INTRODUCTION
Seasonal respiratory infections present a major burden on primary healthcare [Reference Nair1–Reference Fowlkes5], with northern temperate countries, such as England, experiencing increased activity in the winter each year between weeks 40 and 20 [Reference Bollaerts6]. Understanding how different respiratory pathogens contribute to this seasonal burden and how the burden can vary from year to year is very important for public health, both for planning interventions and for interpreting routine surveillance.
Syndromic surveillance is the near real-time collection, analysis, interpretation and dissemination of health-related data to enable the early identification of the impact (or absence of impact) of potential human or veterinary public health threats that require effective public health action [Reference Triple7]. Syndromic surveillance plays a key role in the early detection and monitoring of seasonal outbreaks of respiratory illness and of non-seasonal outbreaks, like the influenza A(H1N1) 2009 pandemic [Reference Chu8].
The Public Health England (PHE) Real-time Syndromic Surveillance Team established a telehealth syndromic surveillance system in 2001 to monitor calls to the National Health Service (NHS) Direct telephone helpline (NHS Direct) [Reference Baker9]. During 2013 NHS Direct was replaced in England by a new service, NHS 111 [Reference Cook10]; PHE subsequently developed a new national syndromic surveillance telehealth system based upon the daily calls received by NHS 111 [Reference Harcourt11].
Across the world there are several examples of telehealth syndromic surveillance systems that are used to supplement public health infectious disease programmes [Reference Baker9, Reference Andersson12–Reference Rolland14]. However, compared with other forms of syndromic surveillance, e.g. emergency department, telehealth systems are not commonplace, primarily due to the limited availability of the telehealth services. Telehealth syndromic surveillance systems provide a unique opportunity to monitor community-based healthcare-seeking behaviour, providing valuable pre-diagnostic symptom-based data on patients who may not subsequently present to a healthcare professional [Reference Andersson12, Reference van Dijk15].
The aim of this work was to assess the burden of respiratory infections on a national telehealth system and explore the early warning potential of telehealth syndromic surveillance and specifically to provide key information to service providers and public health organisations about seasonal outbreaks of respiratory disease.
METHODS
Syndromic data
Daily (anonymised) counts of calls received by NHS 111 and used by PHE for syndromic surveillance were analysed over the period 30 September 2013 to 19 July 2015 (week 40 in 2013 to week 29 in 2015). Each call to NHS 111 is allocated a ‘pathway’ (code) by the call handler based on the caller's main presenting symptom, e.g. diarrhoea; these ‘pathways’ were used to aggregate calls into syndromic indicators. The syndromic indicators were chosen to be as consistent as possible with the NHS Direct syndromic surveillance system and other national syndromic surveillance systems coordinated by PHE. The syndromic indicators relevant to respiratory illnesses included cold/flu, cough and difficulty breathing and sore throat. Fever was also included in this study as previous work had shown a strong association between fever in children and influenza laboratory reports [Reference Cooper16]. Daily counts of NHS 111 calls were aggregated into weekly periods to match the reporting frequency available for laboratory reports of pathogens. Total weekly NHS 111 calls were also analysed as an offset, to account for increases in calls as the service usage increased during 2013.
Laboratory data
Laboratory data were collected through PHE's ‘second generation laboratory surveillance system’ (SGSS) which collates information on confirmed pathogens from national and regional laboratories in England [Reference Holleyman17]. Anonymised laboratory data were collated between period 30 September 2013 to 19 July 2015, (week 40 in 2013 to week 29 in 2015), as weekly counts of respiratory pathogens by specimen date. The respiratory pathogens included in the analysis were selected as those being previously found to be responsible for the majority of seasonal variation in NHS Direct calls [Reference Cooper18]; adenovirus, coronavirus, invasive Haemophilus influenzae, influenza (A and B), Mycoplasma pneumoniae, parainfluenza, respiratory syncytial virus (RSV), rhinovirus, invasive Streptococcus pneumoniae and additionally human metapneumovirus (HMPV; which was not included in the earlier study). For the pathogens causing invasive disease (H. influenzae and S. pneumoniae), culture tests using either blood or cerebrospinal fluid were included; for the other pathogens, tests included were cultures, antigen detection, genomic polymerases or ligase chain reaction detection and all specimen types were included in the analysis.
