Introduction
Restless legs syndrome (RLS) is characterized by an overwhelming urge to move the legs, especially at night and associated with unpleasant sensations in the legs that begin or worsen during inactivity or rest. Reference Tan and Ondo1,Reference Silber, Buchfuhrer and Earley2 These unpleasant sensations can be partially or totally relieved by movement. RLS-related sleep disturbances can cause significant impact on patients’ mood, energy, behavior, and cognition. Reference Tan and Ondo1,Reference Silber, Buchfuhrer and Earley2
The prevalence of RLS varies (3%–14%) across different populations, although it appears to have a lower prevalence (<2%) in some Asian populations. Reference Tan, Seah, See, Lim, Wong and Koh3,Reference Kim, Yoon and Park4 This discrepancy in the prevalence among Caucasians and Asians populations maybe due to differences in genetic susceptibility, lifestyle or environmental factors, or underdiagnosis in Asian populations. A positive family history of RLS is present in some patients and may be inherited in an autosomal dominant or recessive pattern. RLS is a complex genetic disorder in which environmental factors and genetic predisposition contribute to the phenotype. Current genetic association studies have identified numerous gene variants to be associated with RLS. Reference Winkelmann, Xiong, Dion, Rye and Rouleau5 First- and second-degree relatives of patients with RLS had a significantly greater risk of RLS than similar relatives of controls. Reference Allen, La Buda, Becker and Earley6 Secondary RLS can be caused by a variety of conditions such as iron deficiency, pregnancy, and end-stage renal disease. Several medications like antidopaminergic medications may also exacerbate the symptoms of RLS. Reference Tan and Ondo1–Reference Kim, Yoon and Park4
In the first genome-wide association study (GWAS) of RLS in 2007, Winkelmann et al. identified several genetic variants that have significant associations with RLS. The genes which were identified to be associated with RLS were MEIS1, BTBD9, and MAP2K5. The combined allelic variants for those genes conferred more than half the risk for RLS. However, after correcting for multiple testing, only MEIS1 rs2300478 was found to reach genome-wide significance (OR = 1.74). Reference Winkelmann, Schormair and Lichtner7
Since then, there have been numerous studies (mostly based on a candidate gene approach) examining the genetic risk factors of RLS. For example, in a cohort of Chinese, Li G et al. reported that the BTBD9 allelic variants rs9296249 and rs9357271 show higher frequency among RLS patients than controls (OR = 1.44 and OR = 1.73, respectively). MAP2K5/SKOR1 rs11635424 allelic variant G also shows higher frequency among RLS patients than controls (OR = 1.49). Reference Li, Tang and Wang8 In a cohort in Québec population, rs9296249 in BTBD9 (OR = 1.71), rs10494048 in PRMT6 (OR = 0.80), rs4776976 in SKOR1 (OR = 1.34), rs3104767 in TOX 3 (OR = 1.28), and rs12962305 in SETP1 (OR = 1.26) modulate RLS risk. Reference Akçimen, Ross and Sarayloo9
Despite several genetic association studies over the past decades, there are still several unanswered questions. First, it is not clear if there are common gene variants linked to RLS risk that can be consistently replicated in independent studies. Second, if there is a publication bias between Caucasians and Asians. Third, if there are differences in genetic susceptibility among different ethnic races. Fourth, if identified genetic variants have potential functional relevance in RLS.
To address some of these gaps in knowledge, we conducted a systemic review of genetic association studies in RLS to summarize the common genetic variants that have been associated with sporadic RLS and highlight the limitations and challenges of genetic association studies in RLS.
Search Strategy
This systematic review was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. We screened related studies and articles on Embase, Cochrane, and PubMED between 2012 and 2022. The search terms used in our search strategy included “restless legs syndrome” and “genetic association studies”. The search strategy included free-text terms and any appropriate subject indexing (eg. MeSH). Figure 1 shows the PRISMA flow chart of our search strategy. The search results were then screened to remove duplicates.

Figure 1: PRISMA flowchart.
