When energy is required, e.g. during periods of fasting, starvation or long-term exercise, NEFA are released from adipose TAG stores into the circulation by lipolysis and are subsequently oxidized, primarily by skeletal muscle, to provide energy(Reference Arner1). Yet, adipose tissue also plays an important role in other physiological processes and it can be considered as an endocrine organ(Reference Trayhurn and Beattie2). Adipocytes through the synthesis and release of peptide hormones by adipocytes, named adipokines(Reference Trayhurn3), are involved in the immune response, blood pressure control, haemostasis, bone mass, thyroid and reproductive function.
Fatty acids, apart from being an energy source, are involved in the transcriptional regulation of gene expression in adipose tissue and other tissues or organs(Reference Duplus, Glorian and Foresti4–Reference Hihi, Michalik and Wahli6). NEFA also modulate gene transcription by exerting a direct, membrane-independent influence on the molecular events that govern gene expression(Reference Armoni, Harel and Karnieli7). Furthermore, it has been demonstrated that, compared to dietary fats rich in linoleic acid, fish oil increases insulin-stimulated glucose transport and metabolism in isolated white adipocytes(Reference Ezaki, Tsuji, Momomura, Kasuga and Itakura8). The genes investigated in the present study are responsible for the trapping, release and storage of fatty acids, as well as being involved in glucose uptake and transduction of the insulin signal. PPARγ is a nuclear receptor with a major role in the regulation of adipocyte differentiation, lipid metabolism and insulin action(Reference Tanaka, Itoh and Doi9, Reference Walczak and Tontonoz10). In pigs the isoforms of PPARγ identified thus far include: PPARγ1 and PPARγ2(Reference Omi, Brenig, Špilar Kramer, Iwamoto, Stranzinger and Neuenschwander11). PPARγ1 has four transcription variants: PPARγ1a, 1b, 1c, 1d(Reference Omi, Brenig, Špilar Kramer, Iwamoto, Stranzinger and Neuenschwander11). The PPARγ coactivator 1 (PPARGC1A in pigs; also known as PGC-1) is a nuclear-encoded transcriptional coactivator that plays a pivotal role in glucose metabolism, mitochondrial biogenesis, muscle fibre specialization and adaptive thermogenesis, and is a co-activator of PPARγ(Reference Knutti and Kralli12–Reference Yoon, Puigserver and Chen14). GLUT4 is an insulin-responsive glucose transporter and affects whole-body glucose homeostasis(Reference Ezaki, Tsuji, Momomura, Kasuga and Itakura8, Reference Kahn15). TNFα is a key modulator of adipocyte metabolism, with a direct role in several insulin-mediated processes, including glucose homeostasis and lipid metabolism(Reference Sethi and Hotamisligil16). Adiponectin is a factor exclusively secreted by adipose tissue that has been shown to exert anti-inflammatory and anti-atherogenic effects and reverse insulin resistance in rodents(Reference Yamauchi, Kamon and Waki17), by increasing insulin sensitivity(Reference Hu, Liang and Spiegelman18) through improvement of glucose and lipid metabolism(Reference Arner1). Leptin is secreted principally, but not exclusively, by adipocytes and acts both centrally and peripherally, with a major role in the regulation of food uptake, body weight and energy balance(Reference Trayhurn, Hoggard, Mercer and Rayne19). It has been shown in gilts that acute changes in feed intake affect leptin secretion(Reference Whisnant and Harrell20). Fatty acid binding protein 4 (FABP4) functions to traffic fatty acids away from the TAG after hydrolysis(Reference Vogel Hertzel and Bernlohr21). Lipoprotein lipase (LPL) mainly has a function in hydrolysis of TAG, and although muscle LPL outweighs that of LPL in adipose tissue, the latter removes relatively more TAG in the postprandial period, making LPL of adipose tissue a prime target for metabolic regulation(Reference Frayn and Snell22). Meanwhile, relationships have been reported between the activation of PPARγ and the function of other genes, such as GLUT4(Reference Dana, Hoener, Bilakovics, Crombie, Ogilvie, Kauffman, Mukherjee and Paterniti23), TNFα(Reference Miles, Romeo, Higo, Cohen, Rafaat and Olefsky24), adiponectin(Reference Maeda, Takahashi and Funahashi25), leptin(Reference Cohen, Novick and Rubinstein26) and LPL(Reference Schoonjans, Peinado-Onsurbe, Lefebvre, Heyman, Briggs, Deeb, Staels and Auwerx27). The relationship between circulating leptin and insulin with leptin mRNA and leptin receptor (LEPR) mRNA (also known as Ob-R1), respectively, have been investigated in growing pigs(Reference Xingjie, Defa, Jingdong, Yuhua, Heliang, Huawei and Ganfeng28). The nature of such relationships in lactating sows during the first days of lactation is not known yet.
