The worldwide prevalence of childhood overweight and obesity is increasing rapidly( Reference de Onis, Blossner and Borghi 1 ), and is expected to reach 9 % in 2020( Reference de Onis, Blossner and Borghi 1 ). In Spain, the prevalence of overweight is about 45 % in children aged 6–9 years( 2 , Reference Moleres, Ochoa and Rendo-Urteaga 3 ). Young people with excessive body weight are more likely to develop obesity-related co-morbid conditions at early ages( Reference de Onis, Blossner and Borghi 1 , Reference Moreno, Gonzalez-Gross and Kersting 4 ), mainly type 2 diabetes, CVD( Reference Baker, Olsen and Sorensen 5 ), hypertension and others( Reference Wabitsch 6 , Reference Reinehr and Wabitsch 7 ). In this sense, obesity is viewed as one of the major current public health problems with a high impact on children, leading to significant morbidity in adulthood( Reference Owen, Martin and Whincup 8 ). Treating obesity in children and adolescents is critical to prevent obesity-related complications in adults( Reference Crocker and Yanovski 9 ). Small reductions in body weight (between 0·25 and 0·50 BMI standard deviation score (BMI-SDS)) can trigger an improvement in metabolic outcomes in obese children( Reference Katz, O'Connell and Yeh 10 – Reference Lavelle, Mackay and Pell 12 ).
The use of blood cells for gene expression studies offers an advantage over the use of other human tissues because they are the most readily accessible tissue and are therefore of great interest for clinical studies and experimental research( Reference Burczynski and Dorner 13 , Reference Caimari, Oliver and Keijer 14 ). Moreover, the use of these cells for gene expression studies is a non-invasive technique, which offers an advantage for clinical studies. Specifically, the possibility of using gene expression measures in blood cells as the markers of metabolic status in obese children has been indicated( Reference Sanchez, Priego and Pico 15 ). Blood cells are being used to assess biological responses, as gene expression profiles may reflect the pathological and physiological states of the organism( Reference Sanchez, Priego and Pico 15 ). Previous microarray studies in human subjects have shown changes in gene expression profiles after a low-energy dietary intervention( Reference Bouchard, Rabasa-Lhoret and Faraj 16 ), aimed to identify target genes associated with obesity that may help to develop personalised dietary treatments( Reference Crujeiras, Parra and Milagro 17 ). Peripheral blood mononuclear cells (PBMC) have been used in other studies investigating gene expression profiles in children( Reference Radom-Aizik, Zaldivar and Leu 18 , Reference Radom-Aizik, Zaldivar and Leu 19 ); however, no results for after weight-loss (WL) intervention programmes in obese children are available.
The analysis of individual gene responses using techniques such as microarrays is becoming a powerful tool that plays a role in understanding the mechanisms that control the physiological response of immune cells to a variety of perturbations, but the enormity of the data generated by such analyses can be perplexing, and at the same time, it is increasingly recognised that the functional and physiological significance of changes in gene expression may be better understood by examining the coordinated modulation of groups of genes acting in discrete pathways. In general, very little is yet known about the individual expression of genes in mononuclear cells in response to the dietary intervention, and even less about specific gene profiles.
In the present study, the effects of a moderate energy-restricted diet on the gene expression profile of PBMC were examined in obese boys. We hypothesised that a moderate energy-restricted diet could promote the changes in the gene expression of PBMC in the population under study. In particular, the aim of the present study was to characterise the changes in the expression of genes in PBMC after a nutritional intervention, specifically between the high-responder (HR) and low-responder (LR) groups at (1) baseline and (2) after the nutritional intervention.
Subjects and methods
Subjects
In the present study, seventy-one children aged 7–15 years who were classified as overweight or obese according to the criteria of Cole et al. ( Reference Cole, Bellizzi and Flegal 20 ) were invited to participate in an informative session. Children were recruited from the Endocrinology Paediatric Units of the University of Navarra Clinic and Navarra's Hospital Complex in Pamplona, Navarra. All of them were Spanish or schooling foreigners for at least 1 year in Spain. Participants with a major psychiatric illness, significant neurological disease, eating disorders, familial hyperlipidaemia or any sort of either major cardiovascular or respiratory complication were excluded. The present study was conducted according to the guidelines laid down in the Declaration of Helsinki, and all procedures involving the subjects were approved by the Ethics Committee of the University of Navarra. Written informed consent was obtained from all the children and their parents.