Study Selection
The studies were reviewed independently by at least two authors and any discrepancies were resolved through discussion. We included any case-control study that investigated the genetic associations between specific gene variants and the risk of RLS, regardless of ethnicity of study participants. Exclusion criteria included: a. studies on secondary RLS, b. studies without frequencies of individual SNPs among case and controls, c. studies in languages other than English. Only full journal articles were included, and conference abstracts, commentaries and editorials were excluded. After thorough screening, we managed to identify 18 case control candidate gene-based studies. Separately there were three studies using a GWAS approach.
Results
A total of 18 candidate gene-based case control studies Reference Li, Tang and Wang8–Reference Jiménez-Jiménez, Gómez-Tabales and Alonso-Navarro25 were examined using a systematic review. Out of these 18 studies, 13 were conducted in Caucasian populations (America and Europe). Reference Akçimen, Ross and Sarayloo9,Reference Jiménez-Jiménez, Esguevillas and Alonso-Navarro14–Reference Jiménez-Jiménez, Gómez-Tabales and Alonso-Navarro25 A total of 10,794 Caucasian subjects, comprising 4984 RLS cases and 5810 controls were studied. The other five studies were conducted in Asian populations (Chinese and Korea), Reference Li, Tang and Wang8,Reference Huang, Wang, Luo and Ma10–Reference Seo, Yeom, Jeon, Cho, Jeong and Lee13 involving a total of 2009 Asian subjects comprising 796 RLS cases and 1213 controls. Many gene loci were analyzed for associations with RLS, further details on the gene loci studied for each study are summarized in Tables 1 and 2.
Table 1: Genetic association studies using candidate gene approach Reference Li, Tang and Wang8–Reference Jiménez-Jiménez, Gómez-Tabales and Alonso-Navarro25
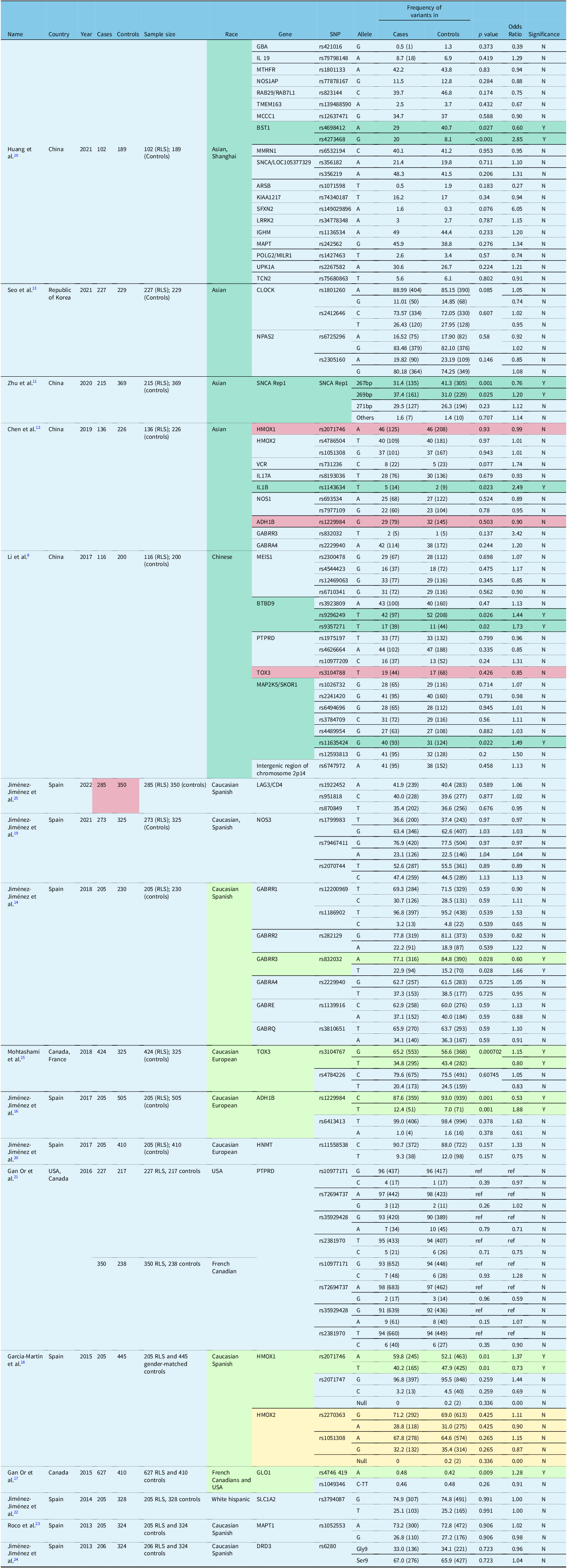
Table 2: Case-control study using candidate gene approach Reference Akçimen, Ross and Sarayloo9

Common gene loci studied in different studies were analyzed regarding their associations with RLS. Out of the 18 studies, only some gene variants were commonly studied, examples include HMOX1, HMOX2, ADH1B, GABRR3, GABRA4, and BTBD9. Reference Li, Tang and Wang8–Reference Jiménez-Jiménez, Gómez-Tabales and Alonso-Navarro25 Each of these gene loci were evaluated by two different studies, one conducted among Caucasian populations and one conducted among Asian populations. There were few consistent findings across the two major ethnic populations Reference Li, Tang and