The peri-parturient period in sows(Reference Nachrein29) ranges from 4 d before to 3 d after parturition. The first days of lactation are of clinical importance, since the incidence of postpartum dysgalactia syndrome may occur until 72 h after parturition(Reference Klopfenstein, Farmer, Martineau, Straw, Zimmerman, Allaire and Taylor30). During pregnancy, sows are in an anabolic status, because they develop fetuses and mammary tissue. After farrowing, sows are in an anabolic (mammary gland) and a catabolic status, because body reserves (mainly adipose tissue) are mobilized to provide nutrients for milk production(Reference Mullan and Williams31). During early lactation in sows body reserves are more important determinants of milk yield than dietary intakes(Reference Mullan and Williams32). Therefore in the present study, samples of backfat adipose tissue of sows were taken at the first day of lactation, as it is the day of lactation closer to the transition period between pregnancy and lactation.
Information is limited regarding the effects of the n-6:n-3 ratio of the lactation diet of the sows and the prepartal feeding strategy on the abundance of expression of genes of backfat adipose tissue during this critical period. In addition, the interrelationships between PPARγ activation and function of genes involved in glucose and lipid metabolism, and between gene expression and levels of hormones in blood reflecting metabolism, during the same period, remain to be elucidated. The aim of the present study was to investigate the effect of two diets differing in n-6:n-3 ratio and fed from either 3 or 8 d before parturition onwards on the abundance of the genes involved in lipid and glucose metabolism in early lactation in sows. The relationships between PPARγ and its target genes, and between mRNA levels of genes and circulating hormones, were also investigated.
Materials and methods
Study population and experimental design
Seventy-two pregnant hybrid sows (Topigs 20 breed: Dutch Landrace × Great York) were included. Until day 107 of gestation, the sows were group-housed in a gestation unit and they were fed twice a day with a conventional gestation diet (4 kg/d). The composition (g/kg) of the gestation feed was: DM 892; ash 67; crude protein 126; crude fat 34; neutral-detergent fibre 197; metabolizable energy 10 400 kJ/kg. Linoleic acid content was 11·53 g/kg, total n-3 PUFA was 1·66 g/kg, and DHA and EPA contents were fairly low: 4·09 × 10− 5 and 3·47 × 10− 5 g/kg, respectively.
At day 107 of gestation, the pregnant sows were moved to the farrowing crates, and they were randomly allocated using a 2 × 2 experimental design to four different treatment groups namely f3, f8, s3, s8. Sows from the f and the s groups received a diet that was supplemented with fish oil or mainly sunflower oil, respectively. The compositions of the diets are shown in Table 1. Half of the sows belonging either to the f group or the s group received the experimental diet from day 107 of gestation (f8 and s8 groups) onwards (8 d before expected farrowing) until the end of lactation, the other half of the sows received the experimental diet from day 111 of gestation (f3 and s3 group) onwards, being 3 d before the expected day of farrowing (onset day groups). The sows of the f3 group continued to receive the conventional gestation feed between days 107 and 111 of gestation. From day 107 of gestation onwards until farrowing, the sows from all four groups received the same amount of feed, namely 3·3 kg/sow per d. A 3-week batch production system was maintained, and the seventy-two sows farrowed in three consecutive batches of twenty-four sows each. Within each batch, the sows were randomly assigned to the four experimental groups. The mean parity of the sows between the experimental groups (f3 4·2; f8 4·4; s3 4·6; s8 4·3) was similar and not significantly different between groups (P = 0·570).
Table 1 Chemical composition of the two different feeds for the sows: the feeds were supplemented either with fish oil or with sunflower oil and differed in the n -6:n -3 ratio
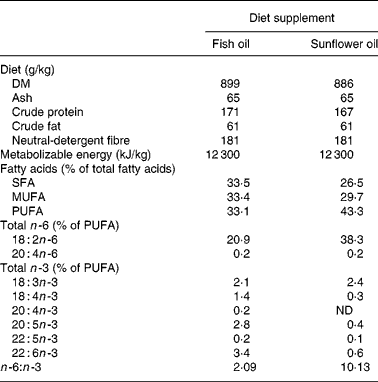
ND, not detectable.
