The arrival of Hurricane Sandy in New York City (NYC) on October 29, 2012 caused flooding, power disruption, and population displacement. Although serious outbreaks of infectious disease have not been reported following previous disasters in more developed countries, their risk may have been affected by floodwater exposure, residence in emergency shelters,1 overcrowding, poor personal hygiene, poor nutritional status, lack of refrigeration or heating, improper food handling or storage, water supply disruption or contamination, or other storm-related conditions.Reference Toole2-Reference Kouadio, Aljunid, Kamigaki, Hammad and Oshitani5
Communicable disease surveillance efforts following hurricanes have often focused on monitoring the proportion of visits to health care facilities for various syndromesReference Lee, Fonseca and Brett6-Reference Baer, Elbert, Burkom, Holtry, Lombardo and Duchin8 and detecting disease among displaced persons residing in temporary housing.9-Reference Ghosh, Patnaik and Vogt13 Postdisaster efforts frequently depend on active surveillance, such as the construction of ad hoc surveillance systems in shelters during the immediate postdisaster period.Reference Landesman14
To our knowledge, no previous reports in the public health disaster literature have compared the risk of a comprehensive set of passively reported diseases across areas within a jurisdiction that were differentially affected by a disaster. Our primary objective was to assess whether residents of areas more severely affected by Hurricane Sandy had a higher incidence of any selected disease compared with lesser affected areas of NYC, after adjusting for both baseline differences across areas and posthurricane changes in health care seeking and disease reporting. A secondary objective was to develop methods for such an assessment that could be adapted for use in the aftermath of future emergencies in NYC and other jurisdictions.
Methods
Data Sources
Diseases of Interest
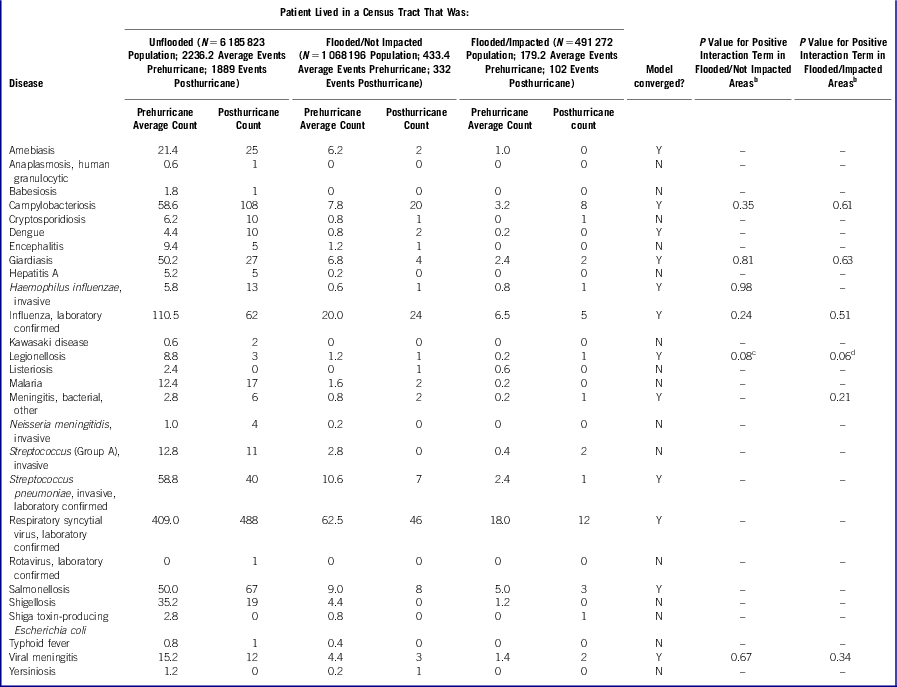
Of the more than 70 diseases reportable by providers or laboratories to the Bureau of Communicable Disease (BCD) of the NYC Department of Health and Mental Hygiene (DOHMH),15 we identified a subset of 42 diseases (Table) that had relatively short incubation periods and, based on mode of transmission, could have been affected by Hurricane Sandy-related exposures. Cases were assigned a status according to disease-specific criteria for diagnostic certainty (ie, confirmed, probable, suspected, pending, or not a case) or other feature (ie, chronic carrier, unresolved, or case contact).