Wang8–Reference Jiménez-Jiménez, Gómez-Tabales and Alonso-Navarro25 [Table 1]. HMOX2 rs1051308 and GABRA4 rs2229940 were found to have no significant associations with RLS by the studies that analyzed these two gene loci. In the Asian population, the gene variants in BST1, SNCA Rep1, IL1B, BTBD9, and MAP2K5/SKOR1 were associated with risk of RLS (odds ratio range 1.2–2.8). Gene variants in GABRR3 TOX3, ADH1B, HMOX1, GLO1, DCDC2C, BTBD9, SKOR1, and SETBP1 were associated with an increased risk of RLS (odds ratio range 1.1–1.9) in Caucasian populations Reference Li, Tang and Wang8–Reference Jiménez-Jiménez, Gómez-Tabales and Alonso-Navarro25 (Tables 1, 2).
In addition, three recent large GWAS studies in Asian and Caucasian populations have identified other risk loci for RLS. Reference Cho, Choi and Kang26–Reference Didriksen, Nawaz and Dowsett28 In the Korean GWAS study, 325 RLS patients and 2603 non-RLS subjects are investigated in initial analysis, followed by a replication study with 227 RLS and 229 control subjects. Reference Cho, Choi and Kang26 Based on the results from the initial GWAS and replication meta-analysis, rs9390170 in UTRN gene, was identified to be a novel genetic variant to be associated with RLS. Reference Cho, Choi and Kang26 There was a borderline association with rs3923809 and rs9296249 in BTBD9 in the replication cohort. The detailed GWAS results are illustrated in Table 3. In the GWAS study using three GWAS datasets (EU-RIS GENE, INTERVAL, and 23andMe) with diagnosis data collected from European cohorts from 2003 to 2017, 13 new risk loci for RLS were identified and replicated. Identified genes and pathways are associated with neurodevelopment, axon guidance, synapse formation, and neuronal specification. Reference Schormair, Zhao and Bell27 Among these, rs113851554 in MEIS1 gene was found to be the strongest genetic factor for RLS. Reference Schormair, Zhao and Bell27 Others include variants in BTBD9, MAP2K5, TOX3, and novel variants in MYT1, DCDC2C, etc. The detailed GWAS data were summarized in Table 4. In 2020, a new GWAS analysis was conducted based on data from more than 500,000 Caucasian subjects. Reference Didriksen, Nawaz and Dowsett28 Besides 20 previously reported RLS sequence variants, three novel RLS associated gene variants (rs112716420-G, rs10068599-T, and rs10769894-A) were identified. Reference Didriksen, Nawaz and Dowsett28 Variants in MEIS1 and BTBD9 have the strongest association with RLS. Reference Didriksen, Nawaz and Dowsett28 The detailed GWAS results are summarized and highlighted in Table 5.
