The first line of defence against bacterial infections is mediated by the cells of the innate immune system (including monocytes, macrophages and neutrophils), which involves the secretion of cytokines, resulting in inflammation and further activation of the adaptive immune system( Reference Sánchez-Margalet, Martín-Romero and Santos-Alvarez 1 ). Adaptive immune systems, which are mediated by lymphocytes, recognise specific pathogens and protect against recurrent infections( Reference de Mello, Kolehmanien and Schwab 2 , Reference Jin, Park and Jo 3 ). Peripheral blood mononuclear cells (PBMC) is a general term encompassing any blood cells having a round nucleus, including lymphocytes, monocytes and macrophages. PBMC have been widely used in research related to the immune response by micro-organisms like bacteria, viruses, fungi and parasites. The lipopolysaccharide (LPS), which is a known component in the outer membranes of some gram-negative pathogens such as enterotoxigenic Escherichia coli, triggers the production of the cytokines that may contribute to the inflammation during the infection( Reference Shimazu, Villena and Tohno 4 – Reference Murofushi, Villena and Morie 6 ).
Toll-like receptors (TLR), as important initiators of the innate immunity, play a critical role in pathogen recognition and host defence. It is well accepted that the innate immune response, recognising the bacterial LPS, recruits a signalling pathway through the TLR4 to activate the NF-κB, which leads to inflammatory gene expression and the clearance of the infectious agent by pro-inflammatory cytokines( Reference Moue, Tohno and Shimazu 7 , Reference Doyle and O’Neill 8 ). Lactic acid bacteria (such as Lactobacillus), known as probiotics, play a beneficial role in the immune response by balancing pro- and anti-inflammatory cytokines( Reference Shimazu, Villena and Tohno 4 , Reference Tsai, Cheng and Pan 9 , Reference Taranu, Marin and Pistol 10 ). In a previous report, Shimazu et al. ( Reference Shimazu, Villena and Tohno 4 ) demonstrated that Lactobacillus jensenii TL2937 attenuated the expression of pro-inflammatory cytokines, caused by an LPS challenge, by down-regulating the TLR4-dependent NF-κB and the mitogen-activated protein kinase (MAPK) activation in a porcine intestinal epithelial cell line in vitro. An understanding of the precise mechanism that underlies the modulation of immune responses in porcine PBMC by Lactobacillus is important, not only from the perspective of fundamental research in cell biology and immunology but also for the more practical use in porcine nutrition and animal science. Despite the precise mechanism of modulating the immune responses through TLR4-dependent NF-κB regulation by Lactobacillus, bacterial infection is a critical subject in porcine health that has not yet been investigated in porcine PBMC in vivo.
Therefore, to characterise the regulation of the immune response by Lactobacillus, we evaluated the effect of feeding with Lactobacillus acidophilus on the immune responses after LPS challenge, and analysed the level of pro- and anti-inflammatory cytokines (as well as their related genes) by ELISA in blood serum and quantitative real-time PCR (qRT-PCR) in the PBMC.
Methods
The animal care and protocol used in the present study were approved by the Animal Care and Use Committee of Dankook University.
Experimental design, feeding, Lactobacillus acidophilus and lipopolysaccharide challenge
In the present study, we used dried L. acidophilus fermentation product derived from L. acidophilus in an anaerobic fermentation technology platform to produce beneficial microbial metabolites. Dried L. acidophilus fermentation product was kindly provided by a commercial company (Diamond V, Cedar Rapids, IA).
A total of forty weaned pigs ((Landrace×Yorkshire)×Duroc, 24–25 d aged) with an average initial body weight (BW) of 7·10 (sem 0·22) kg were used to evaluate the effects of feeding with L. acidophilus after LPS challenge in a 14-d feeding trial. Each pig was kept in individual pens (ten pens per dietary treatment) and was housed in an environmentally controlled nursery facility with slatted plastic flooring and a mechanical ventilation system. The temperature of the room was maintained approximately at 27°C and humidity at 60 %. Each pen was equipped with a one-sided, stainless steel self-feeder and a nipple drinker that allowed the pig access to feed and water ad libitum. Experimental treatments were as follows: (T1) control diet+saline challenge; (T2) control diet with 0·1 % L. acidophilus+saline challenge; (T3) control diet+LPS challenge; and (T4) control diet with 0·1 % L. acidophilus+LPS challenge. The control diet was based on a maize and soyabean meal and was formulated to meet or exceed the nutrient requirements (Table 1) recommended by the Nutrient Requirements of Swine ( 11 ). Dietary Ca, P and amino acids (lysine and methionine) were analysed according to the procedures described by the official methods of analysis. Dietary Ca was assayed by atomic absorption spectrophotometry after wet ash procedures, and P was determined by colorimetry. Lysine and methionine were measured using an amino acid analyser (Beckman 6300; Beckman Coulter Inc.) after 24 h 6 n-HCl hydrolysis at 110°C.