Among the initial seventy-one volunteers, fifty-four successfully underwent baseline anthropometric measurements, of which forty-four (twenty-two boys and twenty-two girls) concluded the 10-week dietary intervention (dropout rate 18·5 %; see Fig. 1), as published elsewhere( Reference Rendo-Urteaga, Garcia-Calzon and Martinez-Anso 21 , Reference Rendo-Urteaga, Puchau and Chueca 22 ). The BMI-SDS was calculated as a function of the degree of obesity of the subjects when compared with BMI local reference standards( Reference Santacruz, Marcos and Warnberg 23 ). The response of the participants to the intervention programme was based on the changes in BMI-SDS. After dichotomising the sample at the median (equal to 0·5 BMI-SDS), participants were divided into two groups: HR group (n 6), who had a more decreased BMI-SDS; LR group, who either maintained or had an even increased BMI-SDS (Fig. 1).

Fig. 1 Flow diagram describing the recruitment, study flow and follow-up of the participants. We chose six boys who had a decreased BMI-standard deviation score (SDS) as the high-responder (HR) group, and six boys who either maintained or had an increased BMI-SDS as the low-responder (LR) group.
Previous studies have shown that an improvement in body composition and cardiometabolic risk can be achieved with a BMI-SDS reduction of ≥ 0·25 in obese adolescents, but greater benefits occur when losing at least 0·5 BMI-SDS( Reference Ford, Hunt and Cooper 11 ).
Dietary intervention
Dietary follow-up, weight control and nutritional education were performed weekly through individual sessions with a registered dietitian. Obese boys from the HR and LR groups – with their father, mother or tutor – did similarly attend the appointments. Subjects were prescribed a fixed full-day meal diet, calculated according to the BMR. Energy expenditure was estimated taking into account basal metabolism using the Schofield equation, according to sex( Reference Schofield 24 ) and body growth rate( Reference Moreno, Ochoa and Warnberg 25 ). Moderate energy restriction was calculated according to the children's obesity degree( Reference Moreno, Mesana and Gonzalez-Gross 26 ), not taking into account the body growth rate. In all cases, diets were not lower than 5439·2 kJ/d (1300 kcal/d) and not higher than 9204·8 kJ/d (2200 kcal/d). Participants and their parents received personal training in nutritional and physical education throughout the whole intervention period. A semi-quantitative FFQ, previously validated in Spain( Reference Marques, Moleres and Rendo-Urteaga 27 , Reference Moleres, Rendo-Urteaga and Zulet 28 ), containing 132 food items was filled in to evaluate dietary patterns in the obese children under study.
Anthropometric, clinical and biochemical measurements
Weight and height were measured with an electronic scale (Type SECA 861; Seca) and a telescopic height-measuring instrument (Type SECA 225; Seca), respectively. Waist and hip circumferences were measured with a flexible non-stretchable measuring tape (Type SECA 200; Seca). Body fat mass was estimated by bioelectrical impedance analysis (TBF-410; Tanita, Inc.). Blood pressure measurement was obtained from the left arm after a 15 min rest using a blood pressure monitor (OMRON M6; OMRON Healthcare Europe B.V.). Glucose, insulin and lipid profiles were determined by standard autoanalyser techniques. Insulin resistance was calculated from fasting glucose and insulin levels by using the homeostasis model assessment of insulin resistance( Reference Matthews, Hosker and Rudenski 29 ). Also, the quantitative insulin sensitivity check index method was used for the calculation of insulin sensitivity( Reference Katz, Nambi and Mather 30 ). All parameters were measured at baseline and after the nutritional intervention.
Extraction and isolation of RNA from peripheral blood mononuclear cells
Gene expression analysis was carried out in PBMC from twelve obese boys (six HR and six LR) at baseline and after the nutritional intervention. PBMC were obtained from 10 ml EDTA anti-coagulated peripheral blood by density centrifugation at 450 g for 30 min at 21°C using a Polymorphrep procedure (Axis-Shield PoC AS).
Total RNA was isolated from PBMC using the TRIzol method according to the manufacturer's instructions (Invitrogen). Extracted RNA was purified using the RNeasy Mini kit (Qiagen). RNA concentrations and purity were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). RNA quality was assessed on an Agilent 2100 Bioanalyser, yielding a number of RNA integrity higher than 8·0 in all the samples.