Table 3: Gene variants associated with RLS in GWAS analysis in Korean cohortsReference Cho, Choi and Kang 26

Table 4: Gene variants associated with RLS in GWAS analysis in European cohorts Reference Schormair, Zhao and Bell27
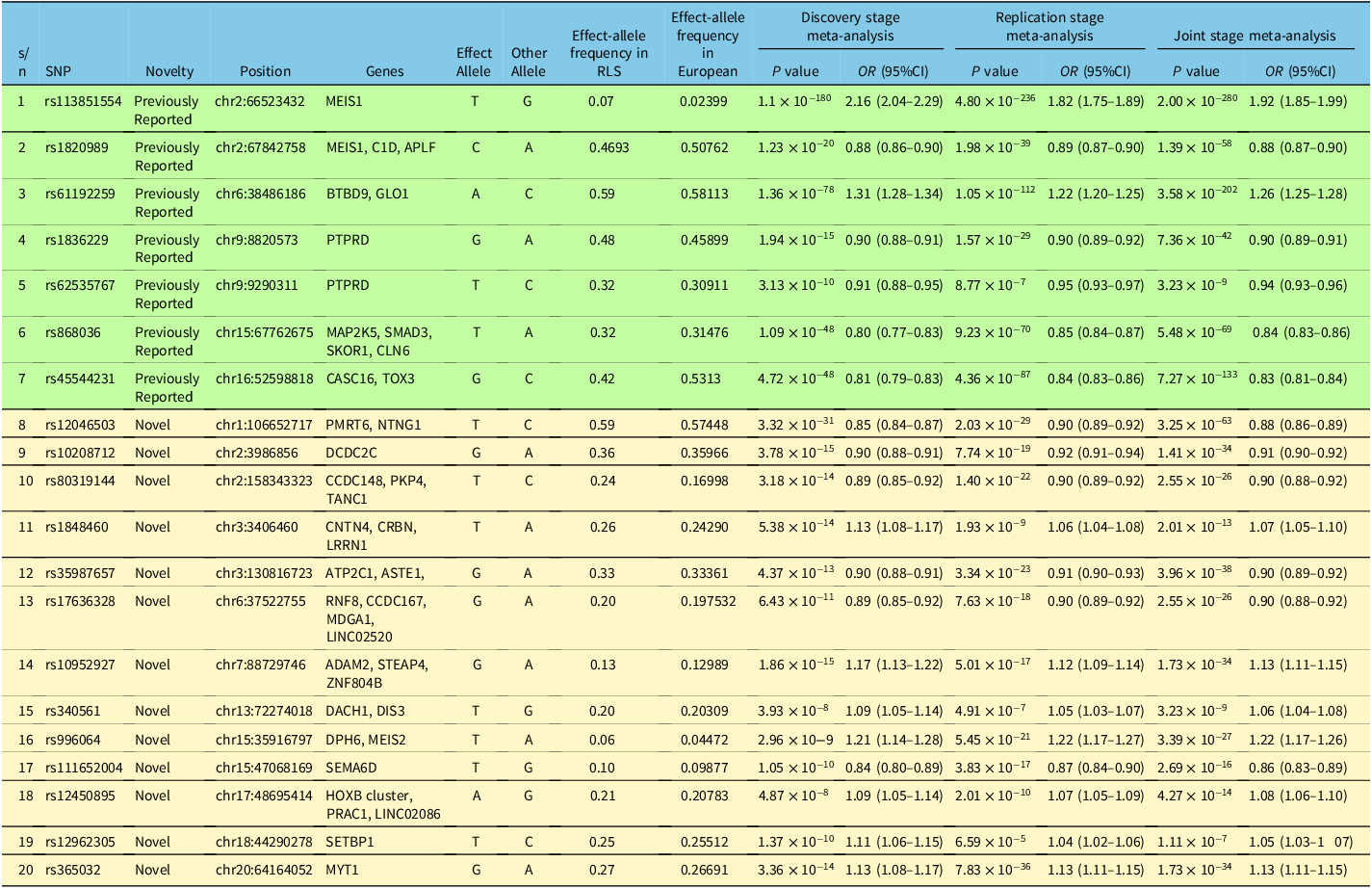
Table 5: Gene variants associated with RLS in GWAS analysis of Caucasian cohorts Reference Didriksen, Nawaz and Dowsett28

Discussion
Based on the studies using a candidate gene approach, we highlight variants of several genes (GABA receptor, ADH1B, TOX3, BST1, HMOX1, alpha synuclein Rep1, and MAP2K5/SKOR1) that have been found to be significantly associated with RLS. Gabaminergic dysregulation has been implicated in RLS, such as the association with deficient Gamma-aminobutyric acid (GABA)-mediated inhibitory control. Reference Lanza and Ferri29 GABA levels have also been found to be negatively correlated with severity of RLS in the cerebellum and positively correlated in the thalamus of RLS patients. Reference Winkelman, Schoerning, Platt and Jensen30 Furthermore, drugs that target GABA receptors have been used as a form of treatment for some RLS patients such as benzodiazepines. Reference Burke and Faulkner31 In a Spanish study, the frequency of GABRR3 allelic variant rs832032T was higher in RLS patients than controls (p = 0.028, OR = 1.66). Reference Jiménez-Jiménez, Esguevillas and Alonso-Navarro14 GABRA4 rs2229940TT genotype has also been found to be associated with earlier age of onset of RLS. Separately the association between the GABRR3 rs832032T and RLS was also higher among RLS patients compared to controls in the Chinese population, though with borderline significance (p = 0.137, OR = 3.42). Reference Chen, Luo, Li, Huang and Ma12