For both syndromic and laboratory data, separate weekly counts were collated for all ages, children under 5-years old, children aged 5–14 and adults aged 65 years and over, from de-identified data.
Regression models
A methodology of multiple linear regression was used as described elsewhere [Reference Cooper18]. Separate models were created with each syndromic indicator as the dependant variable for each age band and for all ages combined. Syndromic data were expressed as the proportion of total calls to NHS 111 that were assigned to each indicator. Using proportions allowed for any variation in counts due to changes in system usage. The independent (predictor) variables were the specific pathogen laboratory reports. Whether or not public holidays occurred within the week was included as a confounder in the analysis. Backwards stepwise regression was used to remove pathogens which were not significantly positively correlated to the NHS 111 calls. Model fit was compared with actual data including an analysis of the impact on coefficients for existing pathogens in the models when additional pathogens were added or subtracted.
In addition, new models were constructed incorporating lags where syndromic call data preceded laboratory specimen dates by between 1 and 4 weeks [Reference Cooper16]. These models were then compared with the original zero-lag models to identify whether they provided a better fit (as measured by the adjusted R 2) and therefore could explain more of the seasonal variation. Finally, model coefficients were used to estimate the burden on the telephone advice service from specific pathogens. In order to quantify the burden on the telehealth service, we calculated the average number of respiratory calls each week during the two winter seasons (weeks 40–20) which were associated with different pathogens, stratified by age band and syndromic indicator.
Results
Syndromic data
In the 21 months between September 2013 and July 2015 there were 18·2 million calls made to NHS 111 transferred to PHE and used in the syndromic surveillance system, of which 2·0 million (11%) related to cough, cold/flu, difficulty breathing, sore throat and fever calls, designated here as respiratory calls (Table 1).
Table 1. NHS 111 calls for respiratory indicators in England (week 40 in 2013 to week 29 in 2015)

Cough, cold/flu and difficulty breathing calls all peaked during week 49 in 2013 and week 52 of 2014, however the peak was much higher in the 2014/15 season for cough and cold/flu calls (Fig. 1). In both 2014 and 2015, cough and cold/flu calls had a second peak around week 6. Fever and sore throat calls had less clear winter peaks, although both peaked around week 51 in 2014.

Fig. 1. Seasonality of NHS 111 respiratory calls in England, week 40 in 2013–week 28 in 2015.
Laboratory data
Between week 40 in 2013 and week 29 in 2015 there were 36 711 positive laboratory reports for the respiratory pathogens extracted from SGSS. The number of weekly reports varied widely between pathogens, the most common pathogen reported being RSV (Table 2).
Table 2. Laboratory-confirmed samples by respiratory pathogen (week 40 in 2013–week 29 in 2015)
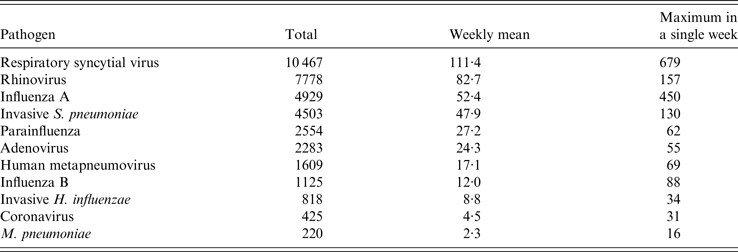
The largest seasonal peaks were seen in laboratory reports for influenza and RSV, with influenza B only showing a large peak in the 2014/15 season (Fig. 2). Reports for RSV peaked during week 52 in both seasons, whilst influenza A peaked during week 10 in 2014 and week 2 in 2015. Influenza B peaked later, in week 13 in 2015. There were also seasonal peaks in HMPV and coronavirus reports in both seasons, and M. pneumoniae peaked around week 5 in 2015 (Fig. 2).

Fig. 2. Small multiple charts showing seasonality of weekly counts of laboratory reports in England week 40 in 2013–week 20 in 2015. Each chart shows the same respiratory pathogens but with one of six selected pathogens highlighted. RSV, respiratory syncytial virus; HMPV, human metapneumovirus.