Synthesis of complementary DNA
RNA (about 300 ng) was converted into complementary DNA following the manufacturer's instructions (Ambion® WT Expression kit; Affymetrix UK Ltd). Then, each aliquot of complementary DNA was fragmented using UDG (Uracil DNA Glycosylase) and APE1 (apurinic/apyrimidinic endonuclease 1), and labelled with terminal transferase using the Whole-Transcript Terminal Labelling kit of Affymetrix.
Hybridisation and scanning
Gene expression was analysed by microarray hybridisation in Progenika Biopharma, S.A. The Human Gene 1.1 ST 24-Array Plate (Affymetrix) was used the microarray, which analyses 28 869 genes with about twenty-six probes per gene.
The hybridisation, wash, developing and scanning of the samples was performed using a GeneTitan® platform (Affymetrix), according to the manufacturer's recommendations.
Quantitative assessment of gene expression by real-time PCR
To confirm the findings on gene expression microarray data, a real-time PCR (RT-PCR) protocol was performed with the TaqMan Universal PCR Master Mix and using an ABI PRISM 7900HT Fast Real-Time PCR System (Applied Biosystems). The expression of both target and housekeeping (cyclophilin A) genes for each sample was quantified based on their respective threshold cycle value (ddC t). The threshold cycle value of target genes was normalised to that of cyclophilin A, and then expressed as the fold change (FC). Primer references obtained from Applied Biosystems are indicated as follows: signal-regulatory protein β1 (SIRPB1; NM_006065; Hs03645221_m1); leptin receptor (LEPR; NM_002303; Hs00174497_m1); epidermal growth factor (EGF; NM_001963; Hs00196731_m1); tissue factor pathway inhibitor (TFPI; NM_006287; Hs01099999_m1); junctional adhesion molecule 3 (JAM3; NM_032801; Hs00230289_m1); latent transforming growth factor β-binding protein 1 (LTBP1; NM_206943; Hs00386448_m1); multimerin 1 (MMRN1; NM_007351; Hs00201182_m1); polycystic kidney and hepatic disease 1-like 1 (PKHD1L1; NM_177531; Hs00415429_m1); selectin P (SELP; NM_003005; Hs00927900_m1); Igκ constant (IGKC; BC073772; Hs02384840_gH); amphiregulin (AREG; NM_001657; Hs00950669_m1); heparin-binding EGF-like growth factor (HBEGF; NM_001945; Hs00181813_m1); nuclear receptor subfamily 4, group A, member 2 (NR4A2; NM_006186; Hs00428691_m1); TNFα-induced protein 3 (TNFAIP3; NM_006290; Hs00234713_m1). A comparative study of the following genes was performed, with cyclophilin A being used as a stable internal marker: RNA; 28S ribosomal 1 (RN28S1); actin β (ACTB); glyceraldehyde 3-phosphate dehydrogenase (GAPDH); phosphoglycerate kinase 1 (PGK1). These genes were measured by RT-PCR in the samples obtained from the obese children under study before and after the WL programme. The expression levels of cyclophilin A mRNA (Hs04194521_s1) were not found to be affected by the experimental conditions and thus was used as the housekeeping gene. In this sense, cyclophilin A has previously been adopted as a housekeeping gene in PBMC experiments( Reference Cinar, Islam and Pröll 31 , Reference Wang, Liang and Sandford 32 ).
Statistical analyses
Statistical analyses were performed using SPSS for Windows 15.0 software (SPSS, Inc.). For all the analyses, the significance level was set at α = 0·05, and all the tests were two-sided. The sample size calculation indicated that five subjects per group were required to be included in the study. This estimation was based on the following assumptions: an α error of 5 %, a power of 80 %, and a mean difference of 0·50 (sem 0·27) units in BMI-SDS after the nutritional intervention.
The Shapiro–Wilk test was used to determine variable distributions. Non-normally distributed variables (insulin levels) were log-transformed before an appropriate application of parametric statistical tests. An unpaired t test was used to assess the differences in anthropometric and metabolic characteristics between the HR and LR groups at baseline. A paired t test was used to compare the changes in both groups based on the nutritional intervention (before v. after intervention). A two-way ANOVA (response and time) was performed to compare the changes in the expression levels of genes from the microarray data between the HR and the LR groups at baseline and after 10 weeks of the nutritional intervention. In addition, a linear regression model was fitted to assess the changes in anthropometric variables according to basal gene expression levels. The respective value of the metabolic covariate was adjusted at baseline (after controlling for metabolic variables), and B regression coefficient values and 95 % CI were calculated.