The link between alcohol consumption and risk of RLS has been reported by several authors. In an observation study, Mackie et al. reported the presence of RLS symptoms in 21.7% of primary alcohol use disorder subjects in the first few days following alcohol withdrawal. Reference Mackie, McHugh, McDermott, Griffin, Winkelman and Weiss32 A lower risk of RLS among subjects who had some consumption of alcohol has been highlighted. Reference Batool-Anwar, Li, De Vito, Malhotra, Winkelman and Gao33 These correlations suggest the possibility of alcohol metabolic genes modulating RLS risk.
A study among Caucasians reported an increased risk of RLS in carriers of rs1229984T (p = 0.001, OR = 1.88). Reference Jiménez-Jiménez, Gómez-Tabales and Alonso-Navarro16 This allele codes for the most active form of the alcohol dehydrogenase 1B (ADH1B) enzyme, which correlates with higher rates of metabolism of alcohol into acetaldehyde, Reference Jiménez-Jiménez, Gómez-Tabales and Alonso-Navarro16 suggesting that alcohol has protective effects against developing RLS.
TOX high mobility group box family member 3 (TOX3) gene variants (rs3104767) have been found to be associated with painful RLS, which is a sub-phenotype of the condition. Reference Karroum, Saini, Trotti and Rye34 Hence, there might be a possible correlation between TOX3 gene variants and risk of developing RLS. In a Caucasian population case-control study, rs3104767 minor allele (p = 0.0007, OR = 0.80) have been associated with reduced risk in RLS. Reference Mohtashami, He and Ruskey15
In a Chinese study, the BST1 gene variant (rs4273468) has been associated with increased risk of RLS (p value= < 0.001, OR = 2.85). Reference Huang, Wang, Luo and Ma10 BST1 has a role to play in the brain oxytocin system and is also found to be associated with Parkinson’s disease (PD) in some populations. Reference Peeraully and Tan35 The possible etiological similarities between PD and RLS point to underlying pathophysiologic links to dopaminergic disorders. Reference Peeraully and Tan36 Similar risk genes such as BST1 raise the possibility of shared pathophysiology mechanisms in both conditions.