Table 1 Composition of the control diet (as-fed basis)

CP, crude protein.
* Provided per kg of complete diet: vitamin A, 11 025 IU; vitamin D3, 1103 IU; vitamin E, 44 IU; vitamin K, 4·4 mg; riboflavin 8·3 mg; niacin, 50 mg; thiamine, 4 mg; pantothenic acid, 29 mg; choline, 166 mg; and vitamin B12, 33 μg.
† Provided per kg of complete diet: Cu, 12 mg; Zn, 85 mg; Mn, 8 mg; I, 0·28 mg; and Se, 0·15 mg.
For the challenging assay, all pigs from each dietary treatment were injected intraperitoneally with E. coli LPS (T3 and T4) and saline solution (T1 and T2). The LPS (serotype O55:B5; Sigma-Aldrich) was diluted in sterile saline solution and injected at 0·01 % (0·1 g/kg) of BW at the end of the 14-d feeding trial. The dosage of LPS used was based on the results of our previous study( Reference Li and Kim 12 ). No vaccines or antibiotics were used in this experiment.
Growth performance and analysis of blood characteristics
To evaluate growth performance, a total of forty weaned pigs were allocated to control (T1 and T3) and L. acidophilus (T2 and T4) treatment (n 20). The BW of each pig was recorded at the beginning and at d-14 (before challenge), and feed consumption was recorded on an individual pig basis during the experiment to calculate the average daily weight gain (ADWG), the average daily feed intake (ADFI) and weight gain:feed intake (WG:FI).
Collection of blood samples and analyses were performed according to our standard protocol( Reference Li and Kim 12 ). Briefly, blood samples were collected from all pigs via jugular venepuncture at 0, 2, 4, 6 and 12 h after challenge. Blood samples were collected via anterior vena cava puncture from all pigs and were collected into a non-heparinised and K3EDTA vacuum tube (Becton Dickinson Vacutainer Systems) to get serum and whole blood. Leucocytes and lymphocyte counts were determined using an automatic blood analyser (ADVIA120; Bayer).
For flow cytometry, erythrocytes were removed by erythrocyte lysis buffer according to the manufacturer’s instruction (Sigma-Aldrich). Cells were then incubated with monoclonal CD4+ and CD8+ antibodies for 30 min (Abcam). The cells were washed three times with cold washing buffer (PBS containing 0·5 % bovine serum albumin) and analysed for CD4+, CD8+ and CD4+/CD8+ using FACS Calibur flow cytometry (BD Bioscience).
Whole blood sample was subsequently centrifuged at 3000 g for 15 min at 4°C and the serum was harvested. Thereafter, samples were frozen and stored at –20°C until further analysis. The concentration of IgA, IgM and IgG in blood serum was measured using an automatic biochemistry analyser (HITACHI 747; Boehringer Mannheim). Serum IGF-1 (Abcam), cortisol (Endocrine Technologies), TNF-α (R&D Systems) and IL-6 (R&D Systems) were determined by ELISA.