Microarray data analysis
For the analysis, microarray raw files were imported into Partek GS (Partek, Inc.). Feature intensity values were normalised and reduced to expression summaries using the Robust Multichip Average method( Reference Irizarry, Hobbs and Collin 33 ). Probe sets were filtered based on raw intensity values by setting an arbitrary threshold of 100 fluorescence units to remove those with near-to-background values. In addition, probe sets that remained unchanged across the experiment were also removed from further analysis using the standard deviation of normalised intensity. A total of 10 173 genes passed these filters. Differentially expressed genes were detected using repeated-measures ANOVA, setting the significance threshold at P <0·05 (n 2173 genes). Multiple testing was controlled after the application of Bonferroni (data not shown) and false discovery rate corrections. The results were annotated using DAVID (Database for Annotation, Visualization and Integrated Discovery) software( Reference Dennis, Sherman and Hosack 34 – Reference Huang da, Sherman and Lempicki 36 ), and pathway analysis was conducted using the Kyoto Encyclopaedia of Genes and Genomes (KEGG) metabolic pathway database( Reference Bouwens, Afman and Muller 37 ). Raw gene expression data obtained from the microarray data analysis are available at the Gene Expression Omnibus database( Reference Rendo-Urteaga, Garcia-Calzon and Martinez-Anso 21 ) (accession no. GSE41505).
Results
The characteristics of the six obese boys who had a more decreased BMI-SDS (HR group), and the six boys who either maintained or had an even increased BMI-SDS (LR group) are summarised in Table 1. No differences in the characteristics, including the Tanner stage (data not shown), were found between the two groups at baseline. Food intake at baseline was found to be similar in both groups (HR v. LR), and after the 10-week intervention programme, a trend was observed towards the lower intake of total energy and macronutrients (Table 1), which was statistically significant only in the HR group (P= 0·021).
Table 1 Characteristics of the obese boys at baseline and after 10 weeks of the nutritional intervention according to the diet response (Mean values with their standard errors)

HR, high responders; LR, low responders; SDS, standard deviation score; DBP, diastolic blood pressure; SBP, systolic blood pressure; HOMA-IR, homeostasis model assessment of insulin resistance; QUICKI, quantitative insulin sensitivity check index.
* P value for the comparison between baseline and after the nutritional intervention in subjects distributed according to the response.
† P value for the comparison between the HR and LR groups at baseline.
‡ P value for the comparison between the HR and LR groups after the nutritional intervention.
§ Non-normally distributed variables were log-transformed before analysis.
The 10-week intervention programme had a beneficial effect on anthropometric and biochemical measurements (Table 1). Obese boys in the LR group did not achieve statistically significant WL, but were able to maintain their body weight. A significant decrease in serum HDL-cholesterol concentration was observed in the LR group after the nutritional intervention. In the HR group, the mean WL was found to be 4·47 (sem 0·23) kg (5·7 % loss of initial body weight, P< 0·001). Specifically, these individuals showed a significant reduction in BMI-SDS ( − 20·6 %, P= 0·001) and waist circumference ( − 6·5 %, P= 0·001) after the nutritional intervention. During the moderate energy-restricted diet intervention, WL was associated with an improvement in insulin sensitivity, as assessed by the quantitative insulin sensitivity check index (6·7 %, P< 0·05) and the homeostasis model assessment index ( − 44·3 %, P= 0·001).
With regard to the microarray data, forty-eight genes were overexpressed and seven genes repressed, after a statistical comparison of the expression profiles of PBMC between the LR and HR groups at baseline (FC ± 1·5, P< 0·05; see online supplementary Table S1(A)). Similarly, nine genes were up-regulated and thirty-three down-regulated in the HR group after the nutritional intervention (FC ± 1·5, P< 0·05; see online supplementary Table S1(B)).
In addition, genes implicated in metabolism and obesity-related alterations were carefully examined according to the literature, since the aim of the study was to characterise the response to WL in obese boys.