Table Confirmed, Probable, and Suspected Case Counts Pre- and Posthurricane for 42 Diseases of InterestFootnote a by Hurricane-Affected Area

a Fifteen diseases of interest had no posthurricane cases: brucellosis; cholera; ehrlichiosis, human monocytic; ehrlichiosis, not otherwise specified; hemolytic uremic syndrome; leptospirosis; norovirus, laboratory-confirmed; paratyphoid fever; rickettsialpox; Rocky Mountain Spotted Fever; Staphylococcus aureus with reduced susceptibility to vancomycin; Streptococcus (Group B), invasive in ≤7-day-old infants; trichinosis; Vibrio species (non-cholera); and West Nile viral neuroinvasive disease and West Nile fever.
b For each disease, P value is shown if the model for the primary analysis using an offset term of the log of the total population converged and the parameter estimate for the interaction term was positive.
c In the sensitivity analysis using an offset term of the log of total cases, P = .06.
d In the sensitivity analysis using an offset term of the log of total cases, P = .04.
Posthurricane cases were defined as confirmed, probable, or suspected cases with event dates during the 4-week period from October 30 through November 26, 2012. To best approximate the disease-onset date, event dates were determined using the following hierarchy of available data: (1) onset date, (2) diagnosis date (defined as the earlier of the physician-reported diagnosis date or the collection date of the first diagnostic specimen), (3) report date, and (4) date case was created in Maven (Consilience Software), the database system used by BCD for disease surveillance.
The selection of a 4-week observation period was consistent with the period routinely used by the Centers for Disease Control and Prevention16, Reference Stroup, Wharton, Kafadar and Dean17 and BCD in weekly analyses of reportable diseases for aberration detection. In addition, a 4-week period was consistent with the postimpact phase of a disaster, between the immediate impact phase and the recovery phase, in which the first waves of person-to-person transmission (including respiratory [airborne and droplet] and fecal-oral), foodborne, and/or waterborne infections may occur.Reference Kouadio, Aljunid, Kamigaki, Hammad and Oshitani5 For many of the diseases of interest, a period shorter than 4 weeks would have been inadequate to detect an increase in incidence, as it can take weeks for patients to be exposed, develop symptoms, seek medical attention, and receive a clinical or laboratory-confirmed diagnosis.
We analyzed data soon after Hurricane Sandy with the goal of identifying potential elevations in diseases amenable to rapid public health investigation and possible intervention. For this analysis, we used data for the 4-week period of interest that were reported via routine passive surveillance by providers or laboratoriesReference Nguyen, Thorpe, Makki and Mostashari18 as of December 13, 2012 (first data pull). As reportable disease case counts during an established period of interest can dynamically fluctuate (eg, case counts can increase due to lags between specimen collection and laboratory confirmation and reporting; conversely, case counts can decrease due to investigation and reclassification of possible cases as “not a case”), we later repeated analyses to assess any effect of fluctuating case counts, using data reported as of January 22, 2013 (second data pull).
For most diseases, prehurricane cases were identified in 5 previous years (2007-2011) for the same 4-week period (to account for seasonality and to provide an adequate historical sample size).Reference Stroup, Wharton, Kafadar and Dean17 Less than 5 years of baseline data were used for respiratory syncytial virus, norovirus, and rotavirus (as these became reportable in 2008); for paratyphoid fever (because it became separately reportable from salmonellosis in 2009), and for influenza (to exclude data prior to and including the 2009 influenza (A) H1N1 pandemic).
The residential address at the time of case report was geocoded and classified according to census tract (n = 2168, per 2010 boundaries). This assessment was conducted in the course of routine public health practice, so institutional review board approval was not required.
Total Diseases
Total disease reporting volume posthurricane was affected by health care facility and laboratory closures, reporting lags, and population displacement, which could have led to systematic under-ascertainment of cases in hurricane-affected areas. To minimize this potential bias, we identified the total number of cases reported for residents of each census tract with event dates during each 4-week period of interest, pre- and posthurricane, for use as an offset term in a sensitivity analysis. To reflect reporting volume, these cases were not limited to the cited diseases of interest; all diseases currently under surveillance by BCD were included.15 We used all case status options except “not a case.” Cases were geocoded to the census tract of residence at the time of being reported.
Total Population
To account for the population size in each census tract, we used total population denominators from the 2010 Census.19
Neighborhood Poverty
To adjust for baseline socioeconomic differences in disease incidence between hurricane-affected and unaffected areas, we included neighborhood poverty as a covariate.Reference Krieger, Chen, Waterman, Rehkopf and Subramanian20 Consistent with DOHMH's agency-wide approach to defining poverty in the absence of individual-level poverty measures,Reference Toprani and Hadler21 neighborhood poverty was defined as the percentage of residents in a census tract with incomes below the federal poverty level, per the American Community Survey, 2006-2010.22 Census tracts were categorized into 4 levels: 0 to less than 10% of the population with incomes below the federal poverty level (low poverty areas); 10% to less than 20% (medium poverty areas); 20% to less than 30% (high poverty areas); and 30% to 100% (very high poverty areas).