Peripheral blood mononuclear cell preparation and quantitative real-time PCR
For PBMC isolation, a total of twenty blood samples from randomly selected five pigs per each treatment were collected into a K3EDTA vacuum tube at 0, 6 and 12 h after saline or LPS challenge. The collected blood sample was diluted with an equal volume of balanced salt solution. The PBMC were immediately isolated by a histopaque density gradient according to the manufacturer’s instruction (Sigma-Aldrich). Briefly, the collected blood (with an equal volume of balanced salt solution) was mixed with a half volume of Histopaque solution and was then centrifuged at 400
g
for 35 min at room temperature. The PBMC were carefully aspirated from the Histopaque solution-plasma interface. The RNA was isolated using a TRIzol reagent (Invitrogen). For the qRT-PCR, total RNA (100 µg) was used for the complementary DNA synthesis with a Maxima First Strand cDNA Synthesis Kit (Life Technologies). The primers for the qRT-PCR of each gene transcript were designed using the program Primer3 (http://frodo.wi.mit.edu/) (online Supplemental Table S1). The qRT-PCR analysis was performed using the 7500 Fast Real-Time PCR System (Applied Biosystems). The qRT-PCR conditions were as follows: 94°C for 3 min, followed by 40 cycles at 94°C for 30 s, 59–61°C for 30 s and 72°C for 30 s; the melting curve profiles were analysed for the amplicons. The qRT-PCR data were normalised relative to the expression of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an endogenous control and calculated using the
![]() $$2^{{\Delta \Delta C_{t} }} $$
method, where
$$2^{{\Delta \Delta C_{t} }} $$
method, where
![]() $$^{{\Delta \Delta C_{t} }} $$
=(C
t
of the target gene−C
t
of GAPDH) treatment – (C
t
of the target gene−C
t
of GAPDH) control(
Reference Livak and Schmittgen
13
).
$$^{{\Delta \Delta C_{t} }} $$
=(C
t
of the target gene−C
t
of GAPDH) treatment – (C
t
of the target gene−C
t
of GAPDH) control(
Reference Livak and Schmittgen
13
).
Statistical analysis
The individual pig was considered the experimental unit. Growth performance was analysed by means of the Student’s t test with SAS software (SAS Institute). To evaluate the significance between treatments for immune cells and cytokine production, the data were analysed with the general linear model (GLM) in SAS. Differences among all treatments were separated by Duncan’s multiple range tests. For qRT-PCR analysis, to evaluate the significance between the treatment and control groups, the data were analysed with the GLM in SAS. Significant differences between the control and treatment groups are indicated as * P<0·05 and ** P<0·01. A P value <0·05 was considered to indicate statistical significance.
Results
The effects of Lactobacillus acidophilus on growth performance
The effects of L. acidophilus on growth performance are indicated in Table 2. Feeding with L. acidophilus increased the BW at 14-d, as well as the ADWG and ADFI, compared with the control diet (P<0·05). No significant impact on the GW:FI ratio was found (P>0·05).
Table 2 The effects of the Lactobacillus acidophilus treatment on growth performance in weaned pigs for 14-d feeding trial (Mean values with their standard errors; n 20)
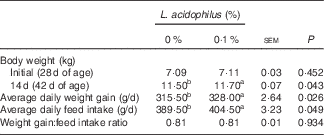
a,b Mean values within a row with unlike superscript letters were significantly different between groups (P<0·05).
The effects of Lactobacillus acidophilus on IGF-1, Cortisol, IgA, IgM, and IgG in serum after a saline or lipopolysaccharide challenge
The effects of L. acidophilus treatment on IGF-1 in serum after a saline or LPS challenge are indicated in Table 3. With the control diet, the LPS challenge (T3) decreased the IGF-1 concentration compared with the saline challenge (T1) at 4, 6 and 12 h. Whereas with saline challenge, L. acidophilus treatment (T2) showed no effect compared with the control diet (T1), with the LPS challenge L. acidophilus treatment (T4) increased the IGF-1 concentration at 4, 6 and 12 h compared with the control diet (T3).
Table 3 The effects of the Lactobacillus acidophilus treatment (T) on IGF-1 and leucocytes in blood after saline or lipopolysaccharide (LPS) challenge (Mean values with their standard errors; n 10)Footnote *
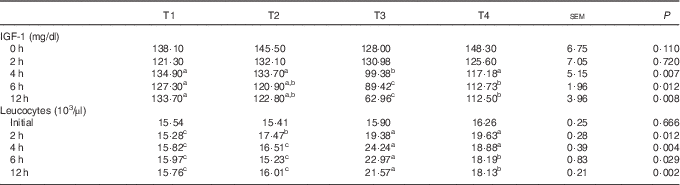
a,b,c Mean values within a row with unlike superscript letters were significantly different between groups (P<0·05).
* Experimental treatments were as follows: (T1) control diet+saline challenge; (T2) control diet with 0·1 % L. acidophilus+saline challenge; (T3) control diet+LPS challenge; and (T4) control diet with 0·1 % L. acidophilus+LPS challenge.