No feed was provided on the day of parturition (day 0). From the first day of lactation until weaning, the sows were fed twice a day at 08.00 and 18.00 hours. The amount of feed that was provided daily was 3·0 kg on days 1 and 2, 3·5 kg on day 3, 4·0 kg on days 4 and 5, 4·5 kg on days 6 and 7, 5·0 kg on days 8 and 9, and 5·5 kg from day 10 until day 21 (weaning). The amount of feed consumed by every sow was recorded daily from day 1 until day 21.
The study was conducted at the Provimi Research Farm ‘De Viersprong’, Velddriel, The Netherlands and the experimental protocol of the study was approved by the Animal Sciences Group in Wageningen, The Netherlands.
Sample collection and cDNA synthesis
A biopsy of the backfat white adipose tissue of each of the seventy-two sows was taken on the first day of lactation, by using a spring-loaded biopsy instrument (Biotech PPB-U, Nitra, Slovakia) using a 10 mm cannula set at a depth of 2 cm, as described previously(Reference Craven, Hernessy and Friis33). The site of the biopsy was located 5 cm distal to the last rib to the side of the midline. Biopsies were collected immediately after the blood sampling procedure and before the morning meal from each sow at the first day of lactation.
For RNA preservation matters, the samples were immediately submerged in RNAlater (Sigma-Aldrich) according to the manufacturer's protocol and stored at − 20°C until they were crushed to powder with liquid nitrogen. The samples were subdivided per 80–100 mg and stored at − 80°C. Total RNA was extracted with the Aurum Total RNA Fatty and Fibrous Tissue Kit (Bio-Rad) according to the instructions manual, which also included an on-column DNase treatment. In order to check the RNA integrity by evaluating the 28S and 18S ribosomal RNA bands, RNA was loaded on to a 0·8 % agarose gel. Next, a minus reverse transcription control PCR (with the YWHAZ primers(Reference Erkens, Van Poucke, Vandesompele, Goossens, Van Zeveren and Peelman34)), which included a positive porcine genomic DNA control and a no-template control, was performed to check if any DNA contamination was still present. After this, the purity and concentration of the RNA were measured with the ND-1000 spectrophotometer (NanoDrop). The samples showed an optical density 260/280 ratio between 1·75 and 2·10 and an RNA concentration of 24–40 ng/μl with a total yield of 0·72–1·2 μg RNA. By means of a 20 μl reverse transcription reaction with the iScript cDNA synthesis kit (Bio-Rad), approximately 0·8 μg RNA from each sample was converted to cDNA. A verification of the reverse transcription reaction was performed through a control PCR (with the same YWHAZ primers as for the minus reverse transcription control) using 2·5 μl cDNA (diluted ten times with 2-amino-2-hydroxymethyl-1,3-propanediol hydrochloride buffer, pH 8, 10 mm). To check for DNA contamination, every PCR included a no-template control.
Primers
Primers for reference gene selection (ACTB, B2M, GAPDH, HMBS, HPRT1, RPL13A, SDHA, TBP, TOP2B, YWHAZ) for normalization of real-time PCR mRNA expression data were used from Erkens et al. (Reference Erkens, Van Poucke, Vandesompele, Goossens, Van Zeveren and Peelman34). The National Center for Biotechnology Information database(35) and literature was searched for available porcine sequences and primers of the genes under investigation(Reference Jacobs, Rohrer, Van Poucke, Piumi, Yerle, Barthenschlager, Mattheeuws, Van Zeveren and Peelman36, Reference Jung, Park, Kim and Hong37) (Table 2). Primers were designed using Primer3(Reference Rozen, Skaletsky, Krawetz and Misene38), while amplicon specificity of the primers was checked with Blast(Reference Altschul, Gish, Miller, Myers and Lipman39) and possible secondary structures of the amplicon were verified with Mfold(Reference Zuker40). During optimization, the optimal concentration and annealing temperature for each primer were determined. Also, each amplicon was verified by sequencing.
Table 2 Forward and reverse primers used for PPARγ1a/1b, γ1c/1d and PPARγ2, PPARGC1A, lipoprotein lipase (LPL), GLUT4, fatty acid binding protein 4 (FABP4), TNFα, adiponectin, leptin and leptin receptor (LEPR), amplicon length, annealing temperatures determined for each primer, and GenBank accession number or reference

Reference gene selection and mRNA expression analysis
Determination of the most stable reference genes and their number to be used for normalization of real-time mRNA expression data was done as described previously(Reference Erkens, Van Poucke, Vandesompele, Goossens, Van Zeveren and Peelman34) by using the geNorm algorithm(Reference Vandesompele, De Preter, Pattyn, Poppe, Van Roy, De Paepe and Speleman41), on twenty-four randomly selected samples from all four groups of sows. PCR efficiencies for the reference genes ranged between 90 and 104 %. The mRNA expression for each of the genes in Table 2 was determined by real-time PCR with Platinum SYBR Green qPCR Supermix UDG (Bio-Rad) on the iCycler iQ Real-Time PCR Detection System (Bio-Rad) and subsequently normalized, as described in Erkens et al. (Reference Erkens, Van Poucke, Vandesompele, Goossens, Van Zeveren and Peelman34). PCR efficiencies were between 91 and 103 %.