Areas Flooded and Impacted
Using 2010 Census data, we determined the proportion of the population in each census tract living in a census block inundated by flooding immediately after the hurricane (Figure 1). Block inundation was established by whether a block centroid appeared in the inundation zone depicted in an operational inundation map developed by the NYC Office of Emergency Management.

Figure 1 Percentage of Census Tract Population Residing in Census Blocks Flooded by Hurricane Sandy.
Operational teams involved in NYC's emergency response to Hurricane Sandy also determined the subset of flooded tracts that were severely impacted (Figure 2), as defined by the need for sustained emergency response services in the weeks after the hurricane, involving rescue, food, water, sanitation, heating, and needs assessments. This measure reflected flooded areas that also had prolonged service outages, displacement of populations from their homes, or physical damage. Only populated areas were considered to be flooded or impacted; unpopulated areas (eg, parks) were excluded. Impacted areas included parts of Staten Island, lower Manhattan, Coney Island, and the Rockaway Peninsula.

Figure 2 Census Tracts Severely Impacted by Hurricane Sandy.
These two variables—proportion flooded and impacted—were highly collinear, in that the more severely flooded areas also tended to be the impacted areas. We presumed that residents of flooded but not impacted areas would have lower risk of communicable disease than residents of flooded and also impacted areas. Thus, tracts were grouped into 3 categories, according to whether they were unflooded, flooded but not impacted, or flooded and impacted. To improve model interpretability, we derived 2 new variables, 1 for impacted tracts and 1 for not impacted tracts. The first variable (flooded/impacted) was assigned the value of proportion flooded if the tract was impacted (else, zero). The second variable (flooded/not impacted) was assigned the value of proportion flooded if the tract was not impacted (else, zero).
Primary Analyses
A separate multivariable regression model was constructed for each disease using PROC GENMOD in SAS 9.2 (SAS Institute Inc). The outcome was case count, modeled using a negative binomial distribution. The independent variables were neighborhood poverty; whether cases were before or after the hurricane (time); the proportions flooded in impacted and in not impacted tracts; and interaction terms between the flood/impact variables and time. Models used repeated measures to adjust for correlation of observations from the same tract using an exchangeable correlation structure. The offset term was the log of the total population.
We identified diseases for which interaction terms were positive and statistically significant (α = .05), indicating that more severely flooded areas had a significantly greater increase in cases after the hurricane, adjusting for baseline differences.
Sensitivity Analyses
Sensitivity analyses assessed the effects of including pending cases (ie, preliminary posthurricane cases that may later have been ruled out or reclassified as confirmed, probable, or suspected) and of accounting for variations in reporting volume by using an offset term of the log of total cases. As noted, the effect of fluctuation in case counts was also assessed by analyzing data for the period of interest from 2 data pulls more than 1 month apart.
Results
Of the 20 823 total disease cases with onset or diagnosis during the 4-week period of interest before (2007-2011) and after the hurricane (2012) and reported as of the second data pull, 16 960 (81%) were successfully geocoded to a NYC census tract. No clear systematic pattern in the rate of geocoding failure by disease was noted. Of the total number of cases, 467 (2%) were not geocoded because they resided outside NYC, 2263 (11%) could not be geocoded but had a known NYC borough (Brooklyn: 665 [29%], Manhattan: 539 [24%], Bronx: 504 [22%], Queens: 468 [21%], and Staten Island: 87 [4%]), and 1133 (5%) could not be geocoded for other reasons (eg, missing city). Analyses were restricted to cases that were successfully geocoded.
Minor case count fluctuations occurred for the period of interest (October 30 through November 26, 2012) between the first data pull on December 13, 2012 and the second data pull on January 22, 2013. As of the second data pull, additional cases had been reported for the posthurricane period of interest for 14 diseases (amebiasis, babesiosis, campylobacteriosis, cryptosporidiosis, dengue, influenza, legionellosis, malaria, invasive Neisseria meningitidis, invasive Streptococcus pneumoniae, respiratory syncytial virus, salmonellosis, shigellosis, and typhoid fever). In contrast, fewer cases were observed for 3 diseases (giardiasis, hepatitis A, and Shiga toxin-producing Escherichia coli [STEC]). The giardiasis case count after the hurricane decreased from 38 to 33 because patient interviews determined that onset dates were before the period of interest. The hepatitis A case count decreased from 6 to 5 and the STEC case count decreased from 5 to 1 due to reclassification of cases to “not a case” or to deduplication. As of the second data pull, no cases from the period of interest were still pending. Because findings were similar for analyses using data from each data pull, only results from analyses using the second data pull are presented.