In total, the expression levels of ten target genes (EGF, TFPI, LEPR, JAM3, LTBP1, MMRN1, PKHD1L1, SELP, SIRPB1 and IGKC) were found to be similar in both groups at baseline, as assessed by the RT-PCR and microarray analysis. Specifically, the microarray data were summarised after the comparison between (1) the LR and HR groups at baseline and (2) before and after the nutritional intervention in the HR group.
Comparison of low responders v. high responders at baseline
At baseline, the gene expression profile of PBMC in the LR group compared with the HR group showed the overexpression of forty-eight genes and the repression of seven genes (FC ± 1·5, P< 0·05; Fig. 2(a); see also online supplementary Table S1(A)). Notably, three of the seven underexpressed genes in the LR group did correspond to IGKC ( − 1·75-fold), encoding the constant domain of κ-type light chains for antibodies. Several overexpressed genes in the LR group compared with the HR group were as follows: SIRPB1 (NM_006065; 4·09-fold); LEPR (1·50-fold); EGF (1·52-fold); TFPI (1·82-fold); JAM3 (2·09-fold); LTBP1 (1·69-fold); MMRN1 (1·87-fold); PKHD1L1 (1·87-fold); SELP (1·82-fold).

Fig. 2 Heat map analysis of microarray data showing (a) hierarchical clustering of fifty-five differentially expressed genes between the low-responder (LR) group and the high-responder (HR) group at baseline (LB and HB, respectively), and (b) hierarchical clustering of forty-two differentially expressed genes in the HR group before (HB) and after (HA) the 10-week nutritional intervention programme. The red or green colours indicate differentially up- or down-regulated genes, respectively (1·5-fold change, P< 0·05), in the samples from twelve obese boys from the two groups (HR and LR), (b) before and (a) after the intervention. The comparison of (a) HB v. LB and (b) HA v. HB is shown. For a list of gene names and abbreviations, see online supplementary Table S1. (A colour version of this figure can be found online at http://www.journals.cambridge.org/bjn).
Moreover, a comparison of gene expression levels from the microarray data was made between the HR and LR groups at baseline and after 10 weeks of the nutritional intervention using the two-way ANOVA (response, time; Fig. 3). There were statistically significant changes in the expression levels of the ten target genes between the LR and HR groups (P response). The transcript levels of IGKC were significantly higher in the HR group, whereas those of the other nine genes were underexpressed in the HR group compared with the LR group (Fig. 3). However, the effect of the intervention (before v. after, P time) did not reach statistical significance for any of the genes studied.

Fig. 3 Comparison of gene expression levels from the microarray data between the high-responder (HR, ■) group and low-responder (LR, □) group at baseline and after 10 weeks of the nutritional intervention. Values are means, with their standard errors represented by vertical bars. Two-way ANOVA: P response, differences between the HR and LR group; P time, differences between after and before the nutritional intervention; P interaction, interactive effect between diet response and time. For a list of gene names and abbreviations, see online supplementary Table S1.
To confirm the microarray results, the expression levels of the ten target genes (EGF, TFPI, LEPR, JAM3, LTBP1, MMRN1, PKHD1L1, SELP, SIRPB1 and IGKC) were measured using RT-PCR. The genes chosen were among the top ten genes with a higher FC of up-regulation. The only exceptions were LEPR and EGF, which were chosen because of their key regulatory role in energy homeostasis. Additionally, within the top ten up-regulated genes, three (glutathione S-transferase mu 4 (GSTM4), lectin, galactoside-binding, soluble, 2 (LGALS2) and major histocompatibility complex, class II, DP beta 1 (HLA-DPB1)) were not selected for quantitative PCR validation because of their higher P value and false discovery rate, which offered less guarantee for the validation by the other technique. Among the down-regulated genes, IGKC was selected because of its highest change in the expression levels (FC − 1·7). The changes in the expression levels of down-regulated genes were more modest (see online supplementary Table S1(A)) than those of the up-regulated genes, supporting the selection of only one gene in this category.
For all the genes, the direction and magnitude of the FC obtained from RT-PCR were comparable to those obtained from the microarray technique (Fig. 4(a)). Moreover, RT-PCR showed significant differences in five of the genes between the LR and HR groups (P< 0·05), and two of them almost reached statistical significance (P< 0·1). Overall, most of the genes tested were validated by RT-PCR, suggesting that the microarray data represent true expression changes.