CLOCK genes are one of a few circadian genes that control our body’s circadian rhythmicity, others include NPAS2 and BMAL1. Reference Franken37 As RLS often worsens at night, there is a clear circadian rhythm to the condition, Reference Guo, Huang and Jiang38 suggesting a biological link between circadian genes and the development of primary RLS. A case-control study in Korea reported a lower frequency of the G allele of CLOCK rs1801260 (p = 0.085, OR = 0.74) among RLS patients. Though the association was borderline, it suggests potential protective effects of the allele on RLS risk. Reference Seo, Yeom, Jeon, Cho, Jeong and Lee13
Heme oxygenase (HMOX) enzymes are involved in the initial steps of heme catabolism and they break down heme into carbon monoxide, iron and biliverdin. The HMOX1 and HMOX2 genes, respectively, code for the two isozymes which are an inducible HMOX-1 and constitutive HMOX-2. Reference Poon, Calabrese, Scapagnini and Butterfield39 HMOX is known to be protective against aging of the brain due to free radical oxidative stress. Reference Zhu, Wu and Wang40 It is also interesting to note that peripheral hypoxia has been associated with RLS symptoms, and dopaminergic therapy led to improvement of hypoxia and symptoms. Reference Salminen, Rimpilä and Polo41 Since Iron deficiency anemia (IDA) is a well-studied cause of secondary RLS, genes involved in the iron metabolism pathways are hypothesized to play a role in primary RLS as well. Reference García-Martín, Jiménez-Jiménez and Alonso-Navarro18 A case-control study in Chinese showed no significant association between HMOX genes and RLS Reference Chen, Luo, Li, Huang and Ma12 though a weak association was fund between HMOX1 rs2071746T allele (p = 0.010, OR = 0.73) and decreased risk of RLS. Reference García-Martín, Jiménez-Jiménez and Alonso-Navarro18
Alpha synuclein (SNCA) Rep 1 allele variants (265-, 269-, 271-bp alleles) have been associated with increased risk of developing PD, Reference Tan, Matsuura, Nagamitsu, Khajavi, Jankovic and Ashizawa42 probably through its effects on striatal dopaminergic pathways. Reference Lahut, Vadasz and Depboylu43 In contrast with PD, there was a decrease in Rep 1 271-bp allele frequency among RLS subjects in Caucasian populations. Reference Lahut, Vadasz and Depboylu43 However, a study in Chinese found an increase in the Rep1 269-bp allele frequency (p = 0.025, OR = 0.650) and decrease in Rep1 267-bp allele frequency (p = 0.001, OR = 0.650) among RLS patients. Reference Zhu, Wang and Wu11 Ethnicity differences may contribute to the variance in allele frequencies in the two studies.
MAP2K5/SKOR1 gene variants have been implicated with RLS risk in genome wide association studies. Reference Winkelmann, Schormair and Lichtner7 It is suggested that MAP2K pathway has an important role in the protection of dopaminergic neurons, which can contribute to dopaminergic disorders leading to RLS. Reference Winkelmann, Schormair and Lichtner7 An earlier study in America showed an association of MAP2K5 rs1026732 with RLS. Reference Yang, Li, Chen, Foldvary-Schaefer, Ondo and Wang44 Marginal associations with RLS in the Chinese cohort have been reported with MAP2K5/SKOR1 rs11635424 (p = 0.022, OR = 1.49) and rs12593813 (p = 0.2, OR = 1.50). Reference Li, Tang and Wang8 These observations suggest a possible role of these variants in RLS.