Of the 42 diseases of interest, 15 had 0 cases after the hurricane. The Table shows case counts posthurricane for the remaining 27 diseases of interest. Models to assess whether a significantly greater change was seen in case counts posthurricane in affected areas did not converge for 15 of these diseases due to sparse counts (generally <3 cases in flooded areas posthurricane for each disease). For the 12 diseases for which the model converged in the primary analysis using an offset term of the log of the total population, any increase in disease rates in the hurricane-affected areas was not statistically significantly greater than in other areas. In the sensitivity analysis to account for variations in reporting volume by using an offset term of the log of total cases, legionellosis was the only disease with a statistically significant increase in cases: the model convergence was questionable, but legionellosis had a significantly greater increase in cases posthurricane in more severely flooded/impacted areas (P = .04), and a marginally significantly greater increase in cases in more severely flooded/not impacted areas (P = .06). However, only 5 legionellosis cases were diagnosed posthurricane, with no clusters in hurricane-affected areas: cases were distributed across unflooded areas (n = 3), flooded/not impacted areas (n = 1), and flooded/impacted areas (n = 1).
Discussion
Areas affected by Hurricane Sandy did not appear to experience larger increases in the incidence of selected reportable communicable diseases than other areas in NYC. The multivariable analysis described in this report complemented other posthurricane surveillance activities that focused on early detection and monitoring of health issues related to immediate disaster effects. Those activities included focal shelter surveillance that was implemented in response to a cluster of cases of gastrointestinal illness in a shelter for hurricane evacuees,Reference Bernstein23, Reference Ridpath, Bregman and Jones24 syndromic surveillance (eg, for hurricane-related injuries, carbon monoxide poisoning, and mental health syndromes),Reference Paladini25 and weekly analyses of reportable diseases for aberration detection. This report establishes an additional methodology that health departments can use to test hypotheses about whether reportable disease incidence increased in the weeks following a disaster, while adjusting both for baseline differences across areas and also for postdisaster changes in disease reporting. Such an approach should be considered when the infrastructure that supports passive disease surveillance remains sufficiently intact, disease reports are not very rare, and capable staff and resources are available.
Although inherently subject to reporting lags, it should be possible to use passive disease surveillance data to detect elevated disease transmission within a couple months of a disaster. Such increases could otherwise remain undetected, as it may not be feasible to conduct active surveillance on the entirety of an affected population, and intensive disaster response efforts may conclude before disease incubation periods fully elapse. Even if an increase in disease incidence is not first detected until a couple months after a disaster, transmission could potentially be ongoing and warrant public health action. In addition, recognition of increased disease incidence associated with a disaster, even if delayed, could inform preparedness efforts for future disasters.
Limitations
These analyses are subject to at least 6 limitations, with the greatest challenge being the appropriate measurement of exposures of interest. We identified census tracts that were generally severely impacted by the hurricane via prolonged service outages or physical damage, but this measurement neither separated the effects of flooding, lack of electricity, and lack of heat, nor reflected short-term or time-varying service outages, nor reflected areas that experienced service outages in spite of no flooding. Furthermore, population denominators may have shifted between the 2010 Census and the hurricane in 2012. Population displacement after the hurricane was dynamic, as affected individuals moved away from and returned to affected areas, according to damage severity and service disruption and restoration. As population movement was not directly measured over the 4-week assessment period, we could neither account for this dynamism in the disease-affected numerator nor the population-based denominator. Cases were analyzed according to the census tract in which they resided at the time of report, which may have resulted in differential misclassification of cases as unexposed; persons who were displaced by the hurricane may have reported their permanent residence in a hurricane-affected area or their temporary residence (eg, an emergency shelter or a home of a friend or relative) in a not affected area.
Second, 11% of cases had a known borough but were not successfully geocoded and were excluded from analysis. This reduced power and potentially introduced a bias, if cases that were not successfully geocoded were located differentially in space. However, this bias did not appear to be strong, as the distribution of cases with geocoding errors across boroughs was similar to that of the NYC population.26
Third, reporting lags could have resulted in case under-ascertainment. The first and second data pulls were 2 and 8 weeks, respectively, after the end of the period of interest. Sensitivity analyses using the log of the total cases as an offset term did adjust for variations in reporting volume, but would not have been able to identify elevated incidence among cases that were not diagnosed and reported by the time of the second data pull. The stability of our findings between the first and second data pulls suggests that reporting lags did not strongly bias the findings.