Fig. 4 Gene expression levels from the microarray (■) and real-time PCR (□) data. (a) Comparison between the low-responder (LR) and high-responder (HR) groups at baseline, and (b) comparison between baseline and after the moderate energy-restricted diet intervention in the HR group. Mean change compared with baseline was significant: * P< 0·05, ** P< 0·01, *** P< 0·001. Mean change compared with baseline was marginally significant (trend): † P< 0·10. For a list of gene names and abbreviations, see online supplementary Table S1.
DAVID software( Reference Dennis, Sherman and Hosack 34 ) was used to identify Gene Ontology biological processes and KEGG pathways. At baseline, nineteen biological processes showed significant differences between the LR and HR groups (P< 0·05; see online supplementary Table S2), with platelet activation and blood coagulation being the most divergent processes. Moreover, nine KEGG pathways were significantly overexpressed in the LR group when compared with the HR group at baseline (Table 2), including extracellular matrix–receptor interaction, focal adhesion, cell adhesion molecules and IgG Fc receptor II (FcγR)-mediated phagocytosis.
Table 2 Overexpression of Kyoto Encyclopaedia for Genes and Genomes (KEGG) pathways in the low-responder (LR) group compared with the high-responder (HR) group at baseline, and down-regulation in the HR group after the nutritional intervention
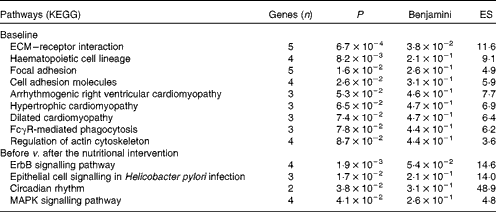
ES, enrichment score; ECM, extracellular matrix; FcγR, IgG Fc receptor II; ErbB, erythroblastoma virus B receptor family; MAPK, mitogen-activated protein kinases.
A linear regression model was fitted to predict the changes in anthropometric variables after the 10-week nutritional intervention programme, according to the baseline expression levels of the ten target genes (Table 3). Importantly, baseline transcript levels of EGF, TFPI, LEPR, JAM3, LTBP1, MMRN1, PKHD1L1, SELP, SIRPB1 and LEPR were associated with the changes in body weight, BMI-SDS and body fat mass after the nutritional intervention, indicating that the lower the expression levels, the higher the decrease in adiposity indices.
Table 3 Association between baseline gene expression levels of the ten selected target genes and changes in adiposity indices after the 10-week nutritional intervention* (B regression coefficient values and 95 % confidence intervals, n 12)

SDS, standard deviation score; EGF, epidermal growth factor; TFPI, tissue factor pathway inhibitor; LEPR, leptin receptor; JAM3, junctional adhesion molecule 3; LTBP1, latent transforming growth factor β-binding protein 1; MMRN1, multimerin 1; PKHD1L1, polycystic kidney and hepatic disease 1-like 1; SELP, selectin P; SIRPB1, signal-regulatory protein β1; IGKC, Igκ constant.
* Adjusted for the corresponding anthropometric variable at baseline.
Comparison of before v. after the nutritional intervention in the high-responder group
The nutritional intervention did alter the gene expression profile of PBMC, with a total of 2173 genes showing differential transcript levels (P< 0·05) in obese boys (Fig. 5). Specifically, approximately 28 % of them (390 genes) were changed in both groups (HR and LR). In addition, 35·2 % (880 genes: 555 up-regulated and 325 down-regulated) were significantly modified in the HR group; meanwhile, 36·8 % (903 genes: 527 up-regulated and 376 down-regulated) were altered in the LR group.

Fig. 5 Comparison of the gene expression profile of peripheral blood mononuclear cells (PBMC) in obese boys according to the response (high-responder (HR) and low-responder (LR) groups) to a weight-loss (WL) programme, showing the relative magnitude of the effects (circles) and the size of the overlap (shaded area). A total of 390 PBMC genes were significantly altered by the WL programme in both groups.
Specifically, nine genes were up-regulated and thirty-three down-regulated in the HR group (FC ± 1·5, P< 0·05) after the nutritional intervention (Fig. 2(b); see online supplementary Table S1(B)). AREG ( − 3·21-fold), HBEGF ( − 2·54-fold), NR4A2 ( − 2·49-fold) and TNFAIP3 ( − 2·13-fold) were among the down-regulated genes.