The effects of L. acidophilus treatment on cortisol, IgA, IgM and IgG in serum after a saline or LPS challenge are indicated in online Supplemental Table S2. There were no significant effects on cortisol, IgA, IgM and IgG in serum after a saline or LPS challenge (P>0·05).
The effects of Lactobacillus acidophilus on immune cells after a saline or lipopolysaccharide challenge
The effects of L. acidophilus on lymphocytes, CD4+ and CD8+ after a saline or LPS challenge are indicated in Fig. 1. For lymphocytes, under the control diet, the LPS challenge (T3) increased the number of lymphocytes compared with the saline challenge (T1) at 2, 4, 6 and 12 h (Fig. 1(A)). Whereas with the saline challenge L. acidophilus treatment (T2) showed no effect compared with the control diet (T1), with the LPS challenge L. acidophilus treatment (T4) decreased the number of lymphocytes compared with the control diet (T3) at 4 h.
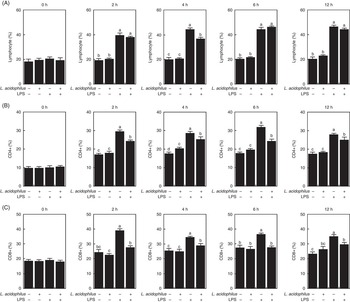
Fig. 1 The effects of Lactobacillus acidophilus on immune cells, lymphocytes (A), CD4+ (B) and CD8+ (C) in blood at 0, 2, 4, 6 and 12 h after the lipopolysaccharide (LPS) challenge. Weaned pigs were randomly allocated into four groups: (T1) control diet+saline challenge; (T2) control diet with 0·1 % L. acidophilus+saline challenge; (T3) control diet+LPS challenge; and (T4) control diet with 0·1 % L. acidophilus+LPS challenge. Lymphocyte (A), CD4+ (B), and CD8+ (C) counts were determined by ELISA (n 10). Differences between the treatments were determined by means of Duncan’s multiple range tests. A P value <0·05 was considered to indicate statistical significance. Values are means, with standard errors respresented by vertical bars.
For CD4+, with the control diet the LPS challenge (T3) increased the number of CD4+ cells compared with the saline challenge (T1) at 2, 4, 6 and 12 h (Fig. 1(B)). Whereas with the saline challenge L. acidophilus treatment (T2) increased the number of CD4+ cells compared with the control diet (T1) at 4 h, with the LPS challenge L. acidophilus treatment (T4) decreased the number of CD4+ cells compared with the control diet (T3) at 2, 4, 6 and 12 h.
For CD8+, with the control diet the LPS challenge (T3) increased the number of CD8+ cells compared with the saline challenge (T1) at 2, 4, 6 and 12 h (Fig. 1(C)). Whereas with the saline challenge L. acidophilus treatment (T2) showed no effect compared with the control diet (T1), with the LPS challenge L. acidophilus treatment (T4) decreased the number of CD8+ cells compared with the control diet (T3) at 2, 4, 6 and 12 h.
The effects of L. acidophilus on leucocytes after a saline or LPS challenge are indicated in Table 3. For leucocytes, with the control diet the LPS challenge (T3) increased the amount of leucocytes compared with the saline challenge (T1) at 2, 4, 6 and 12 h. Whereas with the saline challenge L. acidophilus treatment (T2) increased the amount of leucocytes compared with the control diet (T1) at 2 h, with the LPS challenge L. acidophilus treatment (T4) decreased the amount of leucocytes compared with the control diet (T3) at 6 and 12 h.
The effects of Lactobacillus acidophilus on the inflammatory cytokine production in serum after a saline or lipopolysaccharide challenge
The effects of L. acidophilus on TNF-α production after a saline or LPS challenge are indicated in Fig. 2(A). With the control diet, the LPS challenge (T3) increased the TNF-α production compared with the saline challenge (T1) at 2, 4, 6 and 12 h. Whereas with the saline challenge L. acidophilus treatment (T2) showed no effect compared with the control diet (T1), with the LPS challenge L. acidophilus treatment (T4) decreased the TNF-α production compared with the control diet (T3) at 6 and 12 h.