Regression models
Regression models explained the vast majority of seasonal variation in cold/flu, cough, difficulty breathing and sore throat calls in terms of the association with respiratory pathogens, with adjusted R 2 values for all-ages models being 89%, 89%, 83% and 72%, respectively (Table 3). However, only 34% of the variation in fever calls could be explained by the regression models.
Table 3. Selected regression models with associated burden from significantly correlated pathogens

* Blank cells indicate no significant positive association.
† Lag shows the number of weeks offset between laboratory specimen dates and NHS 111 call dates that exhibit the strongest association.
Regression models for children under 5 years and adults aged 65 years also showed strong associations with respiratory pathogens for cold/flu, cough and difficulty breathing but not for sore throat calls (Table 3). Associations were less clear for children aged 5–14 years, with the best-fitting model being for cold/flu calls (adjusted R 2 value of 67%).
Unsurprisingly, all syndromic indicators were associated with more than one pathogen, i.e. having a statistically significant (P < 0·05) positive result. The strongest associations were with the two pathogens that have the greatest seasonal variation, influenza and RSV. In addition, HMPV was associated with cough and sore throat calls, and difficulty breathing calls in the elderly. Whilst parainfluenza was associated with cough and difficulty breathing calls, coronavirus was only associated with cold/flu calls in children under 5-years old and rhinovirus only with cold/flu calls in adults aged 65 years and over. S. pneumoniae was only associated with sore throat calls.
NHS 111 call numbers for all syndromic indicators rose prior to the rise in positive laboratory reports (Figs 3 and 4). Model fit was improved by introducing a lag, with specimen date 1 week after the syndromic call in all models except for cold/flu calls to children aged 5–14 years (Table 3). Further improvements in model fit occurred when a lag of 2 weeks was included in models for difficulty breathing calls in all ages and for under 5-years old.

Fig. 3. NHS 111 cold/flu calls in England with estimated pathogen burdens from regression model, week 40 in 2013–week 28 in 2015.

Fig. 4. NHS 111 cough calls in England with estimated pathogen burdens from regression model, week 40 in 2013–week 28 in 2015.
Model coefficients were used to calculate the percentage of calls that were associated with specific pathogens (Table 3). Although increases in influenza and RSV reports were associated with increases in cold/flu, cough and difficulty breathing calls, their relative impact varied. For instance 34% of cold/flu calls were associated with influenza and 14% with RSV. By contrast 17% of cough calls were associated with influenza and 22% with RSV.
Cold/flu calls had peaks that corresponded to peaks in both RSV and influenza A (Fig. 3). Cold/flu calls started to rise from week 36, when children returned to school after summer holidays before the larger rise that coincided with the RSV and influenza peaks.
During the two winter seasons (weeks 40–20) an average of 948 respiratory calls per week were associated with respiratory pathogens, including 408 for RSV and 331 for influenza (Fig. 5). The greatest burden was for young children, with 405 calls related to children under 5-years old. The majority of the total respiratory calls associated with respiratory pathogens were cough calls (n = 604).

Fig. 5. Modelled burden of respiratory pathogens to NHS 111 in England during weeks 40–20, 2013/14 and 2014/15.
DISCUSSION
Main findings
This study explored the burden placed on a national telehealth syndromic surveillance system from a range of commonly circulating winter respiratory pathogens. We find a greater burden from influenza and RSV compared with the other pathogens included in the model, however, individually RSV was found to contribute the greatest burden on the telehealth service during the winter. Historically syndromic surveillance has been promoted as providing early warning over existing ‘traditional’ public health surveillance systems [Reference Baker9, Reference Cooper16, Reference Hughes19, Reference Zheng20]. The results we present here seem to further support this. We used laboratory reports and found the strongest associations when laboratory specimen dates lagged 1 week behind the telehealth calls. When it is considered that prospective surveillance of laboratory data has to take into account delays for transporting, testing and reporting results, it is likely that this period of early warning is more likely to be in excess of 2 weeks.
