority of the Sixth Framework Programme (contract no. FP6-2005-513946). The data analyses have been conducted under the Danish Strategic Research Program of Gene–Diet Interactions in Obesity (GENDINOB, grant no. 09-067111). A. A. is a member of the Scientific Advisory Board for Pathway Genomics, La Jolla, USA. No other potential conflict of interest relevant to this article was reported. J. H., R. J. F. L., T. I. A. S., D. L., N. V., K. S. V., A. A. and W. H. M. S. contributed to the conception and design of the study. T. H.-D., S. A. J., P. H., T. M. L., J. A. M., A. P., A. F. H. P., M. A. v. B., A. A. and W. H. M. S. were involved in the acquisition of the data. L. Ä. and C. H. performed the statistical analyses. L. K. B. and L. H. L. were responsible for the drafting of the manuscript. All authors contributed to the analysis and interpretation of the data, revised the manuscript critically for important intellectual content and approved the final version of the manuscript.