In addition to gene variants that have been found to modulate RLS risk, our systemic review also identified some differences in the findings between Asians and Caucasians. As an illustration, the gene variants that increased the risk of RLS in Asian populations include BST1 rs4273468, SNCA Rep1 269bp, IL1B rs1143634, BTBD9 rs9296249, BTBD9 rs9357271, and MAP2K5/SKOR1 rs11635424. Reference Li, Tang and Wang8,Reference Huang, Wang, Luo and Ma10–Reference Chen, Luo, Li, Huang and Ma12 However, the gene variants associated with an increased risk of RLS in Caucasian populations include GABRR3 rs832032, TOX3 rs3104767, ADH1B rs1229984, HMOX1 rs2071746, and GLO1 rs4746419. Reference Jiménez-Jiménez, Esguevillas and Alonso-Navarro14–Reference Gan-Or, Zhou and Ambalavanan17 Some of the gene variants that are associated with a decreased risk of RLS in Asian populations include BST1 rs4698412 and SNCA Rep1 267bp, Reference Huang, Wang, Luo and Ma10,Reference Zhu, Wang and Wu11 whereas in Caucasians, the GABRR3 rs832032, TOX3 rs3104767, and ADH1B rs1229984, and HMOX1 rs2071746 are associated with reduced RLS risk. Reference Jiménez-Jiménez, Esguevillas and Alonso-Navarro14–Reference García-Martín, Jiménez-Jiménez and Alonso-Navarro18
The apparent differences in the findings between Asians and Caucasians may be due to different ancestral origins, genetic drift (change in frequency of a gene variant due to random chance), or even natural selection. The allele frequency and linkage disequilibrium patterns of genetic loci across populations may vary. For a complex disease like RLS, there may be gene–gene and gene–environmental interactions and other factors that may account for unexplained differences. The differences may also be a result of false positive or negative findings due to the various inherent limitations of genetic association studies (refer to section on limitations below) especially when vast majority of the reported RLS studies have been in Caucasian populations, and it is also unclear if there are any mixed ethnicities in some of the study subjects. Furthermore, most of these candidate gene-based studies were carried out in small cohorts.
During the period of the systematic review, three GWAS studies (one in Asian and two in Caucasian populations) identified several new risk loci/variants for RLS, Reference Cho, Choi and Kang26–Reference Didriksen, Nawaz and Dowsett28 details of which were summarized in Tables 3–5. The risk loci profile appears to be largely different between Asian and Caucasian populations. The rs9390170 variant in UTRN gene was identified to be a genetic marker for RLS in a Korean cohort, whereas rs113851554 in MEIS1 gene was suggested to be a strong genetic factor in Caucasian population. Reference Cho, Choi and Kang26–Reference Didriksen, Nawaz and Dowsett28 BTBD9 and MAP2K5 are two examples of the genes implicated in both Asians and Caucasians when both candidate gene-based and GWAS studies were considered. RLS can be affected by unhealthy lifestyle, such as smoking, alcohol drinking and obesity. However, genetic factors affecting embryonic neurodevelopment, neurogenesis, axon guidance and synapse formation can be the risk factors for RLS. Reference Cho, Choi and Kang26–Reference Didriksen, Nawaz and Dowsett28 Recently, Schormair et al. Reference Schormair, Zhao, Salminen, Oexle and Winkelmann45 was unable to confirm the significant single-variant associations from candidate gene studies conducted in European populations using the GWAS dataset of the International EU-RLS-GENE Consortium, suggesting that some of the candidate gene-based study findings may be false positive or there are other unknown confounding factors to account for the lack of replication. Interestingly, a recent transcriptome-wide association study involving 15,126 RLS cases and 95,725 controls identified 13 genetic associations (in eight independent loci) at the transcriptome-wide significant level. Reference Akçimen, Sarayloo and Liao46 Consistent with the previous GWAS studies, MEIS1, SKOR1, and MAP2K5 genes are associated with RLS reported in transcriptome-wide association study. Reference Akçimen, Sarayloo and Liao46 However, the transcriptome-wide association study identified six new genetic associations with RLS, including SKAP1, SLC36A1, CCDC57, FN3KRP, and NICOA6/TRPC4AP genes, which have not been identified in the previous GWAS studies. Reference Akçimen, Sarayloo and Liao46
Limitations & Future Directions
The litmus test of any genetic association studies is the ability to replicate the positive or negative finding. In this regard, most of the reported studies using a candidate gene approach either did not have an independent replication cohort or the findings have not been consistently replicated. In addition, the small sample size has been a major limitation. This is further compounded by the low prevalence of RLS in Asian populations. The sample sizes may have limited the ability to uncover more modest genetic associations with RLS and small effect size differences will not be identified. In addition, publication bias towards positive studies and against negative studies will invariably limit the detection of multiples small gene effects of many variants. This is particularly so for complex disorders such as RLS.