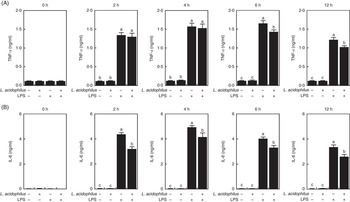
Fig. 2 The effects of Lactobacillus acidophilus on the production of inflammatory cytokines TNF-α (A) and IL-6 (B) in blood serum at 0, 2, 4, 6 and 12 h after the lipopolysaccharide (LPS) challenge. Weaned pigs were randomly allocated into four groups: (T1) control diet+saline challenge; (T2) control diet with 0·1 % L. acidophilus+saline challenge; (T3) control diet+LPS challenge; and (T4) control diet with 0·1 % L. acidophilus+LPS challenge. TNF-α (A) and IL-6 (B) levels were determined by ELISA (n 10). Differences between all treatments were determined by means of Duncan’s multiple range tests. A P value <0·05 was considered to indicate statistical significance. Values are means, with standard errors respresented by vertical bars.
The effects of L. acidophilus on IL-6 production after a saline or LPS challenge are indicated in Fig. 2(B). With the control diet, the LPS challenge (T3) increased the IL-6 production compared with the saline challenge (T1) at 2, 4, 6 and 12 h. Whereas with the saline challenge L. acidophilus treatment (T2) showed no effect compared with the control diet (T1), with the LPS challenge L. acidophilus treatment (T4) decreased the IL-6 production compared with the control diet (T3) at 2, 4, 6 and 12 h.
The effects of Lactobacillus acidophilus on gene expression related to inflammatory cytokines in peripheral blood mononuclear cell after a saline or lipopolysaccharide challenge
We next examined whether L. acidophilus plays a role in the immune defence against bacterial infections that were mediated by the signalling pathway through TLR4 to NF-κB, which leads to inflammatory gene expression in the PBMC. L. acidophilus treatment decreased the TLR4 expression at 6 and 12 h and decreased the NF-κB expression at 12 h after the LPS challenge compared with the control diet (P<0·05) (Fig. 3). To validate whether L. acidophilus regulated the gene expression related to the pro- and anti-inflammatory cytokines, we analysed the expression pattern of TNF-α, IFN-γ, IL-6, IL-8, IL1B1, IL-4 and IL-10 after the LPS challenge. The expression of TNF-α, IFN-γ, IL-6, IL-8 and IL1B1 as pro-inflammatory cytokines was decreased by the feeding with L. acidophilus at 12 h after the LPS challenge (Fig. 4(a)). On the contrary, the expression of IL-4 and IL-10 as anti-inflammatory cytokines was increased by the feeding with L. acidophilus at 12 h after the LPS challenge (Fig. 4(b)).

Fig. 3 Quantitative gene expression of TLR4 and NF-κB in peripheral blood mononuclear cell (PBMC) at 0, 6 and 12 h after the lipopolysaccharide challenge. The quantitative real-time PCR data were normalised relative to the expression of the GAPDH as an endogenous control and calculated using the
![]() $2^{{\Delta \Delta C_{t} }} $
method, where
$2^{{\Delta \Delta C_{t} }} $
method, where
![]() $^{{\Delta \Delta C_{t} }} $
=(C
t
of the target gene – C
t
of GAPDH) treatment−(C
t of the target gene – C
t
of GAPDH) control (n 5). ** Significantly different between the control and treatment groups (P<0·01). Values are means, with standard errors respresented by vertical bars.
$^{{\Delta \Delta C_{t} }} $
=(C
t
of the target gene – C
t
of GAPDH) treatment−(C
t of the target gene – C
t
of GAPDH) control (n 5). ** Significantly different between the control and treatment groups (P<0·01). Values are means, with standard errors respresented by vertical bars. ![]() , Control;
, Control; ![]() , L. acidophilus
, L. acidophilus

Fig. 4 Quantitative gene expression of pro- and anti-inflammatory cytokines in peripheral blood mononuclear cells (PBMC) at 0, 6 and 12 h after the lipopolysaccharide (LPS) challenge. (a) Quantitative gene expression of TNF-α, IFN-γ, IL-6, IL-8 and IL1B1 in PBMC at 0, 6 and 12 h after LPS challenge by feeding with Lactobacillus acidophilus. (b) Quantitative gene expression of IL-4 and IL-10 in PBMC at 0, 6 and 12 h after LPS challenge by feeding with L. acidophilus. The qRT-PCR data were normalised relative to the expression of the GAPDH as an endogenous control and calculated using the
![]() $2^{{\Delta \Delta C_{t} }} $
method, where
$2^{{\Delta \Delta C_{t} }} $
method, where
![]() $^{{\Delta \Delta C_{t} }} $
= (C
t
of the target gene – C
t
of GAPDH) treatment−(C
t
of the target gene – C
t
of GAPDH) control (n 5). ** Significantly different between the control and treatment groups (P<0·01). Values are means, with standard errors respresented by bars.