Blood parameters analysis
Blood was collected on the first day of lactation by jugular venepuncture. Parameters such as leptin, insulin and thyroid hormones were analysed in serum. Leptin was determined using a commercially available RIA test kit (Multi-Species Leptin RIA Kit, catalogue number XL-85K; Linco Research Inc., St. Charles, MO, USA). Insulin was analysed by the Biosource INS-IRMA KIP1251–KIP1254 method. Thyroid hormones (T3 and T4) were analysed by using a specific RIA.
Statistical analyses
All examined parameters were analysed for the four experimental groups with two-way ANOVA. The type of feed (f v. s) and the duration of providing the experimental feed (onset day 3 v. onset day 8 before farrowing) were included as fixed factors in the models. When the interaction of diet type and onset day was significant, further comparisons between the four experimental groups were performed using a post-hoc Duncan test. Mean values and their standard errors were calculated for each experimental group separately (f3; f8; s3; s8). Correlations between the examined parameters (expression of genes, blood parameters) were investigated using Spearman rank correlations. The level of significance was at P ≤ 0·05 (two-sided test). All statistical analyses were performed using SPSS 15.0 (SPSS Inc., Chicago, IL, USA).
Results
Reference gene selection
Analysis of the real-time data indicated that RPL13A was not suitable as a reference gene because it could not be detected in all samples. The application of the geNorm algorithm to the mRNA expression data of the other nine reference genes indicated that it was sufficient to use a set of the three most stably expressed reference genes for normalization. As can be seen in Fig. 1, these genes are hydroxylmethylbilanesynthase (HMBS), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin (ACTB).
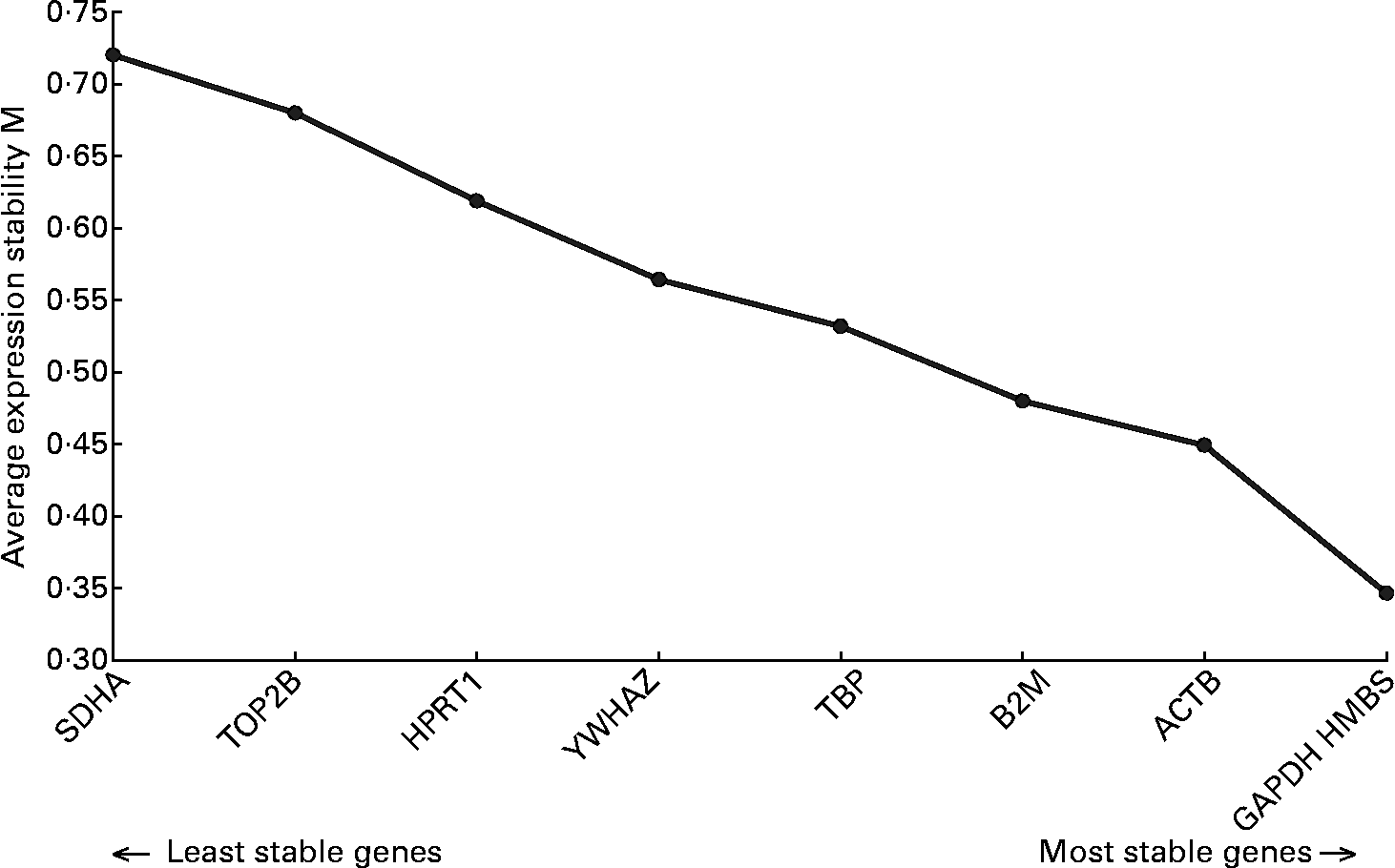
Fig. 1 Stability of the reference genes, calculated by geNorm. M value indicates the expression stability of the reference genes(Reference Erkens, Van Poucke, Vandesompele, Goossens, Van Zeveren and Peelman34). Higher M corresponds with lower stability. ACTB, β-actin; B2M, β-2-microglobulin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HMBS, hydroxylmethylbilanesynthase; HPRT1, hypoxanthinephosphoribosyltransferase 1; SDHA, succinate dehydrogenase complex subunit A; TBP, TATA box binding protein; TOP2B, topoisomerase IIβ; YWHAZ, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta polypeptide.