Population stratification can also complicate analysis especially in small sample sizes Reference Zhu, Wang and Wu11 and frequently documentation of ethnicity has been based on self report which may not be accurate. The recruitment of RLS patients were frequently carried out in tertiary centers and the gender ratios between studies may differ. Inclusion of RLS patients with mild peripheral neuropathy into the case population may also be a confounder in some cases. Control subjects are usually not selected based on a thorough physical examination and detailed history taking and invariably not follow up longitudinally. It is possible that some of them may develop RLS symptoms subsequently. Some studies tried to minimize this by choosing control subjects with mean ages above the age of onset of RLS in case subjects. The definition of RLS is based on key clinical criteria which are primarily based on history taking. Without a clear biological diagnostic marker and gold standard diagnosis, there is a risk of selection of a non homogenous group of patients. Most published studies thus far have utilized a candidate gene approach, which may be biased in the selection of certain gene variants and missing out on testing a large portion of genomic variants.
Large scale multicenter genetic association studies with a standardized recruitment, diagnostic and evaluation protocol will be needed to address some of the major limitations of current studies. When there are sufficient independent studies, meta-analysis to increase the power of analysis will further help to identify more gene variants. Genome wide association approaches using large single nucleotide polymorphism arrays, and if cost not an issue, whole genomic analysis, are more likely to uncover novel variants. Reference Tam, Patel, Turcotte, Bossé, Paré and Meyre47,Reference Foo, Liu and Tan48 Recent GWAS studies Reference Cho, Choi and Kang26–Reference Didriksen, Nawaz and Dowsett28 (one study in Asian and two studies in Caucasians) with larger sample sizes and with validation cohorts have attempted to address some of the limitations and also managed to identify additional gene variants and provided useful functional insights into potential pathophysiology. Reference Cho, Choi and Kang26–Reference Didriksen, Nawaz and Dowsett28 The use of contemporary bioinformatic tools to study population structure, ancestry, and significance of structural variants will be useful. It is important to determine if the association signals reflect variants and genes with direct biological relevance to disease. Determination of polygenic scores based on selected variants will add to the data for risk prediction and personalized medicine. For example, a RLS polygenic score has been shown to correlate negatively with duration of education and cognitive scores. Reference Didriksen, Nawaz and Dowsett28
The identification of specific biomarkers for diagnosis or disease progression will be particularly useful in risk stratification of patients or in subset analysis. Genotype and phenotype correlation studies can potentially provide clinical value as RLS is a common sleep-related disorder. Reference Tan and Ondo1,Reference Silber, Buchfuhrer and Earley2,Reference Pavlova and Latreille49
Conclusions
Our systemic review demonstrates that multiple genetic variants modulate risk of RLS in Caucasians and in Asians. While there are a few common genetic loci, genetic susceptibility in sporadic RLS appears to be largely different between the two races, though this interpretation is potentially confounded by the limited studies in Asians. There is a need to expand RLS genetic association studies in multi-ancestry and admixed cohorts to identify potential shared or unique genetic factors. Current identified gene variants are linked to functions affecting embryonic neurodevelopment, neurogenesis, axon guidance, and synapse formation. Functional studies of identified gene variants in both in vitro and in vivo models will help shed further light and identify novel pathophysiologic clues that may lead to development of new therapeutic targets.
Acknowledgements
We thank the National Medical Research Council for their support to EK-Tan (OF LCG 000207 and STAR) and ZD-Zhou.
Funding statement
EK-Tan has received honoraria from Esai for lectureship, EK-Tan and ZD-Zhou have received grant support from National Medical Research Council, Singapore.
Competing interests
None.
Statement of authorship
Brendan Jen-Wei Tan, Xin-Ler Pang, and Sarah Png contributed equally.
B Tan, XL Pang, S Pang, ZD Zhou searched literature and extracted the data. All authors involved in the analysis and drafting of the manuscript and approved the final version. EK Tan supervised the study.