A further finding of this work was that different syndromic indicators demonstrated differing sensitivities to individual pathogens. Of note, telehealth calls for cough in young children was particularly sensitive to changes in RSV laboratory reporting which is supported by a number of recent international studies confirming cough as a predominant presenting symptom of RSV in children [Reference Nyawanda21–Reference Cui23]. Cold/flu calls in all ages was sensitive to influenza activity. One reported weakness of syndromic surveillance is the lack of association to specific pathogens and therefore this information is a useful tool for prospective surveillance, which can provide further reassurance and evidence of pathogen activity.
The results of this study were generated using a national telehealth system based in England. However, telehealth systems are used in different countries across the world and therefore the results from our study are applicable to a wider context outside of England [Reference Meyer13–Reference van Dijk15, Reference van Dijk, McGuiness and Moore24]. Likewise, the methodology and results from the telehealth data can be applied to different syndromic surveillance settings, e.g. emergency department attendance data, and will therefore provide a valuable tool internationally to determining the burden from specific pathogens on other data sources.
Comparison with previous studies
There is a growing evidence base internationally describing the burden placed on community health services by key winter respiratory pathogens, and in particular the ability of syndromic surveillance systems to provide situational awareness of this activity [Reference Fleming4, Reference Bollaerts6, Reference van Dijk15, Reference Andersson25, Reference Fleming26]. Two studies, coordinated in Belgium and Canada, both reported similar results for the association of influenza with increased telehealth calls using regression models in the current study [Reference Bollaerts6, Reference van Dijk15]. Additionally, the Belgium study was able to identify under-reporting in laboratories for specific pathogens and age bands [Reference Bollaerts6] whilst the Canadian study showed early warning of up to 15 days. The similarity of our results to these international groups highlight the applicability of the methods and results to other countries.
The work presented here supersedes a previous study undertaken using the former telehealth system used in England, NHS Direct [Reference Cooper16, Reference Cooper18]. In general, there was good comparability between the results from the two studies, however there were some differences, e.g. fever calls and difficulty breathing calls were recorded at different levels between the two systems. It is possible that NHS 111 fever calls were lower than NHS Direct due to intrinsic differences in the way that patients presenting with fever symptoms are triaged in the new system; under NHS 111, the fever ‘pathway’ is only used when the caller does not report with another main presenting symptom [Reference Harcourt11]. The percentage of NHS 111 difficulty breathing calls was found to be higher; NHS 111 is a 24/7 urgent care system and therefore when compared with NHS Direct is used by a greater representation of the elderly population, who present more frequently with symptoms of difficulty breathing [Reference Harcourt11]. These differences highlight the problems in interpretation that can be caused by changes in the underlying healthcare system that underpins a syndromic surveillance system and should be considered on an ongoing basis.
We found a greater impact of RSV than was found for influenza on telehealth calls. RSV is known to be an important community respiratory pathogen, causing significant morbidity and mortality particularly in the young and old [Reference Hall2, Reference Fleming26–Reference Schindeler28]. Moreover, in recent years, there is a growing body evidence illustrating the relative importance and impact of RSV when compared with influenza [Reference Hardelid, Pebody and Andrews27, Reference Taylor29]. Our results demonstrate RSV to be associated particularly with cough calls in young children, however, this burden was also found to be associated across other age groups. Other studies have illustrated that RSV impacts on all age groups, and not just young infants, the age group that is predominantly sampled for laboratory diagnosis [Reference Bollaerts6, Reference Fleming26, Reference Hardelid, Pebody and Andrews27]. In particular, Fleming et al. [Reference Fleming26] found a similar burden of RSV across all age groups using GP (general practice) consultations linked to respiratory sampling in England. Therefore, using syndromic surveillance indicators associated with RSV activity, we have supported results from previous groups that have used specific RSV outcomes, i.e. laboratory data, which again illustrates the applicability and usefulness of syndromic surveillance for monitoring pathogen burden.
There were however some differences in the other respiratory pathogens found to be associated with an increased burden reported in this study and work elsewhere; a study in Belgium [Reference Bollaerts6], did not find an association with M. pneumoniae, whilst those in Canada found an association with adenovirus and rhinovirus [Reference van Dijk15, Reference van Dijk, McGuiness and Moore24]; rhinovirus was only found to be associated with calls to patients aged over 65 years in this study.