Effect of n-6:n-3 ratio and prepartal feeding strategy on gene expression
The mRNA expression of the genes in the adipose tissue of the sows during the first day of lactation is shown in Figs. 2 and 3. No significant differences were found in mRNA levels of PPARγ1c/d, PPARγ1a/b, GLUT4, adiponectin, leptin, LEPR, LPL, FABP4 and TNFα. A significant interaction of diet type × onset day was found for PPARγ2 and PPARGC1A mRNA levels (P = 0·019 and P = 0·012, respectively), with the f3 sows showing lower mRNA expression than s3 and f8 for PPARγ2, and lower than s3 for PPARGC1A.

Fig. 2 PPARγ1c/d (A), PPARγ1a/b (B), PPARγ2 (C) and PPARγ coactivator 1A (D) mRNA levels in white adipose tissue from sows fed two lactation diets differing in the n -6:n -3 ratio: low, supplemented with fish oil (f diet type); high, supplemented with sunflower oil (s diet type). The diets were administered from two time-points onwards: 8 or 3 d before farrowing (onset day group 8 or 3) (f3, f diet type, onset day 3; f8, f diet type, onset day 8; s3, s diet type, onset day 3; s8, s diet type, onset day 8). The mRNA abundance for each of the genes was measured by real-time PCR on the iCycler iQ Real-Time PCR Detection System (Bio-Rad) and subsequently normalized. Values are means with their standard errors depicted by vertical bars. a,b Mean values with unlike superscripts were significantly different (P < 0·05).

Fig. 3 GLUT4 (A), adiponectin (B), leptin (C), leptin receptor (D), TNFα (E), lipoprotein lipase (F) and fatty acid binding protein 4 (G) mRNA levels in white adipose tissue from sows fed two lactation diets differing in the n -6:n -3 ratio: low, supplemented with fish oil (f diet type); high, supplemented with sunflower oil (s diet type). The diets were administered from two time-points onwards: 8 or 3 d before farrowing (onset day group 8 or 3) (f3, f diet type, onset day 3; f8, f diet type, onset day 8; s3, s diet type, onset day 3; s8, s diet type, onset day 8). The mRNA abundance for each of the genes was measured by real-time PCR on the iCycler iQ Real-Time PCR Detection System (Bio-Rad) and subsequently normalized. Values are means with their standard errors depicted by vertical bars. No significant differences were found.