RT-PCR was performed to quantitatively measure the differences in the transcript levels of a select group of genes that were identified by the microarray technique. Similar to the microarray data, the decreased gene expression levels of AREG, HBEGF, NR4A2 and TNFAIP3 were also found by the RT-PCR technique, but without any statistical significance (Fig. 4(b)).
The Gene Ontology analysis revealed that twelve biological processes were significantly altered by the nutritional intervention in the HR group (P< 0·05; see online supplementary Table S3). Among these biological processes, four were equally affected in the HR and LR groups: cellular response to stress; lymphocyte homeostasis; macromolecule catabolic process; negative regulation of NF-κB import into the nucleus. Interestingly, in the LR group, biological processes associated with immune, inflammatory and defence responses were significantly up-regulated after the nutritional intervention. According to the KEGG analysis, four pathways including several signalling pathways – erythroblastoma virus B receptor family (ErbB), epithelial cell, mitogen-activated protein kinases (MAPK) and circadian rhythm – were down-regulated after the nutritional intervention in the HR group (Table 2).
Discussion
In the present study, we examined the differences in gene expression levels and pathways in PBMC from obese boys, comparing the HR and LR groups at baseline and after a moderate energy-restricted diet intervention. To the best of our knowledge, the present study is the first to report on the microarray analyses of PBMC in obese boys. Although blood is usually not considered to be a target tissue for obesity, PBMC are relatively easily accessible in humans( Reference Bouwens, Afman and Muller 37 ) and are used to assess biological responses as their gene expression profile may reflect the pathological and physiological states of the organism( Reference Burczynski and Dorner 13 ). It is important to point out that differences in gene expression levels due to nutritional interventions are usually small. However, changes in blood transcript levels in the HR and LR groups were in agreement with the RT-PCR results and informative enough to consider their analyses( Reference Bouchard, Rabasa-Lhoret and Faraj 16 ). Interestingly, at baseline, the HR group showed a lower expression of inflammation and immune response-related pathways, which suggests that the LR group could have a more developed pro-inflammatory phenotype. This group presented higher expression levels of LEPR and SIRPB1, which indicates a tendency towards an impaired immune response and leptin resistance.
Moreover, the baseline expression levels of six genes (TFPI, LEPR, LTBP1, MMRN1, PKHD1L1 and SIRBP1) were consistently associated with the changes in adiposity indices (BMI-SDS).
There has not been much research on gene expression levels after a WL programme in obese children. However, in obese adults, there have been studies investigating the effect of energy restriction on the changes in gene expression. Most of these studies were performed in adipose tissue biopsies (eleven studies), but some in skeletal muscle (two studies) and PBMC (five studies)( Reference Moleres, Ochoa and Rendo-Urteaga 3 ). Unfortunately, they did not mention any changes in the fourteen genes that we characterised in the present study. Differences in study design, type of intervention, characteristics of subjects (age, sex, lifestyle factors, etc.), analytical procedure and so on may explain the discrepancies.
There are some limitations in the present study. The first limitation is the small sample size due to the significant costs involved in the microarray experiment. Second, participants were only males; in addition, there was no sample available to investigate the changes in the gene expression levels of PBMC after a WL programme in obese children. However, the present study reveals novel features regarding transcriptional changes in obese boys after a moderate energy-restricted diet intervention.
Comparison of low responders v. high responders at baseline
The KEGG analysis identified several pathways related to the overexpression of genes in the LR group at baseline, such as extracellular matrix–receptor interaction, focal adhesion and cell adhesion molecules. The first two pathways are associated with cell motility, proliferation, differentiation and survival; meanwhile, cell adhesion molecules are linked to haemostasis, immune response and inflammation.
Another pathway related to the overexpression of genes in the LR group at baseline is ‘FcγR-mediated phagocytosis’, which plays an essential role in host defence mechanisms through the uptake and destruction of infectious pathogens. Specialised cell types, including PBMC subsets such as macrophages, neutrophils and monocytes, take part in this process. In this sense, obesity has been associated with elevated leucocyte and lymphocyte subset counts and higher monocyte and granulocyte phagocytic activities( Reference Oana, Takeda and Hayakawa 38 ).