$^{{\Delta \Delta C_{t} }} $
= (C
t
of the target gene – C
t
of GAPDH) treatment−(C
t
of the target gene – C
t
of GAPDH) control (n 5). ** Significantly different between the control and treatment groups (P<0·01). Values are means, with standard errors respresented by bars. ![]() , Control;
, Control; ![]() , L. acidophilus.
, L. acidophilus.
Discussion
Because of the ban on antibiotics as a growth promoter in animal farming, various natural materials such as probiotics, prebiotics, organic acids and plant extracts have been evaluated as alternatives to antibiotics( Reference Vondruskova, Slamova and Trckova 14 ). Lactobacillus, a genus of the gram-positive facultative anaerobic bacteria, is a major part of the lactic acid bacteria group that plays a role in inhibiting the growth of some harmful bacteria by environmental acidic conditioning through the production of lactic acid. Among Lactobacillus, L. acidophilus is a homofermentative microaerophilic species, fermenting sugars into lactic acid, and it occurs naturally in the animal gastrointestinal tract( Reference Anjum, Maqsood and Masud 15 ). The present study used L. acidophilus as an alternative to antibiotics to evaluate the dietary effect on growth performance in weaned pigs. Growth performance parameters such as ADWG are widely used for evaluating the beneficial health effects of these feed supplements. To determine whether L. acidophilus has an effect on growth performance, we carried out a feeding trial using L. acidophilus in weaned pigs. In this present study, pigs fed with L. acidophilus showed increased ADWG and ADFI compared with those fed without L. acidophilus. Many other studies have assessed the effect of different probiotic supplements on growth performance in pigs, but with varying results according to the species or strain being used( Reference Cui, Shen and Jia 16 , Reference Lodemann, Hübener and Jansen 17 ). For example, in a previous study, Qiao et al. ( Reference Qiao, Li and Wang 18 ) during a 28-d trial in weaned piglets reported that feeding with 0·1 and 0·2 % L. acidophilus increased the ADWG and WG:FI ratio compared with a control diet. Disagreement between the previous and the present study can be seen in the different results of ADFI. In the present study, IGF-1 concentration was decreased by LPS challenge with the control diet and was increased by L. acidophilus treatment with LPS challenge. This indicates that L. acidophilus treatment could have increased ADWG and ADFI compared with the control diet. Also, growth performance and IGF-1 concentration might be affected by the LPS challenge. However, because IGF-1 concentration was not changed with saline challenge, further research is necessary to establish the relationship of growth performance and IGF-1 concentration in pigs. In a number of previous studies, feeding with Lactobacillus reduced the inflammatory activity challenged with bacterial infection( Reference Kawahara, Nemoto and Nakata 19 – Reference Lee, Awji and Lee 21 ). In a previous report, L. jensenii attenuated the expression of the pro-inflammatory cytokines caused by the LPS challenge by down-regulating the TLR4-dependent NF-κB and the MAPK activation in a porcine intestinal epithelial cell line( Reference Shimazu, Villena and Tohno 4 ). Also in another report, L. acidophilus fed to weaned piglets reduced the TNF-α and IFN-γ concentration in serum challenged with E. coli LPS( Reference Qiao, Li and Wang 18 ). However, the precise mechanism of modulating the immune responses through the TLR4-dependent NF-κB regulation by feeding L. acidophilus to pigs challenged with bacterial infection has not yet been investigated in porcine PBMC in vivo. The PBMC, consisting of lymphocytes, monocytes and macrophages, are good models for immune-related research, because of the representing cells of the immune system. It is well accepted that PBMC are good targets in the field of molecular nutrition and nutrigenomics, because they seem to reflect the effect of dietary modification at the level of gene expression( Reference de Mello, Kolehmanien and Schwab 2 ). Therefore, in the present study, we studied the effect of L. acidophilus on inflammatory cytokine production in the blood serum after LPS challenge, and the expression of the TLR4 and NF-κB genes related to the pro- and anti-inflammatory cytokines. We found that the L. acidophilus treatment decreased the pro-inflammatory cytokine and increased the anti-inflammatory cytokine in the serum by decreasing the TLR4 and NF-κB expression in porcine PBMC after LPS challenge. Our finding contributes to understanding the mechanism that underlies modulating immune responses in porcine PBMC.