Correlations between mRNA expression of PPARγ isoforms and PPARγ coactivator 1A, lipoprotein lipase, GLUT4, fatty acid binding protein 4, TNFα, adiponectin, leptin and leptin receptor
The correlations between the mRNA levels of the genes investigated in the present study are listed in Table 3. PPARγ1c/d levels correlated with the levels of all genes except for PPARGC1A and LEPR. PPARγ1a/b correlated with the levels of all genes except for PPARGC1A. PPARγ2 correlated with the levels of all genes except for LEPR.
Table 3 Correlations (r Spearman rank) between mRNA levels of three isoforms of PPARγ and other genes in the adipose tissue of sows†
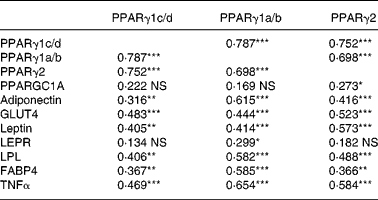
Correlation coefficients were significant: *P < 0·05, **P < 0·01, ***P < 0·001.
† Sows received two lactation diets differing in the n-6:n-3 ratio (a low v. a high) and were administered from two time-points onwards before parturition (8 or 3 d). The mRNA abundance for each of the genes was measured by real-time PCR on the iCycler iQ Real-Time PCR Detection System (Bio-Rad) and subsequently normalized. The correlations represent the results from all treatment groups together (global data).
Correlations between mRNA expression of PPARγ isoforms and PPARγ coactivator 1A, lipoprotein lipase, GLUT4, fatty acid binding protein 4, TNFα, adiponectin, leptin and leptin receptor with circulating leptin, insulin and thyroid hormones
Circulating leptin was negatively correlated with mRNA levels of adiponectin (r − 0·326, P = 0·006), LEPR (r − 0·276, P = 0·027) and LPL (r − 0·341, P = 0·005). T3 was negatively correlated with mRNA levels of FABP4 (r − 0·249, P = 0·047) and T4 with mRNA levels of PPARGC1A (r 0·264, P = 0·038). No significant correlation was found for insulin (e.g. with GLUT4, r 0·083, P = 0·556).
Discussion
The present study investigated the effect of two peripartal diets differing in n-6:n-3 ratio, and fed from either 3 or 8 d before parturition, on the abundance of genes involved in lipid and glucose metabolism in early lactation. Biopsies were collected on the first day of lactation, after blood sampling and before sows received a morning meal. Information regarding sows in this field is scarce. In growing pigs samples of adipose tissue for analysis of leptin and LEPR mRNA expression were collected after blood sampling(Reference Xingjie, Defa, Jingdong, Yuhua, Heliang, Huawei and Ganfeng28). In dogs, biopsies of adipose tissue and skeletal muscle were collected in a fasted state as in the present study(Reference Gayet, Leray, Masayuki, Siliarti and Nguyen42). Fasted state for collection of samples seemed to be the most appropriate as it has been shown in sows that differences in parameters in plasma, such as glucose, NEFA and insulin, occur before and after feeding(Reference Quesnel and Prunier43).
The three most stable reference genes used for normalization of the mRNA expression data of the genes under investigation in the present experiment were HMBS, GAPDH and ACTB. Also in a previous experiment on porcine muscle and fat(Reference Erkens, Van Poucke, Vandesompele, Goossens, Van Zeveren and Peelman34), ACTB belonged to the most stable reference genes. However, in the latter experiment(Reference Erkens, Van Poucke, Vandesompele, Goossens, Van Zeveren and Peelman34), GAPDH proved to be the most unstable reference gene, not suitable for normalization purposes. This shows once again that even in the same tissue type the stability of a reference gene can vary between different experiments and that the expression stability of the reference genes to be used for normalization of real-time relative mRNA expression data should be evaluated for each experiment separately.
The differences observed in PPARγ2 and PPARGC1A mRNA expression might indicate a higher susceptibility to the dietary manipulation at 1 d postpartum of these genes in comparison to the other members of the PPARγ family. The decrease in PPARγ2 and PPARGC1A mRNA expression by the high n-6:n-3 when fed from 3 d prepartum corresponds with evidence from the literature that n-6 and n-3 fatty acids act as ligands for the PPARγ, and as found in the human thromboplast cell line, BeWo, the transcription factors PPARγ were reduced after incubation with n-3 PUFA(Reference Reseland, Haugen, Hollung, Solvoll, Halvorsen, Brude, Nenseter, Christiansen and Drevon44). Other studies also confirmed that PPARγ levels appeared to be increased more by n-6 fatty acids than n-3 fatty acids(Reference Forman, Tontonoz, Chen, Brun, Spiegelman and Evans45, Reference Mater, Pan, Bergen and Jump46). Furthermore, PPARγ2 has been identified as a form that functions in adipocyte differentiation through gene regulation(Reference Rosen and Spiegelman47–Reference Gurnell50) and perhaps this function may explain its sensitivity to the n-6:n-3 ratio and the acute feed changes prior to parturition. Moreover, a critical aspect of the PPARGC1 co-activators is that they are highly versatile and have the ability to interact with many different transcription factors. In doing so, they activate distinct biological programmes in different tissues(Reference Lin, Handschin and Spiegelman51).