Fourth, case counts were low and the model converged for only 12 of the 42 diseases of interest. However, if a large problem resulting in a substantial increase in cases had occurred, it was unlikely to be missed with this approach. The model had questionable convergence for some diseases with very low case counts posthurricane (eg, legionellosis). If this screening approach had identified any diseases with possibly elevated incidence in hurricane-affected areas, then the possible influence of multiple testing would need to be considered, and further analyses would be needed to assess the robustness of the association.
Fifth, we did not account for population distribution and environmental factors that may have changed between the prehurricane baseline (defined as the same 4-week period in the 5 prior years) and the posthurricane period. A more recent baseline (eg, the 4-week period immediately preceding the hurricane) would have restricted the analysis to 8 weeks during the autumn of 2012, thus minimizing the effects of secular changes over time. However, a 5-year historical baseline, which is commonly used in reportable disease aberration detection,16, Reference Stroup, Wharton, Kafadar and Dean17 was considered more appropriate for this analysis than a very recent baseline for the following reasons:
1. Case counts were low and models frequently did not converge. Choosing a baseline with 5 time points improved power and the possibility of model convergence compared with would have been expected using a baseline with a single time point. Also, using 5 time points permitted the estimation of prehurricane case counts to be based on a distribution of case counts in previous seasons instead of a single time point, reducing the possibility that a purely random increase in posthurricane case counts was attributed to the hurricane.
2. Choosing remote instead of recent baseline time points reduced any effect of temporal autocorrelation between pre- and posthurricane disease counts, improving our ability to detect any difference associated with the hurricane.
3. As the many diseases under surveillance exhibit widely variable seasonal patterns, choosing the same dates for the pre- and posthurricane periods (October 30-November 26) likely provided stronger control for fine seasonal effects than the choice of different, earlier dates for the prehurricane baseline (eg, October 1-28). When planning analyses after future disasters, multiple comparison periods may be considered to assess the robustness of results.
Sixth, the model included simplifications, such as an exchangeable (versus, for example, autoregressive) correlation structure for observations within a census tract, and a lack of explicit spatial modeling. Nevertheless, the model specification we used is feasible for a public health department to implement as part of an emergency response and should be adequate to detect a substantial increase in cases. The model can be further refined as needed, for example, to accommodate secular trends in predisaster case counts.
Conclusions
As part of the emergency response to Hurricane Sandy, the NYC DOHMH conducted a novel assessment of reportable disease incidence and found no unusual elevations in hurricane-affected areas. This finding is not surprising, given the absence of serious infectious disease outbreaks after previous disasters in highly developed urban settings. Hurricane-related exposures may have been insufficient to actually increase reportable disease risk. Alternatively, increased disease risk may have been mitigated by response activities such as providing shelter and clean food and water to affected people,Reference Gibbs and Holloway27 and broadly disseminating public health messages about the risks of consuming unrefrigerated perishable food28 and living in buildings without heat.29
Defining and acquiring reliable data and meta-data regarding the geographic distribution and extent of flooding and other storm-related impacts such as the location and duration of electrical grid outages was a challenge. This report highlights the importance during disaster preparedness planning of considering how to define, aggregate, and disseminate disaster impact data as soon as possible in the weeks after an event. The methods described here can be adapted to detect excess disease in affected areas after future emergencies.
Funding and Support
Two of the authors (S.K.G., E.L.W.) were supported by the Public Health Emergency Preparedness (PHEP) Cooperative Agreement (grant: 5U90TP221298-08) from the Centers for Disease Control and Prevention (CDC). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the CDC. The effort on this project for the other authors (K.J.K., A.D.F.) was supported by New York City tax levy funds.
Acknowledgments
Tom Matte, MD, Grant Pezeshki, MA, and Mike Porter, PhD assisted in interpreting the data about flooding and hurricane impact areas; Marci Layton, MD, Jessica Athens, PhD, Steve Schachterle, PhD, and Justin Cohen, PhD contributed to useful discussions regarding model specification and results interpretation; Deborah Kapell, MPH and Alison Levin-Rector, MPH performed geocoding of the disease data; and Deborah Kapell, MPH prepared the maps in the Figures.
Disclaimer
This work was presented in a breakout session on emergency preparedness and response at the annual conference of the Council of State and Territorial Epidemiologists, Pasadena, California, June 9-13, 2013.