Higher expression levels of SIRPB1 and LEPR were found in the LR group at baseline. SIRPB1 belongs to the Ig superfamily and participates in macrophage activation. It is also overexpressed in the adipose tissue macrophages of diet-induced obese mice( Reference Lumeng, Deyoung and Bodzin 39 ), and could be related to low-grade inflammation associated with obesity. This obesity-related inflammation status could impair the response to WL in the LR group. Similar results have been reported in blood cells from overweight children( Reference Sanchez, Priego and Pico 15 ). Moreover, TFPI and EGF genes were also overexpressed in the LR group. It has been reported that the TFPI gene is increased in patients with type 2 diabetes mellitus( Reference El-Hagracy, Kamal and Sabry 40 ), whereas the gene of the EGF receptor is up-regulated in obesity( Reference Skopkova, Penesova and Sell 41 ).
Comparison of before v. after intervention in the high-responder group
The KEGG analysis identified four down-regulated pathways, including ‘ErbB signalling pathway’ and ‘MAPK signalling pathway’, in the HR group after the nutritional intervention. Both pathways are involved in processes that are associated with cell proliferation, differentiation, motility, migration and inflammation. The ‘MAPK signalling pathway’ plays an important role in inflammation( Reference Matsuzawa 42 ), and its inhibition could have anti-inflammatory properties. NR4A receptors may act as important transcriptional mediators of inflammatory signals, which are rapidly stimulated by the MAPK pathway( Reference Rendo-Urteaga, Puchau and Chueca 22 ). These receptors have been associated with obesity in human adipose tissue, which are up-regulated in extreme obesity and normalised after fat loss( Reference Skopkova, Penesova and Sell 41 ). They are mostly expressed in stromal vascular cells, a subset of cells that is rich in macrophages in obese adipose tissue( Reference Skopkova, Penesova and Sell 41 ).
In contrast, AREG, a member of the EGF gene family, is strongly down-regulated in PBMC after WL. AREG is pro-inflammatory( Reference Macias, Gerkin and Macias 43 ), and it may be hypothesised that its expression could be inhibited as a result of the reduction in obesity-linked inflammation. The other gene down-regulated after WL is HBEGF, which has been reported to be hypersecreted in obesity( Reference Matsuzawa 42 ). It plays a role in insulin sensitivity, facilitating the use of peripheral glucose( Reference Fukatsu, Noguchi and Hosooka 44 ), and its expression is stimulated by leptin( Reference Ogunwobi and Beales 45 ). Thus, it could be hypothesised that, after the WL intervention, when leptin serum levels decrease, there may also be a decrease in the gene expression levels of HBEGF.
TNFAIP3, a key regulator of inflammation and immunity involved in the development of various autoimmune diseases( Reference Vereecke, Beyaert and van Loo 46 ), is also down-regulated after WL. TNFAIP3 is a Zn finger protein whose expression is rapidly induced by TNFα( Reference Jenner and Young 47 ). The nutritional intervention decreased the gene expression of TNFAIP3 in the HR group, which could be involved in the amelioration of obesity-induced inflammation and subsequent lower secretion of TNFα.
In conclusion, the present study confirms that changes in the gene expression levels of PBMC in obese boys may help to understand the response to WL. Moreover, the present study suggests that the gene expression levels of TFPI, LEPR, LTBP1, MMRN1, PKHD1L1 and SIRBP1 at baseline could predict the changes in BMI-SDS after the nutritional intervention. However, more studies are needed to confirm these findings.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S0007114514003584
Acknowledgements
The authors thank all the children and their parents who participated in the study.
The present study was funded by grants from the Navarra Government, Departamento de Salud (grant no. PI 54/2009), Linea Especial, Nutrición y Obesidad (University of Navarra), and Carlos III Health Institute (CIBER project, CB06/03/1017). The authors acknowledge the scholarships provided by the Asociación de Amigos de la Universidad de Navarra to T. R.-U. and by the FPU ‘Formación de Profesorado Universitario’ Program of the Spanish Ministry of Education, Culture and Sport to S. G.-C.
The authors' contributions are as follows: J. A. M., M. C. A.-S., M. C. and M. O. designed and conducted the research, and reviewed the paper; T. R.-U. and S. G.-C. conducted the research, analysed the data, and wrote the paper; P. G.-M. and F. I. M. analysed the data and reviewed the paper; A. M. designed and conducted the research, reviewed the paper, and had primary responsibility for the final content. All authors read and approved the final manuscript.
None of the authors has any conflict of interest to declare.