In the human thromboplast cell line (BeWo), n-3 PUFA had a dose- and time-dependent effect on leptin expression(Reference Reseland, Haugen, Hollung, Solvoll, Halvorsen, Brude, Nenseter, Christiansen and Drevon44). Sows in the present study were offered equal amounts of isoenergetic diets. The increase in energy intake at the change of the gestation diet to the experimental diet (type lactation diet) induced changes in circulating leptin levels in the present study (reported elsewhere). Yet, at the day of the biopsies, no significant differences were found in circulating leptin and LEPR mRNA expression. In growing pigs, dietary oil sources did not change plasma leptin and insulin concentrations, whereas the main effect of oil sources and adipose tissue sources were significant for leptin mRNA and LEPR mRNA expression(Reference Xingjie, Defa, Jingdong, Yuhua, Heliang, Huawei and Ganfeng28). This suggests that leptin mRNA expression in white adipose tissue of growing pigs was affected by an insulin-independent mechanism. Furthermore, fish oil supplementation in growing pigs decreased the abundance of leptin mRNA and LEPR mRNA compared with soyabean oil, in the dorsal subcutaneous adipose tissue(Reference Xingjie, Defa, Jingdong, Yuhua, Heliang, Huawei and Ganfeng28). However, in the present study, during the first day of lactation, insulin levels were significantly affected by the type of fat (reported elsewhere), whereas leptin mRNA and LEPR mRNA were not affected by the type of fat source. This difference may indicate different regulations of these parameters between growing pigs and lactating sows, but the lack of effect of the type of diet on leptin mRNA and LEPR mRNA expression might also indicate that these parameters are insulin-independent in lactating sows.
Arachidonic acid, a member of the n-6 PUFA, enhances either the translocation of both major adipocyte glucose transporters (GLUT4 and GLUT1) to the plasma membrane or reduces their rate of internalization(Reference O'Rahilly, Claire, Johannes, Whitehead, Wentworth, Krishna and Chatterjee52), and in 3T3-L1 adipocytes, arachidonic acid regulated GLUT4 gene expression by potentially two independent mechanisms: (1) via oxidative metabolism to the cyclooxygenase metabolite PGE2 and the subsequent elevation of cAMP levels (2) via non-oxidized fatty acid with the potential involvement of a PPAR(Reference Pekala and Long53). A possible explanation for the absence of significant effects on GLUT4 levels in the present study is that in the biopsies of adipose tissue collected from sows, total cellular mRNA expression of GLUT4 was measured, leaving the question whether the n-6:n-3 ratio affected GLUT4 translocation. It is known that reduced glucose transport activity could be the result of fatty acid effects on the GLUT4 transporter directly, i.e. alterations in the trafficking, budding, fusion or activity of GLUT4, or it could result from fatty acid-induced alterations in upstream insulin signalling events, resulting in decreased GLUT4 translocation to the plasma membrane(Reference Kahn54).
Findings from studies in man and in experimental animals suggested that mRNA levels in adipose tissue of adiponectin(Reference Neschen, Morino, Rossbacher, Pongratz, Cline, Sono, Gillum and Shulman55), TNFα(Reference Sethi and Hotamisligil16), FABP(Reference John, Rule, Knabe, Mersmann and Smith56) and LPL(Reference Luo, Salwa, Vidal, Oppert, Colas, Guerre-Millo, Chapuis, Chevalier, Durand and Slama57, Reference Murphy, Zampelas, Puddicombe, Furlonger, Morgan and Williams58) are either influenced by the dietary fat source or they are involved in the regulation of lipid metabolism. In the present study no significant differences were found in the expression levels of these genes. The different metabolism dynamics around parturition may make a major difference too. Yet it cannot be excluded that differences between dietary groups on expression of these genes occurred either earlier or later than 1 d postpartum. Studies in man(Reference Hube, Lietz and Igel59, Reference Masuzaki, Ogawa and Isse60), rodents(Reference Masuzaki, Ogawa and Shigemoto61) and dogs(Reference Gayet, Leray, Masayuki, Siliarti and Nguyen42) provided evidence for differences in mRNA expression between various fat depots of the body. In the present study, adipose tissue was collected only from one position in the backfat adipose tissue in lactating sows, to limit the effect of the induced stress on the metabolism and performance of the sows. The level of inclusion of the fat sources was 2 %, in order to introduce a diet that is closer to the levels of inclusion used in practice, but especially to avoid negative effects associated with higher supplementation of fish oil(Reference Cartwright, Pockley and Galloway62–Reference Weber and Leaf64).