Strengths and limitations
A major strength of our study is the sample size afforded by using a national syndromic surveillance system; NHS 111 is available to the entire population of England (~55 million). The sample size of 1·6 million respiratory calls used within this study was therefore large, particularly when compared with the number of laboratory reports collected (36 700 reports for this study). The selective nature of laboratory sampling and testing, where samples for RSV are also most exclusively sourced from infants admitted to secondary care facilities [Reference Bollaerts6], can potentially introduce bias into the modelling through poor timeliness, however syndromic surveillance is community-based, uses data from a more representative cross-section of the population and is recorded in real-time thus removing bias of sampling delays.
The associations described in this study are between syndromic and laboratory data, and this presents a limitation. In particular, there is a danger of ecological bias because neither NHS 111 calls nor laboratory data will be completely representative of community incidence. The NHS 111 syndromic data are anonymised and based upon the presentation of symptoms. It is therefore not possible to link individual NHS 111 calls to laboratory reports thus verifying the results. However, previous work has demonstrated that despite this limitation, syndromic surveillance data have been shown to be sensitive to specific pathogen activity, and this has been further supported by examples of syndromic surveillance ‘self-sampling’ [Reference Cooper30–Reference Elliot32]. Future special studies using data linkage of patient records with laboratory reports would greatly strengthen the evidence for burden based on specific pathogens.
The timing and intensity of winter respiratory seasons can vary from year to year, with the predominant driving factor the type and strain of circulating influenza [Reference Andersson25, Reference Hardelid, Pebody and Andrews27]. Our study period encompassed two winter seasons (2013/14, 2014/15) and was therefore limited in not including a number of influenza seasons with different characteristics. The reason for the limited analysis period was the launch date of the NHS 111 service (September 2013); we plan to repeat this analysis in future years when the study period will be able to encompass a wider span of winter seasons thus covering differing timing and severity of influenza, and other pathogen activity.
Implications for clinicians/policy makers
Providing early warning and situational awareness in public health is key in enabling timely interventions and treatment. The early warning provided by the NHS 111 system suggests that with respect to influenza surveillance the cold/flu call surveillance data provide added benefit to existing national influenza surveillance programmes. In the UK, the routine use of antiviral medications for influenza is limited to periods when influenza is shown to be circulating in the community [33]. Thus, providing additional awareness of the start of community-based influenza activity will improve the timeliness and confidence for making these policy decisions ultimately allowing improved treatment of patients, particular those at risk of severe complications. Furthermore, the added benefit of understanding the further burden placed on healthcare services by RSV further supports the need for effective RSV antiviral treatments. While candidate vaccines are currently undergoing trials, information on the community burden, including those age groups most affected, will inform policy decisions such as who should be included in vaccination programmes. Data generated by syndromic surveillance systems demonstrated to be reliable indicators of RSV activity will be important in post-RSV vaccine studies to determine how effective the vaccine is at reducing community-level disease, similar to previous work on other vaccine programmes [Reference Pebody34–Reference Bawa36].
SUMMARY
Syndromic surveillance continues to grow as an important public health tool. Internationally the availability of telehealth systems, particularly national systems, is limited compared with other sources of surveillance data, however, the results presented here demonstrate the unique advantages of telehealth surveillance and also illustrate the applicability of the results to other countries and systems. The continuing development of syndromic surveillance in poor-income countries [Reference Perez Garcia-Pando37–Reference Alencar, Lee Ho and Albarracin39] can provide effective public health surveillance systems where the infrastructure to support laboratory networks is lacking. The results presented here support the use of these syndromic systems for enhancing seasonal respiratory surveillance programmes.
ACKNOWLEDGEMENTS
The authors would like to acknowledge support from NHS 111 and NHS Digital and also the help of the Public Health England Real-time Syndromic Surveillance Team, including Sue Smith, Paul Loveridge, Helen Hughes, Amardeep Bains and Leandro Carrilho. R.A.M., G.E.S. and A.J.E. received support from the National Institute for Health Research (NIHR) Health Protection Research Unit in Emergency Preparedness and Response. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, the Department of Health or Public Health England. This research received no specific grant from any funding agency, commercial or not-for-profit sector.
DECLARATION OF INTEREST
None.