In the mammalian immune system, TLR play a critical role as key regulators of both innate and adaptive immunity. The immune system consists of the innate immune system, which is the first line of defence against invading pathogens, and the adaptive system that eliminates pathogens, leading to immunological memory( Reference Carpenter and O’Neill 22 ). TLR, known as pathogen recognition receptors, recognise pathogen-associated molecular patterns that are unique to microbes and essential for their survival( Reference Janeway 23 ). Activated TLR, by bacterial pathogens, lead to activated NF-κB (known as a transcription factor), which binds to a discrete nucleotide sequence in the upstream regions of genes that produce inflammatory cytokines (such as TNF-α, IFN-γ, IL-1 and IL-2), indicating the hallmark of the cellular response to the activation of the innate immune system( Reference Baeuerle and Henkel 24 ). Among the TLR, TLR4 particularly recognises LPS, which is the major component of the outer wall of gram-negative bacteria. LPS is a potent immunostimulant and is the causative agent of endotoxic stimulation( Reference Carpenter and O’Neill 22 ). Challenge with LPS is widely used to study cytokine production, which requires the induction of immune cells, such as macrophages, to produce cytokines as LPS evokes a powerful inflammatory response by stimulating cells to release the inflammatory cytokines such as TNF-α, IFN-γ and IL( Reference Doyle and O’Neill 8 ). In this study, feeding with L. acidophilus decreased TNF-α and IL-6 after the LPS challenge. Also, pigs fed with dietary L. acidophilus supplementation showed decreased immune cells (such as leucocytes, lymphocytes, CD4+ cells and CD8+) after the LPS challenge.
In conclusion, we found that feeding with L. acidophilus attenuated the immune cells (including leucocytes, lymphocytes, CD4+ and CD8+ cells), decreased the pro-inflammatory cytokine and increased the anti-inflammatory cytokine after the LPS challenge. In addition, we confirmed that L. acidophilus decreased the expression of TLR4 and NF-κB in the PBMC on LPS challenge, which leads to the inhibition of gene expression of TNF-α, IFN-γ, IL-6, IL-8 and IL1B1 and induces the gene expression of IL-4 and IL-10 in PBMC (Fig. 5). On the basis of these results, we suggested that L. acidophilus attenuated inflammatory activity by regulating the TLR4 and NF-κB expression in porcine PBMC. These findings can help in the understanding of the precise mechanism that underlies the modulation of immune responses by Lactobacillus in porcine.
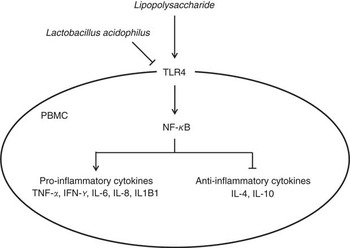
Fig. 5 Schematic illustrating the current working hypothesis regarding the regulation of inflammatory cytokine production by Lactobacillus acidophilus after lipopolysaccharide (LPS) challenge in porcine peripheral blood mononuclear cells (PBMC). L. acidophilus decreased the expression of toll-like receptor 4 (TLR4) and NF-κB in PBMC after LPS challenge, which led to inhibition of gene expression of TNF-α, IFN-γ, IL-6, IL-8 and IL1B1 known as pro-inflammatory cytokines and induced gene expression of IL-4 and IL-10 known as anti-inflammatory cytokines.
Acknowledgements
The authors thank Jae Won Park and Seoung Chul Kim for their assistance in animal care and in the preparation of blood samples.
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
The authors’ contributions are as follows: S. I. L. conceived the study, designed and carried out the experiments, analysed the data and wrote the manuscript; H. S. K. carried out the experiments and analysed the data; J. M. K. analysed the data; I. H. K. designed the experiments and contributed to the writing of the manuscript.
None of the authors has any conflicts of interest to declare.
Supplementary Material
For supplementary material/s referred to in this article, please visit http://dx.doi.org/doi:10.1017/S0007114515004857