Significant correlations between the mRNA expressions of the genes investigated in the present study were also observed in other animal models. It is known that the target genes of PPARγ encode proteins involved in the trapping, release and storage of fatty acids, such as FABP(Reference Pelton, Zhou, Demarest and Burris65), LPL(Reference Schoonjans, Peinado-Onsurbe, Lefebvre, Heyman, Briggs, Deeb, Staels and Auwerx27), leptin(Reference Cohen, Novick and Rubinstein26, Reference Gayet, Leray, Masayuki, Siliarti and Nguyen42), TNFα(Reference Miles, Romeo, Higo, Cohen, Rafaat and Olefsky24), adiponectin(Reference Maeda, Takahashi and Funahashi25, Reference Gayet, Leray, Masayuki, Siliarti and Nguyen42, Reference Neschen, Morino, Rossbacher, Pongratz, Cline, Sono, Gillum and Shulman55, Reference Yamauchi, Kamon and Waki66) and GLUT4(Reference Wu, Xie, Morrison, Bucher and Farmer67). Circulating leptin was inversely related to the LEPR mRNA expression, and no correlation was found between circulating leptin and leptin mRNA expression. Divergences between circulating leptin and leptin gene expression were reported previously in man(Reference Russell, Petersen and Rao68) and in obese women(Reference Lonnqvist, Nordfors, Jansson, Thorne, Schalling and Arner69), in which changes in circulating leptin did not depend on the synthesis of new protein, suggesting that leptin in man was released from a preformed pool within the cells and post-transcriptional regulation of leptin might occur. Another reason for the divergences observed between circulating leptin and leptin mRNA expression in lactating sows is the fact that leptin may be transferred from the maternal circulation to the milk as in rats(Reference Casabiell, Pineiro, Tome, Peino, Dieguez and Casanueva70). In addition, it was found in lactating sows that milk concentrations of leptin were not correlated with backfat thickness or serum levels of leptin(Reference Estienne, Harpera, Barb and Azain71). The existence of a positive relationship between T4 and PPARGC1A may be explained by the fact that thyroid hormones stimulate mitochondrial biogenesis and induce mitochondrial genes encoded in the nucleus(Reference Nelson72). Also the role of PPARGC1A in the regulation of mitochondrial oxidative metabolism is important, as the loss of PPARGC1A leads to significant functional deficits in oxidative metabolism in multiple tissues and renders mice exercise intolerant(Reference Leone, Lehman and Finck73).
In conclusion, adipocyte PPARγ2 and PPARGC1A mRNA expression in sows at 1 d postpartum depends on the prepartum dietary n-6:n-3 ratio in combination with the onset of prepartum feeding plane during late gestation. This suggests that the effect of dietary fatty acid profile on the regulation of these genes on biopsies of backfat adipose tissue of sows taken at the first day of lactation is modulated by the duration of the feeding plane during the last week of gestation. The present findings and the emergence of PPARγ2 as a key modulator of the expression of genes that are critically involved in glucose and lipid metabolism and the role of PPARGC1A as a co-activator may help to elucidate the pathophysiology basis of metabolic-associated problems during the first stages of lactation in sows.
Acknowledgements
Expenses for the laboratory analyses were covered by the scholarship programme awarded to G. A. P. by the Greek State Scholarship Foundation for postgraduate study abroad (www.iky.gr). G. A. P. was responsible for the study design, study performance, parameters analysis, data analysis and manuscript drafting. T. E. contributed equally to this study, and performed the mRNA expression analysis and contributed to manuscript drafting. L. J. P. supervised the mRNA expression analysis and contributed to manuscript drafting. T. A. T. G. v. K. was responsible for supervision of logistic arrangements, experimental diet development and contributed to manuscript drafting. J. B. supervised the analysis of blood parameters and contributed to manuscript drafting. D. G. D. M. and G. P. J. J. evaluated the study design, contributed to data analysis and manuscript drafting, and both are promoters of G. A. P. Maria D. Baucells from the Departament de Ciència Animal i dels Aliments, Universitat Autònoma de Barcelona is acknowledged for the provision of the biopsy device. It is declared by the corresponding author that no conflict of interest exists for this paper.








